Abstract
The constitutive centromere-associated network (CCAN) proteins are central to kinetochore assembly. To define the molecular architecture of this critical kinetochore network, we sought to determine the full complement of CCAN components and to define their relationships. This work identified a centromere protein S (CENP-S)–containing subcomplex that includes the new constitutive kinetochore protein CENP-X. Both CENP-S– and CENP-X–deficient chicken DT40 cells are viable but show abnormal mitotic behavior based on live cell analysis. Human HeLa cells depleted for CENP-X also showed mitotic errors. The kinetochore localization of CENP-S and -X is abolished in CENP-T– or CENP-K–deficient cells, but reciprocal experiments using CENP-S–deficient cells did not reveal defects in the localization of CCAN components. However, CENP-S– and CENP-X–deficient cells show a significant reduction in the size of the kinetochore outer plate. In addition, we found that intrakinetochore distance was increased in CENP-S– and CENP-X–deficient cells. These results suggest that the CENP-S complex is essential for the stable assembly of the outer kinetochore.
Introduction
The centromere is essential for faithful chromosome segregation during mitosis. The kinetochore is assembled on centromeres to form a dynamic interface with microtubules from the mitotic spindle (Cheeseman and Desai, 2008). To understand kinetochore structure and the mechanisms related to chromosome segregation, it is critical to define the identity, organization, and functional roles of the numerous kinetochore proteins.
In recent years, multiple kinetochore proteins have been identified in vertebrate cells using a combination of approaches (Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006; Cheeseman and Desai, 2008; Hori et al., 2008a). These studies have revealed that a constitutive centromere-associated network (CCAN) of proteins associates with centromeres throughout the cell cycle and provides a platform for the formation of a functional kinetochore during mitosis. Other kinetochore proteins, including the KNL1–Mis12 complex–Ndc80 complex (KMN) network, are targeted to kinetochores by CCAN-containing prekinetochores during G2 and mitosis (Cheeseman et al., 2008) to establish a fully assembled kinetochore capable of interacting with spindle microtubules and facilitating faithful chromosome segregation (Cheeseman et al., 2006; DeLuca et al., 2006).
In vertebrates, 15 proteins (centromere protein C [CENP-C], H, I, K to U, and W) have been identified as CCAN components (Hori et al., 2008a). Based on a combination of functional and biochemical analyses, we and others have previously demonstrated that the CCAN is divided into several subclasses (Izuta et al., 2006; Liu et al., 2006; Okada et al., 2006; Kwon et al., 2007; McClelland et al., 2007; Hori et al., 2008a, b).
CENP-S was originally identified as copurifying with CENP-M or -U and was verified as a CCAN component (Foltz et al., 2006). However, CENP-S was not detected as a stoichiometric interacting partner in the CENP-H–containing complex in our biochemical purifications from DT40 or HeLa cells (Okada et al., 2006). Thus, we sought to define the relationship between CENP-S and the other CCAN subcomplexes. In this study, we identify a new CENP-S–interacting protein and define a function for the CENP-S–containing complex in stable outer kinetochore assembly.
Results and discussion
CENP-X is a component of the CCAN
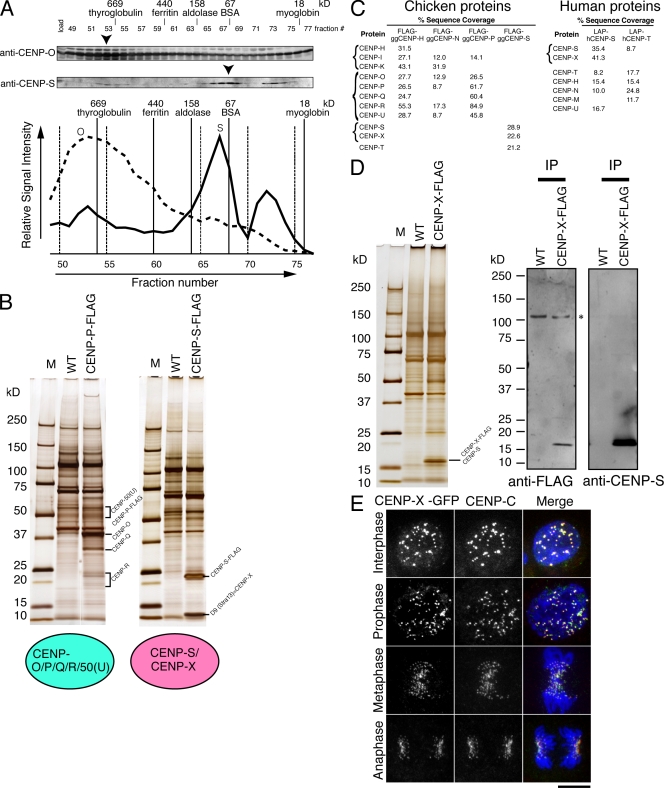
Our previous purifications using epitope-tagged CENP-H, -I, or -O did not isolate CENP-S (Okada et al., 2006), suggesting that CENP-S represents a distinct component of the CCAN from the CENP-H– and CENP-O–containing complexes. To assess this more closely, we fractionated protein extract from DT40 cells by gel filtration chromatography and analyzed each fraction by Western blot analysis with antibodies against CENP-O or -S. The profile of CENP-S was clearly distinct from that of CENP-O (Fig. 1 A), suggesting that the CENP-O–containing complex does not contain CENP-S. To confirm the results of the gel filtration analysis, we performed immunoprecipitation (IP) experiments with cell lines in which endogenous CENP-P (a CENP-O complex protein) or CENP-S was completely replaced with CENP-P–Flag or CENP-S–Flag, respectively (Fig. 1 B). Mass spectrometry indicated that the CENP-P–Flag IPs primarily contained CENP-O, -P, -Q, -R, and -50 (U) but not CENP-S, which is consistent with our previous analysis (Hori et al., 2008b). Similarly, in CENP-S–Flag IPs, we did not observe clear bands at the expected sizes for the CENP-H or -O complex proteins on silver-stained gels (Fig. 1 B). We also confirmed the coprecipitation using high sensitivity mass spectrometry analyses. Finally, we performed IPs with cell lines in which endogenous CENP-H or -N was completely replaced with CENP-H–Flag or CENP-N–Flag, and we similarly did not detect CENP-S in either IP (Fig. 1 C). These results suggest that CENP-S can be separated from the rest of the CCAN and is distinct from the CENP-H– or the CENP-O–containing complex. However, we note that CENP-T was detected in CENP-S IPs using high sensitivity mass spectrometry analyses (Fig. 1 C). Consistent with this, gel filtration chromatography of DT40 extracts revealed two peaks of CENP-S migration, one of which co-migrates with a CENP-T peak, although the proportion of the CENP-S that co-migrates with CENP-T is minor (Fig. S1). CENP-T was also detected by Western blot analysis in CENP-S IPs, but the coprecipitation efficiency of CENP-T with CENP-S is not high (Fig. S1). Considering these data, we conclude that the CENP-S complex is distinct from the CENP-T complex, although CENP-S may associate weakly with the CENP-T complex.
Figure 1.
Identification of the CENP-S–associated protein CENP-X. (A) Immunoblots of DT40 cell extracts fractionated on a Superose 6 gel filtration column using antibodies against anti–CENP-O or –CENP-S. Signal intensities are plotted in the graph, and peak fractions are indicated by arrowheads. (B) SDS-PAGE of proteins isolated by IP with anti-Flag antibodies using cells in which expression of CENP-P or -S was replaced with CENP-P–Flag or CENP-S–Flag, respectively. Wild-type (WT) DT40 cells were also used for IP with anti-Flag antibodies as a control. (C) High sensitivity mass spectrometric analysis of the purifications of chicken and human centromere proteins indicating the percentage sequence coverage for each polypeptide. (D) Co-IP of CENP-S with CENP-X. Immunoprecipitates of CENP-X–expressing and wild-type cells with anti-Flag antibodies were separated by SDS-PAGE and analyzed by Western blotting with anti-Flag or –CENP-S antibodies. (E) Localization of GFP-tagged CENP-X throughout the cell cycle in DT40 cells. Centromeres were costained with anti–CENP-C antibodies. M, molecular mass marker. Bar, 10 µm.
Although we did not observe clear bands of known CCAN proteins in CENP-S–Flag IPs, we detected a strong band with a molecular mass of ∼10.5 kD (Fig. 1 B). We determined the amino acid sequences of the peptides from this band and found that this corresponds to the D9 (Stra13) protein (amino acid sequence from ChEST533l10 clone). We will refer to D9 (Stra13) as CENP-X based on the results of the localization experiments described in the next paragraph. The human homologue of Stra13 (CENP-X) was reported as a protein whose expression is increased upon retinoic acid induction (Scott et al., 1996). To confirm the interaction between CENP-S and -X, we generated DT40 cell lines in which endogenous CENP-X was completely replaced with CENP-X–Flag and used these to conduct IPs with anti-Flag antibodies. Western blot analysis with anti–CENP-S or anti-Flag antibodies indicated clear coprecipitation of CENP-S and -X (Fig. 1 D). To determine whether the interaction between CENP-S and -X is conserved in humans, we conducted similar IPs from HeLa cell lines stably expressing GFPLAP–CENP-S or GFPLAP–CENP-T. Mass spectrometric analysis revealed the presence of CENP-X (Stra13; NCBI Protein database accession no. NP_659435) in CENP-S IPs but not in CENP-T IPs (Fig. 1 C).
To investigate the localization of CENP-X, we generated a DT40 cell line in which endogenous CENP-X was completely replaced with CENP-X–GFP. Colocalization with anti–CENP-C antibodies demonstrated that CENP-X–GFP localizes to centromeres throughout the cell cycle (Fig. 1 E). We also confirmed the colocalization with anti-Flag antibodies in cells in which endogenous CENP-X was completely replaced by CENP-X–Flag (Fig. S1). Based on the copurification with CENP-S and its constitutive localization to centromeres, we conclude that CENP-X is a new component of the CCAN.
Both CENP-S– and CENP-X–deficient DT40 cells are viable but show defects in mitotic progression
To understand the function of the CENP-S–CENP-X complex and its relationship to other kinetochore components, we generated loss-of-function DT40 mutants for CENP-S and -X (Fig. S2). We were able to isolate homozygous CENP-S−/− and CENP-X−/− deletions, indicating that CENP-S– and CENP-X–deficient cells are viable. To determine the consequences of loss of CENP-S and -X, we first examined the growth curves for CENP-S– and CENP-X–deficient cells. Under standard growth conditions, these knockout (KO) cells grew with similar growth rates to wild-type cells (Fig. S3). We also observed a slight accumulation of G2/M cells in both CENP-S– and CENP-X–deficient cell lines compared with wild-type cells by FACS analysis (Fig. S3). In addition, we observed a three- to fourfold increase in the number of apoptotic cells in these mutants compared with control cells based on DAPI staining with microscopic analysis, which may reflect an accumulation of mitotic defects.
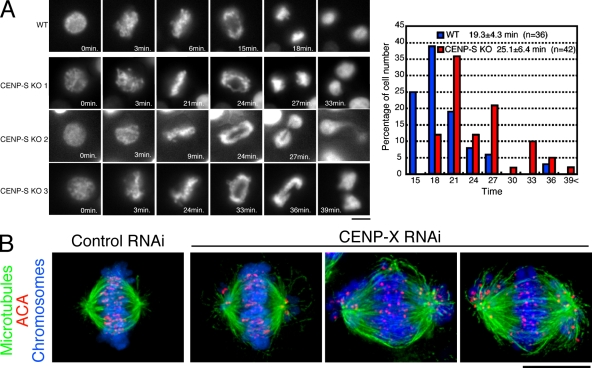
To examine the presence of any errors in mitosis, we visualized the behavior of individual CENP-S–deficient cells expressing histone H2B-RFP. Time-lapse imaging of wild-type and CENP-S–deficient cells (Fig. 2 A) indicated the presence of some mitotic defects in the absence of CENP-S. Although most CENP-S–deficient cells progressed through mitosis, these cells took a mean of 25.1 ± 6.4 min (±SD; n = 42) to progress from prophase to anaphase, which is significantly longer than the time observed in wild-type cells (19.3 ± 4.3 min; n = 36; P = 0.000012). In addition, we observed that >30% of CENP-S–deficient cells displayed anaphase bridging of chromosomes (Fig. 2 A, CENP-S KO 1–3). These results indicate that CENP-S is important for proper mitotic progression and chromosome segregation in DT40 cells even though it is dispensable for viability in a population.
Figure 2.
Both CENP-S– and CENP-X–deficient DT40 cells are viable but show defects in mitotic progression. (A, left) Dynamics of chromosomes in wild-type (WT) or CENP-S–deficient cells visualized by time-lapse observation of living cells. Selected images of chromosomes from prophase to anaphase in these cells are shown. (right) Quantification of the time for progression from prophase to anaphase in wild-type and CENP-S–deficient cells as determined by time-lapse microscopy of living cells. (B) Chromosome morphology and α-tubulin staining (green) in human HeLa cells after siRNA-based knockdown for CENP-X. Human anticentromere antibodies (ACA) were used to detect the position of centromeres (red), and DNA (blue) was stained with Hoechst. Bars, 10 µm.
The phenotype of CENP-S– and CENP-X–deficient cells is similar to that of cells with KOs of CENP-O class proteins, which are viable but show slight mitotic defects (Hori et al., 2008b). Cells with KOs of CENP-O class proteins do not exit mitosis after release from a nocodazole block (Minoshima et al., 2005). Therefore, we tested the ability of CENP-S–deficient cells to recover from spindle damage (Fig. S3 C). We treated cells with nocodazole for 12 h and then removed the drug and examined the entry of the cells into interphase. Wild-type cells progressed to G1 phase immediately, and <10% were mitotic 4 h after release from the nocodazole block. In contrast, ∼60% of CENP-50 (U) (a CENP-O class protein)–deficient cells were mitotic 4 h after release from the nocodazole block, as reported previously (Minoshima et al., 2005). In CENP-S–deficient cells, ∼15% of cells at 4 h were mitotic after release from the nocodazole block. Although this suggests a slight defect in the recovery from spindle damage in CENP-S–deficient cells compared with wild-type cells, the defect is minor compared with CENP-50 (U)–deficient cells. These results suggest that CENP-S–CENP-X function is distinct from the CENP-O class of proteins.
Finally, we also analyzed the mitotic behavior after depletion of human CENP-X by siRNA-based knockdown in HeLa cells (Fig. 2 B). In contrast to DT40 cells, CENP-X RNAi in HeLa cells resulted in numerous mitotic defects. Although we observed a small subset of CENP-X–depleted cells in which chromosomes were largely aligned at a metaphase plate, numerous instances of moderately and highly misaligned chromosomes were observed in these cells (Fig. 2 B). This strong defect in chromosome alignment is qualitatively similar to our previous analyses of CENP-K– and CENP-W–depleted HeLa cells (Okada et al., 2006; Hori et al., 2008a). However, in contrast to CENP-K and -W depletions, we did observe some examples of cells that had apparently entered anaphase based on the presence of separated sister kinetochores.
To determine whether CENP-X has overlapping functions with other CCAN proteins, we also conducted double depletions between CENP-X and CENP-Q, -K, or -W by RNAi. This strategy previously demonstrated a synergy between CENP-K and human KNL1 for assembly of the human kinetochore (Cheeseman et al. 2008). However, double CENP-X/CENP-Q–, CENP-X/CENP-K–, and CENP-X/CENP-W–depleted cells did not show synergistic phenotypes compared with the singly depleted cells (unpublished data). In total, these results suggest that depletion of human CENP-X causes defects that are distinct from those associated with other CCAN components and suggests that CENP-X plays a critical role in proper chromosome segregation.
Kinetochore localization of the CENP-S–CENP-X complex occurs downstream of the CENP-H complex but is distinct from the CENP-O complex
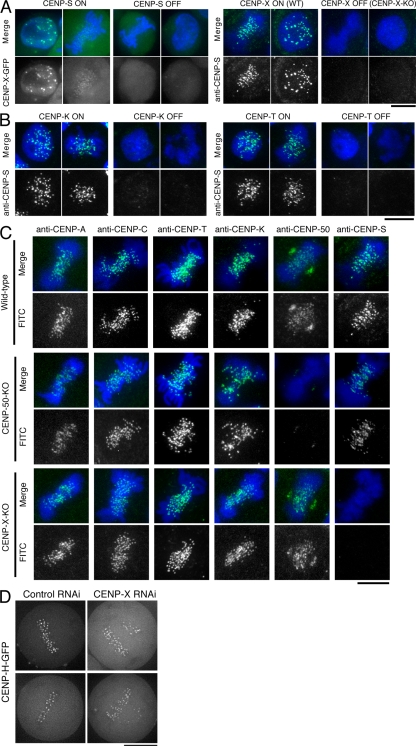
To define the relationship of CENP-S and -X to the other components of the CCAN, we next conducted localization experiments. We found that CENP-X localization was abolished in CENP-S–deficient cells and that CENP-S localization was eliminated in CENP-X–deficient cells (Fig. 3 A). We also observed that CENP-X RNAi in human cells eliminated GFP–CENP-S kinetochore localization (Fig. S3 D). In combination with the biochemical purifications described in the previous section, these results support a model in which CENP-S and -X form a closely associated complex.
Figure 3.
Kinetochore localization of the CENP-S–CENP-X complex occurs downstream of the CENP-H complex but is distinct from the CENP-O complex. (A) CENP-X localization in CENP-S–deficient cells (CENP-S OFF) and CENP-S localization in CENP-X–deficient cells (CENP-X OFF). WT, wild type. (B) CENP-S localization in CENP-K– or CENP-T–deficient cells (CENP-K OFF or CENP-T OFF, respectively). (C) Immunofluorescence analysis in wild-type DT40 and CENP-50– and CENP-X–deficient cells with the indicated antibodies (green). DNA was counterstained with DAPI (blue). (D) Imaging of human HeLa cells stably expressing GFP-tagged CENP-H after siRNA-based knockdown for control or CENP-X. Bars, 10 µm.
We next examined the kinetochore localization of CENP-S in CENP-K (a component of the CENP-H–containing complex)– or CENP-T–deficient cells. The kinetochore localization of CENP-S was eliminated in both CENP-K– and CENP-T–deficient cells (Fig. 3 B). We also examined the kinetochore localization of representative CCAN components, including CENP-C, -T, -K, -50 (U), and -S in CENP-X– and CENP-50 (U)–deficient cells. Our observations indicate that the localization of the tested CCAN proteins did not change in CENP-X– and CENP-50 (U)–deficient cells (Fig. 3 C).
The phenotypes of the CENP-S– and CENP-X–deficient cells are distinct from those of cells with KO of the CENP-O complex proteins based on the recovery from spindle damage assay (Fig. S3 C). Consistent with these distinct functional roles, we observed clear CENP-S signals in CENP-50 (U)–deficient cells (Fig. 3 C), indicating that the kinetochore localization and function of the CENP-S–CENP-X complex is distinct from the CENP-O complex. We have previously shown that CENP-S localization does not change in CENP-C–deficient cells (Hori et al., 2008a), suggesting that CENP-C has distinct function from the CENP-S–CENP-X complex. We also confirmed that the localization of human CENP-H and -O is not affected in human cells depleted for CENP-X (Fig. 3 D and not depicted). In total, these localization experiments demonstrate that kinetochore targeting of the CENP-S–CENP-X complex occurs downstream of the CENP-H complex and is distinct from CENP-C and the CENP-O complex.
Kinetochore outer plates of CENP-S–deficient cells are smaller than those of wild-type cells
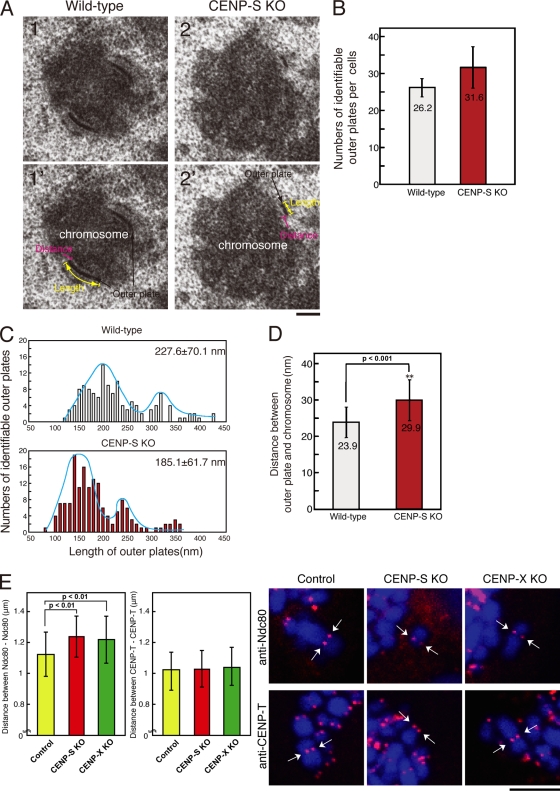
Although we did not observe an apparent reduction in the kinetochore localization of established CCAN proteins in CENP-S– or CENP-X–deficient cells (Fig. 3), it is possible that CENP-S and -X play a role in controlling outer kinetochore assembly. To determine whether CENP-S or -X play a global role in controlling the assembly of non-CCAN proteins, we examined the structural morphology of the kinetochore outer plate by EM. For these experiments, we imaged 30–40 170-nm-thick serial sections for individual mitotic cells treated with nocodazole. In both CENP-S–deficient cells and wild-type DT40 cells, we observed similar numbers of clear electron-dense kinetochore outer plates (Fig. 4, A and B). We also observed a similar number (26.6 ± 8.8) of outer plates in CENP-50 (U)–deficient cells. In contrast, we observed that CENP-H deficiency causes a severe reduction in the number of visible outer plates (unpublished data), as previously shown for CENP-T deficiency (Hori et al., 2008a).
Figure 4.
The length of the outer plate and the distance of the interkinetochore in CENP-S–deficient cells. (A) Electron micrograph showing a typical image of the kinetochore outer plate in wild-type (1 and 1′) or CENP-S–deficient cells (2 and 2′). (B) Numbers of identifiable outer plates per cell (mean ± SD). Approximately 35 serial sections were made for each cell, and the numbers of outer plates were counted. (C) Distribution of the outer plate length in wild-type or CENP-S–deficient cells. In both cell lines, two peaks with distribution were observed. The two peaks in the distribution of the plate length are likely caused by differences in sample orientation. As the orientation of some cells is not plane of the section, the length of plates varies depending on the angle of the section against a chromosome. (D) Distance between outer plate and electron-dense chromatin region in wild-type or CENP-S–deficient cells. (E) Distances between Ndc80 and Ndc80 or CENP-T and CENP-T of control or CENP-S– or CENP-X–deficient metaphase cells in the absence of microtubules. Arrows indicate sister kinetochore pairs. (D and E) Error bars represent SD. Bars: (A) 200 nm; (E) 5 µm.
Although we did not detect differences in the numbers of outer plates between CENP-S–deficient cells and wild-type DT40 cells, we found that the plate length of CENP-S–deficient cells was shorter than that of wild-type cells. As shown in Fig. 4 C, the length of outer kinetochore plates varied even in individual cells. In wild-type cells, we observed a main peak of plate length of ∼200 nm and a shoulder peak of ∼300 nm, with a mean plate length of 227 nm (±70.5 nm; Fig. 4 C). In CENP-S–deficient cells, we observed a main peak of ∼150 nm and a shoulder peak of ∼240 nm, with a mean size of 185 nm (±61.7 nm; Fig. 4 C). The plate length of CENP-50 (U)–deficient cells (261 ± 148.5 nm) was not significantly different from that of wild-type cells. These data suggest that CENP-S is involved in the formation of a functional kinetochore outer plate, which is essential for kinetochore–microtubule attachment and faithful mitotic progression. Liu et al. (2006) have previously reported that outer plate morphology is altered after treatment of siRNA against CENP-I. The contribution of CENP-S to the formation of a functional kinetochore may function coordinately with the CENP-I pathway.
Intrakinetochore distance is increased in CENP-S– and CENP-X–deficient cells in the absence of tension
During our analysis of outer plate structure by EM, we observed that the distance between the outer plate and electron-dense chromatin region in CENP-S–deficient cells was longer than that in wild-type cells (Fig. 4 A). Therefore, we measured the distance in ∼150 kinetochores from our EM analysis for both CENP-S–deficient and wild-type cells and found that the distance in CENP-S–deficient cells (29.9 nm) is significantly longer than that in wild-type cells (23.9 nm; Fig. 4 D). The distance in CENP-50 (U)–deficient cells was 26.3 nm, which is slightly longer than that in wild-type cells but shorter than that in CENP-S–deficient cells. Recent studies suggested that the condensin complex regulates inter- and intrakinetochore distances under tension (Maresca and Salmon, 2009; Uchida et al., 2009). However, inter- and intrakinetochore distances are not changed in condensin-null DT40 cells in the absence of tension (Ribeiro et al., 2009), which suggests that there is a mechanism to prevent the kinetochore from stretching in the absence of external force. Our EM analysis was performed after treatment with nocodazole, which eliminates kinetochore–microtubule attachments and spindle forces. To confirm the EM observations, we stained chromosomes with anti–CENP-T (an inner kinetochore protein) or anti-Ndc80 (Hec1; an outer kinetochore protein) antibodies after nocodazole treatment and measured the distances between sister kinetochores in CENP-S– and CENP-X–deficient and wild-type cells. The distance between Ndc80 at two paired sister kinetochores was increased in CENP-S–CENP-X–deficient cells. In contrast, the distance between CENP-T between two paired inner kinetochores was not changed (Fig. 4 E). These results indicate that the intrakinetochore distance is increased in both CENP-S and –CENP-X–deficient cells and that this occurs structurally between the inner kinetochore protein CENP-T and the outer kinetochore protein Ndc80.
CENP-S– and CENP-X–deficient cells display reduced localization of outer kinetochore proteins
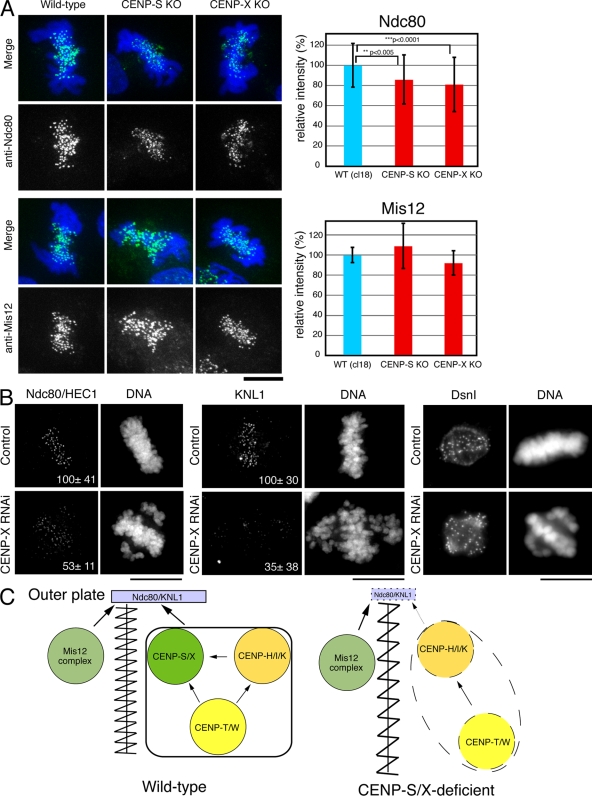
The EM analysis suggests that there are defects in the proper formation of a functional kinetochore outer plate in CENP-S–deficient cells. Therefore, we next analyzed the localization of outer kinetochore proteins in CENP-S– or CENP-X–deficient cells. The KMN network is located at the outer plate (DeLuca et al., 2005) and has a central role in forming kinetochore–microtubule attachments (Cheeseman et al., 2006). Although the KMN network subunits KNL1, Mis12, and Ndc80 all localized to kinetochores in CENP-S– and CENP-X–deficient DT40 cells, we found that the signal intensities of Ndc80 (∼80% relative to control; Fig. 5 A) and KNL1 (not depicted) were reduced in CENP-S– and CENP-X–deficient cells relative to controls. In contrast, we did not detect significant differences of Mis12 signals between the control and mutant cells.
Figure 5.
Localization of outer plate proteins in CENP-S– or CENP-X–deficient cells. (A) Immunofluorescence analysis in wild-type (WT) DT40 or CENP-S– or CENP-X–deficient cells with anti-Ndc80 and -Mis12 antibodies. Signal intensities at each kinetochore were measured in these cells. Error bars represent SD. (B) Immunofluorescence analysis with anti-Ndc80, -KNL1, or -Dsn1 antibodies in HeLa cells treated with control or CENP-X siRNAs. The numbers in the micrographs are relative signal intensities of kinetochore signals. (C) A model for the assembly of kinetochore proteins at mitosis. The CENP-S–CENP-X complex is essential for the stable assembly of outer kinetochore proteins. In addition, the CENP-S–CENP-X complex generates a discrete CCAN structure to prevent the kinetochore from overstretching (left). In CENP-S– and CENP-X–deficient cells, the amount of KNL1 and Ndc80 at kinetochores is reduced, and CCAN structure is less tight, which causes kinetochore to be overstretched (right). Bars, 10 µm.
We also conducted similar experiments to determine whether CENP-X is required for outer kinetochore assembly in human cells (Fig. 5 B). We observed a strong reduction in the levels of human KNL1 (35 ± 38% relative to control) and Ndc80 (53 ± 11% relative to control) at kinetochores, but the level of Dsn1 (a Mis12 complex protein) was not changed. In total, these results demonstrate that the assembly of the outer kinetochore components KNL1 and Ndc80 is compromised in CENP-S– and CENP-X–deficient cells, which causes reduced outer plate formation.
A functional role for the CENP-S–CENP-X complex in the assembly of a kinetochore structure
Based on phenotype analysis of CENP-S– and CENP-X–deficient cells and our previous work on the function and organization of the other CCAN components (Okada et al., 2006; Hori et al., 2008a, b), we propose a model in which the CENP-S–CENP-X complex is essential for the stable assembly of outer kinetochore proteins (Fig. 5 C). Although CENP-S has a potential histone-fold domain, the significance of this domain to the function of CENP-S–CENP-X complex is unclear. As the data in this study indicate, the CENP-S–CENP-X complex functions downstream of other components of the CCAN, and depletion of the CENP-S–CENP-X complex does not affect the localization of other CCAN proteins. Thus, the CENP-S–CENP-X complex may directly allow the outer kinetochore to form efficiently and stably on the CCAN platform.
We also found that the intrakinetochore distance was increased in CENP-S– and CENP-X–deficient cells in the absence of tension (Fig. 4 E). Combined with our EM analysis, the inner kinetochore structure appears to be stretched in CENP-S–deficient cells (Fig. 5 C, right). To our knowledge, these are the first proteins that have been identified with a role in kinetochore assembly related to the structure of the intrakinetochore distance in the absence of tension. Although this function may not be essential under all circumstances, as is seen for the null phenotype in DT40 cells, the ability to assemble a coherent kinetochore structure capable of resisting the force from the spindle microtubules and preventing merotelic attachment to opposing spindle poles is critical to the integrity of this important cell division structure.
Materials and methods
Molecular biology, cell culture, and transfection
Chicken target disruption constructs for each gene were generated such that genomic fragments encoding several exons were replaced with drug resistance cassettes under control of the β-actin promoter. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad Laboratories). All DT40 cells were cultured at 38°C in Dulbecco’s modified medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin. To repress the expression of the tetracycline-responsive transgenes, tetracycline (Sigma-Aldrich) was added to the culture medium to a final concentration of 2 µg/ml. To deplete human CENP-X, a pool of four siRNAs was obtained from Thermo Fisher Scientific (5′-CACCUGCACUUCAAGGAUG-3′, 5′-GACCAAAGAAGCAGCAGUC-3′, 5′-CGCAGCUGCUCCUGGACUU-3′, and 5′-ACGUGGACCAGCUGGAGAA-3′) and transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer’s guidelines.
Biochemical analysis
DT40 whole cell extracts were suspended in lysis buffer (50 mM Na phosphate, pH 8.0, 0.3 M NaCl, 0.1% NP-40, 5 mM β-mercaptoethanol, complete protease inhibitor [Roche], and 20 U/ml DNase I [Takara]) and were centrifuged at 20,000 g for 10 min at 4°C. The soluble supernatant was fractionated on a Superose 6 gel filtration column (GE Healthcare) in 50 mM Na phosphate, pH 8.0, 0.3 M NaCl, 0.3 M imidazole, 0.1% NP-40, and 1 mM β-mercaptoethanol at 4°C.
For IP experiments, DT40 cells expressing Flag-tagged proteins were lysed in 5 ml of lysis buffer and centrifuged at 20,000 g for 10 min at 4°C. Anti-Flag M2 beads (Sigma-Aldrich) were incubated with the supernatant fraction for 2 h at 4°C, washed with lysis buffer, and eluted with lysis buffer in the presence of 3× Flag peptide (Sigma-Aldrich).
Immunofluorescence and image acquisition
Cells were collected onto slides with a cytocentrifuge and fixed in 3% paraformaldehyde in PBS for 15 min at room temperature or in 100% methanol for 15 min at −20°C, permeabilized in 0.5% NP-40 in PBS for 10 min at room temperature, rinsed three times in 0.5% BSA, and incubated for 1 h at 37°C with primary antibody. Binding of primary antibody was then detected with Cy3- or FITC-conjugated goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories) diluted to an appropriate concentration in PBS/0.5% BSA. Affinity-purified rabbit polyclonal antibodies were used against recombinant chicken CENP-C (Fukagawa et al., 1999), CENP-H (Fukagawa et al., 2001), CENP-K (Okada et al., 2006), CENP-50 (Minoshima et al., 2005), CENP-T (Hori et al., 2008a), and CENP-S. Other antibodies used in this study were described previously (Nishihashi et al., 2002; Hori et al., 2003; Regnier et al., 2005). Chromosomes and nuclei were counterstained with DAPI at 0.2 µg/ml in Vectashield antifade (Vector Laboratories). Immunofluorescence images were collected with a cooled EM charge-coupled device camera (Quntem; Roper Scientific) mounted on an inverted microscope (IX71; Olympus) with a 100× NA 1.40 objective lens together with a filter wheel at room temperature. Subsequent analysis and processing of images were performed using MetaMorph software (Roper Scientific). Immunofluorescent human cells were imaged on a Deltavision Core system (Applied Precision, LLC) equipped with a camera (CoolSNAP HQ2; Photometrics). All images were scaled and processed identically.
Living cell observation
For live cell imaging, a histone H2B-RFP plasmid was transfected into CENP-S–deficient cells to visualize chromosomes. Fluorescently stained living cells were observed with an IX71 inverted microscope with an oil immersion objective lens (Plan-Apochromat 60× NA 1.40) in a temperature-controlled box to keep the temperature at 38°C. Time-lapse images were recorded at 3-min intervals with an exposure time of 0.2–0.3 s using a CoolSNAP HQ2 camera. Subsequent analysis and processing of images were performed using IPLab software (Signal Analytics).
EM
DT40 cells were treated with 500 ng/ml nocodazole for 3 h and fixed in 2.5% glutaraldehyde and 0.15% tannic acid in the 0.1 M Na cacodylate buffer for 1 h. Postfixation was performed in 2% OsO4 for 1 h on ice. The cells were dehydrated in ethanol and then infiltrated with Epon 812. Polymerization was performed at 60°C for 48 h. Serial sections were cut with an ultramicrotome (Leica) equipped with a diamond knife (Diatome), and sections were stained with uranyl acetate and lead citrate and examined in an electron microscope (JEM1010; JEOL). The contrast of each image was normalized with the brightness of the membrane structure.
Online supplemental material
Fig. S1 shows the relation of the CENP-S–CENP-X complex with the CENP-T–CENP-W complex. Fig. S2 indicates the strategy for generating KO cell lines for CENP-S and -X. Fig. S3 shows the phenotype for CENP-S– and CENP-X–deficient cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903100/DC1.
Acknowledgments
We are very grateful to K. Suzuki, M. Takahashi, and K. Kita for technical assistance. We also thank E. Suzuki for advice on EM experiments.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T. Fukagawa and awards from the Richard and Susan Smith Family Foundation, the Massachusetts Life Sciences Center, and the Searle Scholars Program to I.M. Cheeseman.
Footnotes
Abbreviations used in this paper: CCAN, constitutive centromere-associated network; IP, immunoprecipitation; KMN, KNL1–Mis12 complex–Ndc80 complex; KO, knockout
References
- Cheeseman I.M., Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface.Nat. Rev. Mol. Cell Biol. 9:33–46. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore.Cell. 127:983–997. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A.. 2008. KNL1 and CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates.Mol. Biol. Cell. 19:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Dong Y., Hergert P., Strauss J., Hickey J.M., Salmon E.D., McEwen B.F.. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites.Mol. Biol. Cell. 16:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1.Cell. 127:969–982. [DOI] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R., III, Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex.Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Pendon C., Morris J., Brown W.. 1999. CENP-C is necessary but not sufficient to induce formation of functional centromere.EMBO J. 18:4196–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T.. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells.EMBO J. 20:4603–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Haraguchi T., Hiraoka Y., Kimura H., Fukagawa T.. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells.J. Cell Sci. 116:3347–3362. [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W., Suzuki E., Okawa K., et al. 2008a. The CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore.Cell. 135:1039–1052. [DOI] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., Fukagawa T.. 2008b. CENP-O class proteins form a stable complex and are required for proper kinetochore function.Mol. Biol. Cell. 19:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K.. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells.Genes Cells. 11:673–684. [DOI] [PubMed] [Google Scholar]
- Kwon M.S., Hori T., Okada M., Fukagawa T.. 2007. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly.Mol. Biol. Cell. 18:2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T., Rattner J.B., Jablonski S.A., Yen T.J.. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells.J. Cell Biol. 175:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T.J., Salmon E.D.. 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity.J. Cell Biol. 184:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S.E., Borusu S., Amaro A.C., Winter J.R., Belwal M., McAinsh A.D., Meraldi P.. 2007. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity.EMBO J. 26:5033–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y., Hori T., Okada M., Kimura H., Haraguchi T., Hiraoka Y., Bao Y.C., Kawashima T., Kitamura T., Fukagawa T.. 2005. The constitutive centromere component CENP-50 is required for recovery from spindle damage.Mol. Cell. Biol. 25:10315–10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W.C., Fukagawa T.. 2002. CENP-I is essential for centromere function in vertebrate cells.Dev. Cell. 2:463–476. [DOI] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R., III, Desai A., Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres.Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W.C., Brown W.. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1.Mol. Cell. Biol. 25:3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P.. 2009. Condensin regulates the stiffness of vertebrate centromeres.Mol. Biol. Cell. 20:2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.M., Mueller L., Collins S.J.. 1996. E3, a hematopoietic-specific transcript directly regulated by the retinoic acid receptor alpha.Blood. 88:2517–2530. [PubMed] [Google Scholar]
- Uchida K.S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T.. 2009. Kinetochore stretching inactivates the spindle assembly checkpoint.J. Cell Biol. 184:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]