Abstract
Actin, a major component of the cytoplasm, is also abundant in the nucleus. Nuclear actin is involved in a variety of nuclear processes including transcription, chromatin remodeling, and intranuclear transport. Nevertheless, the regulation of nuclear actin by posttranslational modifications has not been investigated. We now show that nuclear actin is modified by SUMO2 and SUMO3 and that computational modeling and site-directed mutagenesis identified K68 and K284 as critical sites for SUMOylating actin. We also present a model for the actin–SUMO complex and show that SUMOylation is required for the nuclear localization of actin.
Introduction
Actin is one of the most abundant proteins in cells. It is a major part of the cytoskeleton and an important component of the nucleus. Cytoplasmic actin is involved in a large variety of cellular functions that include cell locomotion, maintenance of cell shape, cell division, intracellular transport, endocytosis, and exocytosis. Nuclear actin is involved in transcription, nuclear export, intranuclear transport, and chromatin remodeling (Hofmann, 2009; Louvet and Percipalle, 2009).
To date, almost 100 actin-binding proteins have been identified (dos Remedios et al., 2003). These proteins regulate the forms and functions of actin in the cell, including the nucleocytoplasmic translocation of actin. For instance, actin, which does not contain a NLS can enter the nucleus complexed to cofilin (Pendleton et al., 2003), a protein with a classical bipartite NLS (Matsuzaki et al., 1988). Moreover, although actin contains two classical leucin-rich nuclear export signals (NESs) that are necessary for the export of actin via exportin 1 (Wada et al., 1998), the association of actin with profilin appears to be necessary for the export of actin via exportin 6 (Stuven et al., 2003).
There is also increasing evidence that posttranslational modifications of actin, including glutathionylation (Wang et al., 2003), nitration (Aslan et al., 2003), nitrosylation (Thom et al., 2008), and arginylation (Karakozova et al., 2006), play important roles in regulating the cellular functions of actin. In addition, actin is modified by ubiquitin in plants (Dantan-Gonzalez et al., 2001), the malaria parasite Plasmodium falsiparum (Field et al., 1993), and mammalian skeletal muscle (Kudryashova et al., 2005). A mono-ubiquitinated form of actin, arthrin, has also been described in insect flight muscle (Ball et al., 1987). Interestingly, ubiquitination appears to lead to rearrangement of the cytoskeleton rather than degradation of the actin.
Several proteomic studies have identified actin as a potential candidate for SUMOylation (Panse et al., 2004; Vertegaal et al., 2004; Rosas-Acosta et al., 2005). Small ubiquitin-related modifier (SUMO) proteins have a molecular mass of ∼11 kD and bind to specific lysine residues of target proteins. This conjugation is covalent and reversible. Importantly, the majority of SUMOylated proteins are found in the nucleus (Johnson, 2004), and SUMOylation has been linked to transcription, cellular translocations, and protein–protein interactions that are often related to nuclear functions (Hay, 2005).
We investigated if actin is indeed SUMOylated and if SUMOylation of actin is connected to its nuclear functions. We found that nuclear actin is modified specifically by SUMO2 and SUMO3. Using computational modeling and site-directed mutagenesis, we identified lysines 68 and 284 as the sites that are important for SUMOylation. Finally, we demonstrated that SUMOylation of actin is important for the retention of actin in the nucleus because mutations that prevent SUMOylation lead to a rapid export of actin from the nucleus through an exportin 1–dependent pathway that can be inhibited by leptomycin B.
Results and discussion
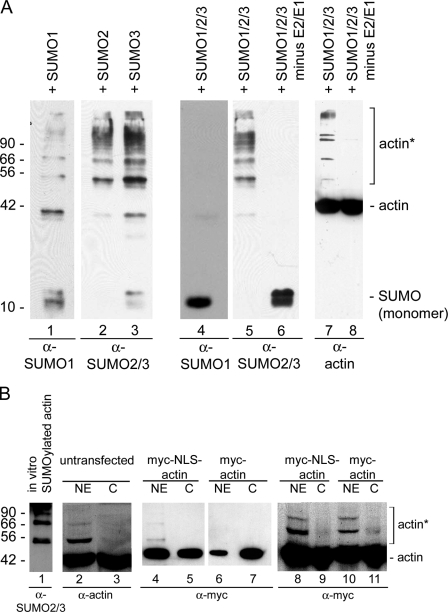
We initially used an in vitro assay to analyze if actin can be SUMOylated. Purified nonmuscle β-actin (>99% purity) was incubated with either SUMO1, -2, or -3, or all three SUMO proteins together in the presence of the SUMO-activating (E1) and the SUMO-conjugating (E2) enzymes. Fig. 1 A shows that actin is indeed modified by all three SUMO proteins when incubated individually. However, when actin was incubated with all three SUMO proteins together, there was no signal with the SUMO1 antibody, which suggests that actin is preferentially modified by SUMO2 and/or SUMO3. Control reactions showed that actin is not modified in the absence of the E1 and E2 enzymes.
Figure 1.
β-Actin is SUMOylated in vitro. (A) Purified β-actin was incubated with SUMO1, -2, or -3 individually (lanes 1–3) or with all three SUMO proteins (lanes 4–8), and probed with SUMO antibodies (lanes 1–6, bottom) and actin antibodies (lanes 7 and 8). SUMO2 and/or -3 modify actin (lanes 5 and 7), but SUMO1 does not (lane 4), when incubated together. Actin is not modified in the absence of the E1 SUMO-activating and E2-conjugating enzymes (lanes 6 and 8). (B) Nuclear (NE) and cytoplasmic (C) extracts were prepared from HeLa cells, and Western blots were probed with the 56-4 antibody to actin (lanes 2 and 3). Actin was recognized by the actin antibody in the C and NE. A strong band at ∼55 kD and weaker, higher molecular weight bands that co-migrate with actin SUMOylated in vitro were also recognized in the NE by the actin antibody. Immunoblot analyses on HeLa nuclear (NE) and cytoplasmic (C) fractions from cells expressing myc-actin and myc-NLS-actin (lanes 4–11) demonstrated the presence of higher molecular weight bands predominantly in the NE. Equal amounts of protein were applied to lanes 4–7, and lanes 8–11 were loaded with equal amounts of myc-tagged actin. Molecular mass markers are indicated in kD; asterisks, SUMOylated actin in A and B. Purified BSA and β-actin were used as markers in B.
The presence of multiple higher molecular weight bands in Fig. 1 A suggested that actin may be mono- and poly-SUMOylated. SUMO proteins have a molecular mass of about ∼11 kD and are covalently bound to their substrate. The higher molecular mass bands increase in units of ∼15 kD, which suggests that the SUMO proteins may form polySUMO chains, as described previously (Matic et al., 2007).
To establish in vivo SUMOylation, we first analyzed HeLa cytoplasmic and nuclear extracts with antibodies to actin. Nuclei were purified using two rounds of centrifugation through a sucrose cushion, a procedure that removes most cytoplasmic proteins (Pestic-Dragovich et al., 2000). Examination of cytoplasmic and nuclear extracts with actin antibodies showed the presence of two bands in the HeLa nuclear fractions that are ∼15 and 30 kD larger than native actin (Fig. 1 B, lane 2). Importantly, these slower migrating bands correlate in size to actin that is mono- and di-SUMOylated in vitro (Fig. 1 B, lane 1). These bands were absent from the cytoplasmic fraction (Fig. 1 B, lane 3).
To verify that the slower migrating bands are nuclear specific, we transfected HeLa cells with two different actin plasmids. One plasmid contained β-actin with an N-terminal myc-epitope (myc-actin), and the other contained β-actin with an N-terminal myc-epitope and an NLS between the actin and the myc-epitope (myc-NLS-actin). The distribution of the myc-actin (Fig. 1 B, lanes 6 and 7; and Fig. S1) is similar to endogenous actin, with the majority of the actin in the cytoplasm, whereas proportionately more of the myc-NLS-actin is targeted to the nucleus (Fig. 1 B, lanes 4 and 5, and Fig. S1). Both actin constructs are functional because they assemble into the cytoskeleton (Fig. S1 A), as described previously (Posern et al., 2002). Moreover, N-terminal fusions do not interfere with actin function (Westphal et al., 1997; Schoenenberger et al., 1999), and N-terminal fusion proteins, both with and without an NLS, have been used successfully to analyze the functions of actin in the cytoplasm and the nucleus (Posern et al., 2002; Chuang et al., 2006; McDonald et al., 2006; Dundr et al., 2007). Importantly, a slower migrating band was detected only in the nuclear extract from cells expressing myc-NLS-actin (Fig. 1 B, lane 4). Because the absence of a higher molecular weight band in lane 6 could be caused by the presence of less actin in the nucleus, immunoblots from gels that were loaded with equal amounts of myc-tagged actin were probed with myc antibodies. These blots demonstrated that the slower migrating bands are still only detected in the nuclear fractions from cells expressing myc-NLS-actin or myc-actin (Fig. 1 B, lanes 8–11).
To further characterize the in vivo SUMOylation of actin, we cotransfected COS-7 cells with myc-actin or myc-NLS-actin and His-tagged SUMO1, SUMO2, or SUMO3 constructs. The cells were lysed 42 h after transfection, and the His-tagged SUMO proteins, which are expressed at equivalent levels under similar conditions (Jacobs et al., 2007), were pulled down using Ni-agarose beads, resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-myc antibodies to detect if the myc-tagged actin is SUMOylated. Fig. 2 (A and B) shows that both the myc-actin and the myc-NLS-actin are modified in cells by SUMO2 and SUMO3 but not to a significant level by SUMO1. This corroborates the results in Fig. 1 A that demonstrate that actin is preferably modified by SUMO 2 and 3 in vitro. Similarly, Ni pull-down experiments on cells expressing myc-actin (Fig. 2 A) and myc-NLS-actin (Fig. 2 B) corroborated the results in Fig. 1 B by demonstrating that actin that is targeted to the nucleus (myc-NLS-actin) is SUMOylated and poly-SUMOylated to a greater extent than the myc-actin, probably because most of the myc-actin is in the cytoplasm.
Figure 2.
Actin is SUMOylated in vivo. (A and B) COS-7 cells were cotransfected with His-tagged SUMO proteins and various myc-tagged actin constructs. His-tagged proteins were purified on a Ni2+ column and analyzed with a myc antibody. Myc-actin (A) and myc-NLS-actin (B) are modified in vivo predominantly by SUMO 2 and 3. Modification of actin by SUMO2 and SUMO3 is increased when the amount of nuclear actin is increased (compare A and B). Vector indicates cells that were only transfected with myc-actin or myc-NLS-actin. (C) SUMOylation of nuclear targeted actin: COS-7 cells were cotransfected with His-SUMO2 and either myc-actin, myc-NLS-actin, or myc-actin-NES-1. Myc-NLS-actin and myc-actin-NES-1 are targeted to the nucleus via the NLS or are retained in the nucleus because of a mutation in the NES. The two actin constructs that are targeted to the nucleus show strong SUMOylation. In contrast, myc-actin, which is predominantly cytoplasmic, shows weaker modification. Molecular mass markers are indicated in kD. Asterisks, SUMOylated actin.
Finally, we cotransfected cells with His-tagged SUMO2 and either myc-actin, myc-NLS-actin, or myc-actin-NES-1 to investigate if SUMOylation is indeed connected to the nuclear localization of actin. Myc-actin-NES-1 contains point mutations in one of the two NESs of actin (Wada et al., 1998). These mutations have been shown to effectively prevent nuclear export, thereby leading to nuclear accumulation of the actin (Wada et al., 1998). Therefore, myc-actin-NES-1 shows an increase in nuclear localization similar to myc-NLS-actin. Ni-agarose pull-down experiments demonstrated increased SUMOylation of myc-NLS-actin and myc-actin-NES-1 that are targeted specifically to the nucleus (Fig. 2 C). Actin with predominantly cytoplasmic distribution (myc-actin-wt) shows distinctly weaker SUMOylation. Collectively, these data demonstrate that actin is SUMOylated in vitro and in vivo by SUMO2 and SUMO3, and that SUMOylation apparently correlates with its nuclear localization.
We next sought to identify the sites that are modified by SUMO. The core consensus motif for SUMOylation is ψKxE/D, where ψ is a bulky hydrophobic residue. SUMO covalently binds through an isopeptide bond between the C-terminal carboxyl group of SUMO and the ϵ-amino group of the lysine residue (Lin et al., 2002). We used the SUMOplotAnalysis program provided by Abgent (see “Plasmids and transfection”) to identify two high probability consensus SUMOylation sites in actin (L67K68Y69P70 and M283K284C285D286). We then applied docking paradigms to explore the possible interactions of SUMO2/3 (PDB code 2IO1; Reverter and Lima, 2006) with actin (PDB code 1HLU; Chik et al., 1996). Because the SUMO structure includes the part of the sequence that is identical between SUMO2 and SUMO3 (the first 14 residues are missing), the model applies for both SUMO2/3. To allow some flexibility in the C-terminal region of SUMO2, the initial rigid-body screening was performed without the Q88QTGG92 C-terminal peptide of SUMO2. The terminal peptide sequence was then added to the structure in the free energy evaluation and ranking steps. Some of the predicted models were not able to accommodate the C terminus and were therefore eliminated. From the remaining models, the docked conformation with the lowest free energy score is shown in Fig. 3 A. This model brings the C-terminal peptide of SUMO2/3 within reach of K284, one of the two actin residues predicted to be conjugated to SUMO.
Figure 3.
Model of the β-actin–SUMO2/3 complex. (A) Predicted model of β-actin (blue) in complex with SUMO2/3 (red). Amino acids involved in complex formation are specified. (B) SUMOylation of actin mutants: myc-tagged wild-type β-actin with an NLS (lane 1) or K68R, K284R, or a K68/284R double mutant β-actin (lanes 2–4), each with a myc tag and NLS, were coexpressed in COS-7 cells with His-SUMO2. His-SUMO2 was purified on a Ni2+ column and probed with anti-myc antibodies. COS-7 cells were cotransfected with His-SUMO2 and myc-NLS-actin constructs containing the indicated amino acid substitutions. Molecular mass markers are listed in kD. Asterisk, SUMOylated actin. (C) Schematic representation of the location of the SUMO protein relative to the location of NES-1 of β-actin. Note that the SUMOylation appears to cover NES-1.
Importantly, the model suggests that the globular domain of SUMO2/3 is stabilized by salt bridges between K68, D80, and D39 on actin and D84, K20, and E80 on SUMO2/3, respectively (Fig. 3 A). Such a bimodal interaction suggests that, after the covalent binding of SUMO2/3 to K284, a salt bridge between D84 on SUMO and K68 on actin plays a critical role in stabilizing SUMO on the actin surface. To test this model, the following single or double point mutations were introduced into the myc-NLS-actin construct: K68R, K284R, and K68/284R. The arginine-for-lysine substitutions specifically target the lysine residues to which SUMO covalently binds. Pull-down experiments using Ni-agarose on COS-7 cells cotransfected with His-SUMO2 and the respective actin constructs demonstrated that substituting lysine for arginine at both K68 and K284 greatly decreased SUMOylation (Fig. 3 B).
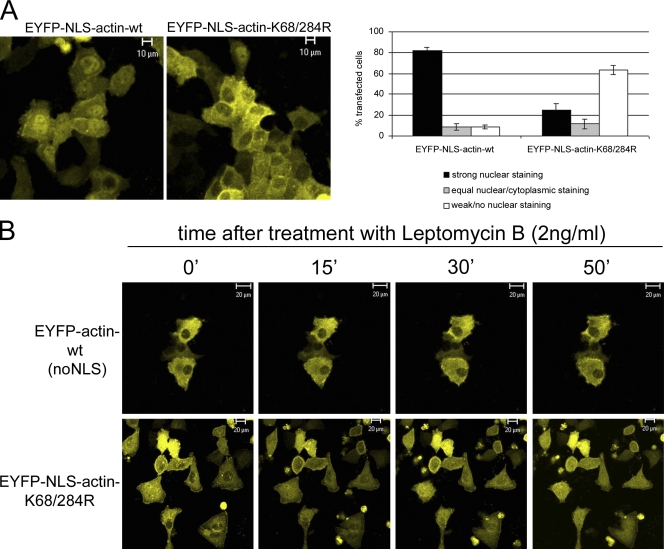
The in vivo effect of SUMOylating actin was analyzed by transfecting HeLa cells with either the EYFP-NLS-actin-wt or EYFP-NLS-actin-K68/284R construct, and taking confocal images of live cells 20 h after transfection (Fig. 4 A). Most of the cells expressing EYFP-NLS-actin-wt (82%) showed a distinctly nuclear localization. In contrast, most of the cells expressing EYFP-NLS-actin-K68/284R, which is not SUMOylated (Fig. 3 B), did not show nuclear localization. Only 25% of these cells showed strong nuclear staining (Fig. 4 A). The exclusion of the EYFP-NLS-actin-K68/284R from the nucleus is quite surprising because it contains a classical nuclear localization signal that is normally sufficient to target proteins to the nucleus (Kalderon et al., 1984).
Figure 4.
Localization of EYFP-actin in live cells. (A) Distribution of EYFP-NLS-actin-wt and EYFP-NLS-actin-K68/284R. Confocal images of live cells taken 20 h after transfection (left). The distribution of the two proteins was quantified in 100 transfected cells per experiment (error bars indicate mean ± SEM, n = 3). (B) Time course of cellular localization of EYFP-actin-wt and EYFP-NLS-actin-K68/284R after leptomycin B treatment (2 ng/ml).
The absence of the NLS-actin-K68/284R construct from the nucleus could be due to two possibilities: it is either not imported into the nucleus or it is imported but then exported immediately. Actin contains two functional NESs (Wada et al., 1998) that are recognized by the export factor CRM1/exportin1 and are leptomycin B (LMB) sensitive (Kudo et al., 1998). To analyze the possibility that the SUMO-deficient actin is rapidly exported from the nucleus, time lapse analysis was performed on HeLa cells transfected with either EYFP-actin-wt (no NLS) or EYFP-NLS-actin-K68/284R and treated with LMB 20 h after transfection (Fig. 4 B). A z stack of cells expressing EYFP-actin-wt did not reveal nuclear accumulation after 50 min of LMB treatment. This is consistent with the previous demonstration that it takes ∼24 h after LMB to detect actin accumulation in the nucleus (Wada et al., 1998). Treatment of cells expressing EYFP-NLS-actin-K68/284R with LMB, in contrast, leads to the accumulation of the modified actin in the nucleus within 30–50 min. Thus, SUMO-deficient actin apparently enters the nucleus through its attached NLS. Because the accumulation only occurs after treatment with LMB, we conclude that the SUMO-deficient actin is rapidly exported out of the nucleus, thereby explaining its absence from the nucleus.
The observation that nuclear actin is modified by SUMO could have far reaching ramifications. Mechanistically, the terminal glycine of SUMO is reported to form a covalent bond with lysine residues on target proteins (Lin et al., 2002). Consistent with this idea, the model with the lowest free energy score predicted that the C-terminal GG motif of SUMO2/3 forms a covalent bond with K284 on actin and salt bridges between Asp84, Lys20, and Glu80 of SUMO2/3 and Lys68, Asp80, and Arg39 on actin. The results presented here support this model by demonstrating that SUMO apparently interacts with K284 and K68 when it modifies actin. Surprisingly, mutating either K68 or K284 into arginine appears to prevent SUMOylation. One plausible explanation for this observation is that both sites are required for the binding of the SUMO enzymes and that K68 stabilizes the SUMO–actin interaction so that it can be covalently linked to actin at K284. Although we cannot establish the binding sequence, our data are consistent with a bimodal interaction between actin and SUMO2, and suggest that cooperativity between K284 and K68 is necessary to stabilize the SUMO2–actin complex.
Functionally, the crystal structure of actin has demonstrated that actin contains four relatively globular subdomains. Our experimental and computational data demonstrate that SUMO2/3 bridges subdomains 2 and 3 (Fig. 3), which may provide insights into the nuclear trafficking of actin. Actin that cannot be SUMOylated enters the nucleus but appears to be rapidly exported because it accumulates in the nucleus only after the export receptor for actin, CRM1/exportin1, is blocked by LMB (Fig. 4 C). Actin contains two functional leucine-rich NESs, with NES-1 and NES-2 spanning residues 170–181 and residues 211–222, respectively, and disrupting either NES blocks nuclear export of actin (Wada et al., 1998). The modeling experiments show that SUMO, by spanning subdomains two and three, lies over NES-1 (Fig. 3). Thus, SUMOylation could prevent the binding of nuclear export factors to NES-1 so that SUMO–actin is retained in the nucleus. Moreover, preventing SUMOylation would make this export signal readily accessible so that actin is rapidly exported from the nucleus.
The demonstration that nuclear actin is SUMOylated raises the intriguing possibility that it also regulates the nuclear structure of actin. The absence of classical actin filaments in the nucleus, as determined by phalloidin staining, has raised questions regarding the form and function of nuclear actin. By spanning subdomains two and three of actin, SUMOylation would interfere with the formation of the classical actin filament as predicted by the Holmes F-actin model (Lorenz et al., 1993). However, SUMOylation might support the formation of unconventional actin structures such as antiparallel lower dimer (LD), a structure possibly adopted by nuclear actin based on the staining of nuclear actin, but not cytoplasmic actin, by a LD-specific antibody that stains actin (Schoenenberger et al., 2005; Jockusch et al., 2006). Although this is a fascinating possibility, further studies are needed to establish the role of SUMOylation in the formation of nucleus-specific actin structures.
In conclusion, we have demonstrated that nuclear actin is SUMOylated. We have also shown that SUMOylation regulates the nuclear trafficking of actin. Modeling and experimental data have also provided important insights into how SUMOylation affects the physiological properties of actin in the nucleus.
Materials and methods
Cell culture and cell extracts
HeLa and COS-7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with10% fetal bovine serum and 1% penicillin/streptomycin, at 37°C with 5% CO2. HeLa nuclear and cytoplasmic extracts were prepared as described previously (Dignam et al., 1983), with a minor modification. To obtain nuclei that are free of cytoplasmic contaminations, isolated nuclei were subjected twice to sedimentation through a 1.9-M sucrose cushion before preparing the nuclear extract (Pestic-Dragovich et al., 2000).
Antibodies
The 56-4 antibody that reacts with β-actin (Hofmann et al., 2004) was used to detect SUMOylated actin. The anti-c-myc antibody was obtained from Santa Cruz Biotechnology, Inc., SUMO1 and SUMO2/3 antibodies were obtained from BIOMOL International L.P., and peroxidase-conjugated secondary anti–mouse or anti–rabbit antibodies were obtained from Jackson ImmunoResearch Laboratories.
Plasmids and transfection
The actin fusion proteins were derivatives of pEFYP-actin (Clontech Laboratories, Inc.) with the addition of the NLS sequence (5′-CCAAAAAAGAAGAAAGGTA-3′) between the EYFP and the actin. Myc-tagged actin constructs were generated by cloning actin or NLS-actin into the pSG5 vector (Agilent Technologies). Plasmids expressing His-tagged SUMO-1, SUMO-2, and SUMO-3 were generated in pcDNA3 and were a gift from R. Hay (University of Dundee, Scotland, UK). Point mutations in actin were generated using the QuickChange site-directed mutagenesis kit (Agilent Technologies). COS-7 cells were seeded in 10-cm plates and transfected 24 h later using FuGENE 6 (Roche) with plasmids encoding the various actin and SUMO proteins (5 µg of DNA, each). Western blot analyses of cell extracts showed that equivalent amounts of actin proteins and SUMO proteins were expressed (Jacobs et al., 2007) within groups.
In vitro SUMOylation assay
In vitro SUMOylation assays were performed using a kit from BIOMOL International L.P. β-Actin (99% pure) was obtained from Cytoskeleton, Inc.
Ni pull-down assays
COS-7 cells were extracted under denaturing conditions 42 h after they were transfected, and His-tagged (SUMOylated) proteins were isolated using nickel (Ni-NTA) agarose beads (QIAGEN) as described previously (Rodriguez et al., 1999). The isolated proteins were separated by SDS–PAGE, and myc-tagged/SUMOylated actin was detected by immunoblotting using standard conditions.
Docking method and predicted model
The scheme used to predict docked conformations between β-actin (PDB code 1HLU) and SUMO2 (PDB code 2CKH) was developed in the Camacho Laboratory. An earlier automated implementation of the method, ClusPro (Camacho and Gatchell, 2003), has been publicly available for several years (http://structure.pitt.edu), and predictions have been validated in blind experiments of protein interactions (Comeau et al., 2004, 2005). The main steps of the method are: a rigid-body screening of surface complementarity between receptor and ligand docked conformations using the fast-Fourier transform–based program DOT (Mandell et al., 2001); a free energy evaluation of docked structures using the FastContact (Camacho and Zhang, 2005) scoring function; clustering (Comeau et al., 2004; Kozakov et al., 2005) the low free energy complex structures; and ranking based on cluster size and free energy score.
Live cell imaging
HeLa cells were plated on Delta T Dishes (0.17 mm, black; Bioptechs) and transfected with the EYFP-actin constructs, as indicated in the Results section. 20 h after transfection, live cells were analyzed at 37°C using a confocal microscope (LSM 510 META; Carl Zeiss, Inc.) equipped with a temperature control circulator (Lauda). Pictures were taken using a Plan-Neofluar 25×/0.80 Imm Korr differential interference contrast objective lens (Carl Zeiss, Inc.). For time-lapse analysis of LMB treatment, cells were treated 20 h after transfection with 2 ng/ml leptomycin B (BIOMOL International, L.P.). Confocal images were taken every 15 min, and after 50 min, z stacks were taken in 0.5-µm steps. Images were processed using LSM 510 software (Carl Zeiss, Inc.).
Online supplemental material
Fig. S1 shows the distribution of N-terminally tagged actin and NLS–actin constructs in HeLa cells by fluorescence microscopy and immunoblotting. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905016/DC1.
Acknowledgments
We thank Dr. Mei Ling Chen, Research Resources Center, University of Illinois at Chicago, for assistance with the confocal microscopy.
We acknowledge support from National Science Foundation (MCB-0444291), the Pittsburgh Supercomputer Center (to C.J. Camacho), the Association for International Cancer Research (AICR06-613 to F.V. Fuller-Pace), and the National Science Foundation (MCB049958P) and National Institutes of Health (GM080587 to P. de Lanerolle). The authors certify that they have no competing financial interests.
Footnotes
Abbreviations used in this paper: LMB, leptomycin B; NES, nuclear export signal; SUMO, small ubiquitin-related modifier.
References
- Aslan M., Ryan T.M., Townes T.M., Coward L., Kirk M.C., Barnes S., Alexander C.B., Rosenfeld S.S., Freeman B.A. 2003. Nitric oxide-dependent generation of reactive species in sickle cell disease. Actin tyrosine induces defective cytoskeletal polymerization.J. Biol. Chem. 278:4194–4204 [DOI] [PubMed] [Google Scholar]
- Ball E., Karlik C.C., Beall C.J., Saville D.L., Sparrow J.C., Bullard B., Fyrberg E.A. 1987. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate.Cell. 51:221–228 [DOI] [PubMed] [Google Scholar]
- Camacho C.J., Gatchell D.W. 2003. Successful discrimination of protein interactions.Proteins. 52:92–97 [DOI] [PubMed] [Google Scholar]
- Camacho C.J., Zhang C. 2005. FastContact: rapid estimate of contact and binding free energies.Bioinformatics. 21:2534–2536 [DOI] [PubMed] [Google Scholar]
- Chik J.K., Lindberg U., Schutt C.E. 1996. The structure of an open state of beta-actin at 2.65 A resolution.J. Mol. Biol. 263:607–623 [DOI] [PubMed] [Google Scholar]
- Chuang C.H., Carpenter A.E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A.S. 2006. Long-range directional movement of an interphase chromosome site.Curr. Biol. 16:825–831 [DOI] [PubMed] [Google Scholar]
- Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. 2004. ClusPro: an automated docking and discrimination method for the prediction of protein complexes.Bioinformatics. 20:45–50 [DOI] [PubMed] [Google Scholar]
- Comeau S.R., Vajda S., Camacho C.J. 2005. Performance of the first protein docking server ClusPro in CAPRI rounds 3-5.Proteins. 60:239–244 [DOI] [PubMed] [Google Scholar]
- Dantan-Gonzalez E., Rosenstein Y., Quinto C., Sanchez F. 2001. Actin monoubiquitylation is induced in plants in response to pathogens and symbionts.Mol. Plant Microbe Interact. 14:1267–1273 [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei.Nucleic Acids Res. 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Remedios C.G., Chhabra D., Kekic M., Dedova I.V., Tsubakihara M., Berry D.A., Nosworthy N.J. 2003. Actin binding proteins: regulation of cytoskeletal microfilaments.Physiol. Rev. 83:433–473 [DOI] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., Matera A.G. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo.J. Cell Biol. 179:1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field S.J., Pinder J.C., Clough B., Dluzewski A.R., Wilson R.J., Gratzer W.B. 1993. Actin in the merozoite of the malaria parasite, Plasmodium falciparum.Cell Motil. Cytoskeleton. 25:43–48 [DOI] [PubMed] [Google Scholar]
- Hay R.T. 2005. SUMO: a history of modification.Mol. Cell. 18:1–12 [DOI] [PubMed] [Google Scholar]
- Hofmann W.A. 2009. Cell and molecular biology of nuclear actin.Int. Rev. Cell Mol. Biol. 273:219–263 [DOI] [PubMed] [Google Scholar]
- Hofmann W.A., Stojiljkovic L., Fuchsova B., Vargas G.M., Mavrommatis E., Philimonenko V., Kysela K., Goodrich J.A., Lessard J.L., Hope T.J., et al. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II.Nat. Cell Biol. 6:1094–1101 [DOI] [PubMed] [Google Scholar]
- Jacobs A.M., Nicol S.M., Hislop R.G., Jaffray E.G., Hay R.T., Fuller-Pace F.V. 2007. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1.Oncogene. 26:5866–5876 [DOI] [PubMed] [Google Scholar]
- Jockusch B.M., Schoenenberger C.A., Stetefeld J., Aebi U. 2006. Tracking down the different forms of nuclear actin.Trends Cell Biol. 16:391–396 [DOI] [PubMed] [Google Scholar]
- Johnson E.S. 2004. Protein modification by SUMO.Annu. Rev. Biochem. 73:355–382 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B.L., Richardson W.D., Smith A.E. 1984. A short amino acid sequence able to specify nuclear location.Cell. 39:499–509 [DOI] [PubMed] [Google Scholar]
- Karakozova M., Kozak M., Wong C.C., Bailey A.O., Yates J.R., III, Mogilner A., Zebroski H., Kashina A. 2006. Arginylation of beta-actin regulates actin cytoskeleton and cell motility.Science. 313:192–196 [DOI] [PubMed] [Google Scholar]
- Kozakov D., Clodfelter K.H., Vajda S., Camacho C.J. 2005. Optimal clustering for detecting near-native conformations in protein docking.Biophys. J. 89:867–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N., Wolff B., Sekimoto T., Schreiner E.P., Yoneda Y., Yanagida M., Horinouchi S., Yoshida M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1.Exp. Cell Res. 242:540–547 [DOI] [PubMed] [Google Scholar]
- Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. 2005. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin.J. Mol. Biol. 354:413–424 [DOI] [PubMed] [Google Scholar]
- Lin D., Tatham M.H., Yu B., Kim S., Hay R.T., Chen Y. 2002. Identification of a substrate recognition site on Ubc9.J. Biol. Chem. 277:21740–21748 [DOI] [PubMed] [Google Scholar]
- Lorenz M., Popp D., Holmes K.C. 1993. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm.J. Mol. Biol. 234:826–836 [DOI] [PubMed] [Google Scholar]
- Louvet E., Percipalle P. 2009. Transcriptional control of gene expression by actin and myosin.Int. Rev. Cell Mol. Biol. 272:107–147 [DOI] [PubMed] [Google Scholar]
- Mandell J.G., Roberts V.A., Pique M.E., Kotlovyi V., Mitchell J.C., Nelson E., Tsigelny I., Ten Eyck L.F. 2001. Protein docking using continuum electrostatics and geometric fit.Protein Eng. 14:105–113 [DOI] [PubMed] [Google Scholar]
- Matic I., van Hagen M., Schimmel J., Macek B., Ogg S.C., Tatham M.H., Hay R.T., Lamond A.I., Mann M., Vertegaal A.C. 2007. In vivo identification of human SUMO polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy.Mol. Cell. Proteomics. 7:132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F., Matsumoto S., Yahara I., Yonezawa N., Nishida E., Sakai H. 1988. Cloning and characterization of porcine brain cofilin cDNA. Cofilin contains the nuclear transport signal sequence.J. Biol. Chem. 263:11564–11568 [PubMed] [Google Scholar]
- McDonald D., Carrero G., Andrin C., de Vries G., Hendzel M.J. 2006. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations.J. Cell Biol. 172:541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast.J. Biol. Chem. 279:41346–41351 [DOI] [PubMed] [Google Scholar]
- Pendleton A., Pope B., Weeds A., Koffer A. 2003. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells.J. Biol. Chem. 278:14394–14400 [DOI] [PubMed] [Google Scholar]
- Pestic-Dragovich L., Stojiljkovic L., Philimonenko A.A., Nowak G., Ke Y., Settlage R.E., Shabanowitz J., Hunt D.F., Hozak P., de Lanerolle P. 2000. A myosin I isoform in the nucleus.Science. 290:337–341 [DOI] [PubMed] [Google Scholar]
- Posern G., Sotiropoulos A., Treisman R. 2002. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor.Mol. Biol. Cell. 13:4167–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D., Lima C.D. 2006. Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.Nat. Struct. Mol. Biol. 13:1060–1068 [DOI] [PubMed] [Google Scholar]
- Rodriguez M.S., Desterro J.M., Lain S., Midgley C.A., Lane D.P., Hay R.T. 1999. SUMO-1 modification activates the transcriptional response of p53.EMBO J. 18:6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Acosta G., Russell W.K., Deyrieux A., Russell D.H., Wilson V.G. 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers.Mol. Cell. Proteomics. 4:56–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger C.A., Steinmetz M.O., Stoffler D., Mandinova A., Aebi U. 1999. Structure, assembly, and dynamics of actin filaments in situ and in vitro.Microsc. Res. Tech. 47:38–50 [DOI] [PubMed] [Google Scholar]
- Schoenenberger C.A., Buchmeier S., Boerries M., Sutterlin R., Aebi U., Jockusch B.M. 2005. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm.J. Struct. Biol. 152:157–168 [DOI] [PubMed] [Google Scholar]
- Stuven T., Hartmann E., Gorlich D. 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes.EMBO J. 22:5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom S.R., Bhopale V.M., Mancini D.J., Milovanova T.N. 2008. Actin S-nitrosylation inhibits neutrophil beta2 integrin function.J. Biol. Chem. 283:10822–10834 [DOI] [PubMed] [Google Scholar]
- Vertegaal A.C., Ogg S.C., Jaffray E., Rodriguez M.S., Hay R.T., Andersen J.S., Mann M., Lamond A.I. 2004. A proteomic study of SUMO-2 target proteins.J. Biol. Chem. 279:33791–33798 [DOI] [PubMed] [Google Scholar]
- Wada A., Fukuda M., Mishima M., Nishida E. 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein.EMBO J. 17:1635–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tekle E., Oubrahim H., Mieyal J.J., Stadtman E.R., Chock P.B. 2003. Stable and controllable RNA interference: Investigating the physiological function of glutathionylated actin.Proc. Natl. Acad. Sci. USA. 100:5103–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal M., Jungbluth A., Heidecker M., Muhlbauer B., Heizer C., Schwartz J.M., Marriott G., Gerisch G. 1997. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein.Curr. Biol. 7:176–183 [DOI] [PubMed] [Google Scholar]