Abstract
Autophagy is a cytoplasmic degradative pathway that can participate in biosynthetic processes, as in the yeast Cvt pathway, but is more commonly known for its functions in removing damaged or surplus organelles and macromolecular complexes. Here, we find that autophagy intersects with human immunodeficiency virus (HIV) biogenesis, mirroring the above dichotomy. Early, nondegradative stages of autophagy promoted HIV yields. HIV Gag-derived proteins colocalized and interacted with the autophagy factor LC3, and autophagy promoted productive Gag processing. Nevertheless, when autophagy progressed through maturation stages, HIV was degraded. This, however, does not occur, as the HIV protein Nef acts as an antiautophagic maturation factor through interactions with the autophagy regulatory factor Beclin 1, thus protecting HIV from degradation. The dual interaction of HIV with the autophagy pathway enhances viral yields by using the early stages while inhibiting the late stages of autophagy. The role of Nef in the latter process enhances yields of infectious HIV and may be of significance for progression to clinical AIDS.
Introduction
Autophagy is a general homeostatic process in eukaryotic cells whereby portions of the cytoplasm, containing cytosol or organelles, are sequestered into double membrane-bound autophagic vacuoles for fusion with lysosomal organelles and subsequent degradation of the captured contents in the resulting autolysosomes (Shintani and Klionsky, 2004; Klionsky, 2007). A functional core autophagy pathway and associated processes are important for cell survival under starvation or growth factor withdrawal conditions, programmed cell death, elimination of aggregated proteins, and removal of surplus or damaged organelles (Levine and Klionsky, 2004; Levine and Kroemer, 2008). Autophagy is regulated by signaling pathways centered around the Ser/Thr protein kinase Tor (target of rapamycin) and phosphatidylinositol 3-kinases (PI3Ks), both type I (inhibitory to autophagy) and type III (essential for execution of autophagy). The type III PI3K hVPS34 acts in a complex with Beclin 1 (yeast Atg6), a factor endowing hVPS34 with its role in autophagy (Pattingre et al., 2005). A detailed picture on these and other autophagy proteins (Atg) in mammalian cells is emerging, with the core pathway resembling that in yeast (Levine and Klionsky, 2004; Shintani and Klionsky, 2004; Klionsky, 2007).
Autophagosome biogenesis and wrapping around autophagic targets is facilitated by the two specialized protein conjugation systems: the Atg5-12/16 complex stimulates a second conjugation system, whereby LC3 (Atg8) undergoes conversion from its free C-terminus state (LC3-I) to its C-terminally lipidated form (LC3-II) covalently modified by phosphatidylethanolamine. The lipidated LC3-II localizes to the membrane of a growing phagophore. Once a phagophore closes, this results in the formation of a double membrane-delimited autophagosome that typically matures into an autolysosome through fusion with multivesicular body (MVB) compartments (Gruenberg and Stenmark, 2004) and other lysosomal organelles (Shintani and Klionsky, 2004). Most cells undergo baseline autophagy to remove protein aggregates and spuriously damaged mitochondria or other organelles, or to adjust the cellular biomass (Levine and Kroemer, 2008). With a broad range of targets, ranging from protein complexes to whole organelles, autophagy is a process affecting a multitude of health and disease states; has been implicated in neurodegeneration, cancer, and aging (Levine and Kroemer, 2008; and has emerged as an important player in inflammatory and infectious diseases (Levine and Deretic, 2007; Deretic and Levine, 2009).
Autophagy is now well recognized as an innate and adaptive immunity mechanism (Levine and Deretic, 2007; Schmid and Munz, 2007). Pharmacologically, physiologically, or immunologically induced autophagy can act as a powerful antimicrobial defense (Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005; Birmingham et al., 2006, 2008; Singh et al., 2006; Levine and Deretic, 2007; Yano et al., 2008; Deretic and Levine, 2009). Autophagy is under the control of immune receptors and cytokine signaling (Levine and Deretic, 2007; Schmid and Munz, 2007), and is stimulated upon microbial recognition by innate immunity pattern recognition receptors (Lee et al., 2007; Sanjuan et al., 2007; Xu et al., 2007; Delgado et al., 2008) or activation with Th1 cytokines (Harris et al., 2007). However, certain pathogens can harness this process to assist their own propagation (Jackson et al., 2005; Orvedahl et al., 2007; Birmingham et al., 2008; Deretic and Levine, 2009). Interestingly, a recent large scale siRNA screen of host cell factors required for human immunodeficiency virus (HIV) type 1 (HIV-1) replication has identified several Atg factors among >250 HIV dependency host genes (Brass et al., 2008). Thus far, no in-depth functional links between Atg proteins or processes and HIV have been established.
Here, we tested mechanistically whether and how autophagy affects HIV yields during de novo virion generation. We found that the Atg proteins LC3 and Beclin 1 (Atg6) are found in complexes with the HIV proteins Gag and Nef, respectively. The latter interaction provides the basis for Nef function in control of autophagy. The Nef protein of HIV-1 and simian immunodeficiency virus (SIV) is required for efficient viral replication and acquired immune deficiency syndrome (AIDS) pathogenicity in HIV-1–infected humans or SIV-infected macaques (Daniel et al., 1992; Deacon et al., 1995; Kirchhoff et al., 1995). The methods by which the Nef protein acts as a pathogenic factor in vivo are not fully understood, but involve several mechanisms (Geleziunas et al., 2001; Swingler et al., 2003; Peterlin, 2006; Roeth and Collins, 2006). Recent findings suggest that the inability of lentivirus Nef to suppress CD4+ T cell activation correlates with viral pathogenesis (Schindler et al., 2006; Schindler et al., 2008). Our findings presented here uncover an additional, previously unappreciated Nef action in control of autophagy. Nef functions in preventing destruction of HIV components in autolysosomes, thus shielding HIV from autophagy in its role of a cell autonomous antimicrobial defense.
Results
Basal autophagy augments HIV yields in macrophages
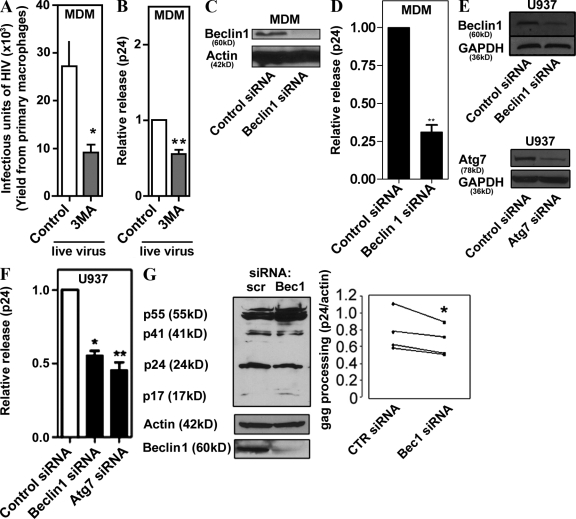
Basal autophagy is operational in all cell types, particularly in mononuclear phagocytic and dendritic cells (Schmid et al., 2007). To test whether basal autophagy can affect HIV yields, we pharmacologically inhibited autophagy in primary human macrophages differentiated from peripheral blood monocytes and infected with the macrophage-tropic HIV strain SF162, and determined yields of the infectious virus released from the macrophages. The macrophages treated with 3-methyl adenine (3MA), a conventional inhibitor of autophagy, yielded threefold fewer infectious virions compared with the untreated control (Fig. 1 A). The magnitude of this effect could be increased with higher inhibitor concentrations, but we used mild pharmacological and other treatments in these and subsequent experiments to avoid nonspecific effects. The autophagy requirement for optimal HIV yields was also assessed by determining extracellular release of the HIV capsid protein Gag p24 (Ono and Freed, 2004). The p24 released from live virus–infected primary macrophages was reduced in cells subjected to inhibition by 3MA (Fig. 1 B). Because pharmacological inhibitors such as 3MA may not affect only the autophagic pathway, we ascertained a role of the bona fide autophagy pathway by knocking down Beclin 1 (Atg6) in primary macrophages (Fig. 1, C and D) and in macrophages differentiated from monocytic U937 cells (Fig. 1, E and F). Knockdown of Beclin 1 diminished p24 yields in both primary monocyte- and U937-derived macrophages (Fig.1, D and F). A knockdown of another essential autophagy factor, Atg7, resulted in a similar effect on p24 yields (Fig. 1 F); Atg7 and Beclin 1 knockdowns affected autophagy in U937 cells, as determined by GFP-LC3 puncta/cell counts (Fig. S1). We next examined the intracellular Gag processing by monitoring Gag-derived p24 band intensity in immunoblots of cell extracts. When U937 cells knocked down for Beclin 1 were infected with vesicular stomatitis virus G (VSV-G)–pseudotyped NL4-3 HIV 1, cellular p24 levels were reduced compared with controls (Fig. 1 G). Collectively, these results indicate that basal autophagy promotes optimal Gag processing and yields of HIV in macrophages.
Figure 1.
Autophagy is required for optimal HIV yields in macrophages. (A) Pharmacological blockage of autophagy inhibits release of infectious virions. Human peripheral blood MDM were infected with SF162 HIV-1 for 10 d, then washed and incubated with control media or 3MA for 4.5 h. Culture supernatants containing HIV particles were used for a MAGI infectivity assay as described in Materials and methods. (B) Relative viral release was calculated as a ratio of extracellular-to-intracellular Gag-derived core antigen capsid protein CA (p24) and normalized to the control. (C) Western blot showing siRNA knockdown of Beclin 1, 7 d after transfection in MDM. (D) Human MDM were transfected with siRNA to Beclin 1 and infected with SF162 HIV-1 for 7 d, then p24 yields were quantified. (E) Western blots showing siRNA knockdown of Beclin 1 and Atg7 48 h after transfection in U937 cells. (F) Knockdown of autophagy regulators Atg7 and Beclin 1 inhibits basal viral yields released from macrophages. U937 cells were cotransfected with Beclin 1 or Atg7 siRNA and pMSMBA (a clone of NL4-3). Data indicate means; error bars indicate ±SEM (n ≥ 3). *, P < 0.05; **, P < 0.01; †, P > 0.05 (analysis of variance [ANOVA]). (G) U937 cells were knocked down for Beclin 1 expression and infected with VSV-G–pseudotyped HIV. Cell lysates were performed for Gag processing analysis. *, P < 0.05, paired t test.
HIV and HIV Gag-derived proteins colocalize with the autophagy marker LC3
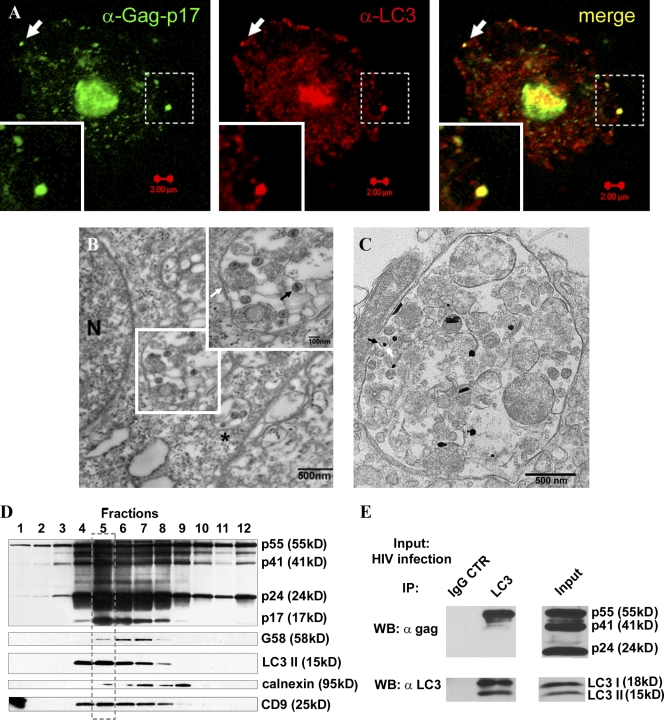
To test whether and how the autophagy pathway intersects with HIV, we examined the relative distribution of HIV virions and Atg proteins. In macrophages, HIV virions are found in membranous domains (Gendelman et al., 1988; Raposo et al., 2002; Pelchen-Matthews et al., 2003; Morita and Sundquist, 2004; Deneka et al., 2007; Jouve et al., 2007) recently shown to be contiguous with plasma membrane (Jouvenet et al., 2006; Deneka et al., 2007; Welsch et al., 2007), which facilitates colocalization studies. The Gag-p17–specific antibody showed colocalization of the budded virus with autophagy marker LC3 (Atg8; Fig. 2 A). By ultrastructural analysis, HIV virions were observed in these compartments (Fig. 2 B), which, based on presence of clathrin-coated pits (Fig. 2 B, asterisk; and Fig. S1 C), were consistent with the previously reported plasma membrane connections (Deneka et al., 2007). These morphologically identified compartments also labeled for LC3 (Fig. 2 C) and p24 (Fig. S1 D). A dual labeling procedure was not practicable, as LC3-enhanced immunogold labeling resulted in globular, oval, and acicular shapes, and precluded clear distinction.
Figure 2.
Autophagy protein LC3 colocalizes, copurifies, and coprecipitates with HIV Gag. (A) MDM were infected with VSV-G–pseudotyped HIV and immunostained for Gag-p17 and LC3. Arrows, a peripheral structure as an example of Gag-p17 and LC3 overlap. (B) Ultrastructural analysis of HIV virions in macrophages infected with HIV. U937 cells were infected with VSV-G–pseudotyped HIV. (inset) Enlarged region boxed in the electron micrograph. White arrow, membrane; black arrow, HIV virion; asterisk, HIV virions in a membranous compartment with a clathrin-coated pit consistent with plasma membrane origin. An enlarged image of this profile is shown in Fig. S1 C. (C) HIV-containing compartments are positive for LC3. Immunoelectron microscopy showing gold particles (enhanced gold particles appear globular, oval, and acicular) of LC3 in HIV-containing compartments. Arrow: virion and LC3 gold particle. See Fig. S1 D for p24 immunoelectron microscopy analysis. (D) HIV Gag precursor and Gag-derived proteins cofractionate with LC3 and the tetraspanin CD9. Subcellular organelle fractionation via isopycnic sucrose gradient separation was performed with lysates from HIV-infected cells (see Materials and methods). 12 fractions starting from the top were immunoblotted for the indicated proteins and organellar markers. The box with the broken line indicates peak band intensity fractions for LC3-II, Gag, and Gag-derived polypeptides, and CD9. (E) HIV Gag coimmunoprecipitates with LC3. U937 cells were infected with HIV and lysates immunoprecipitated for LC3. Immunoblotting with p24 and LC3 antibodies was performed on lysate and immunoprecipitate samples. The p24 antibody recognizes all three Gag proteins, as shown in the input. Note that only the precursor Gag-p55 comes down in immunoprecipitates with LC3 (n = 3).
Biochemical analysis of HIV Gag-derived proteins shows copurification and interactions with the autophagic protein LC3
To determine at the biochemical level whether HIV intersects with the autophagy pathway, we subjected HIV-infected macrophages to subcellular fractionation by isopycnic centrifugation in sucrose gradients. Fig. 2 D shows that membranes containing HIV Gag polypeptides p24 and p17 cofractionated (note the coinciding peaks framed in Fig. 2 D) with the autophagic protein LC3. These membranes were enriched for the lipidated form of LC3, LC3-II, normally generated during engagement in an autophagic conjugation cascade driving autophagy initiation and elongation (Kabeya et al., 2000), and the recently recognized plasma membrane–associated autophagic events in macrophages (Sanjuan et al., 2007). As expected, LC3-I, the soluble cytosolic form of LC3, was not found on these membranes, although it was detectable in whole cell lysates (Fig. S2 A). The LC3-II–positive membranes enriched for Gag p55, Gag processing intermediate p41, and Gag products p24 and p17 did not copurify with the ER marker calnexin, but did cofractionate with CD9, a tetraspanin previously reported to colocalize with HIV virions in monocyte-derived macrophages (MDM; Fig. 2 D; Deneka et al., 2007).
We next tested whether HIV Gag interacted with autophagy proteins in coimmunoprecipitation experiments. Fig. 2 E shows that LC3 is found in protein complexes with the HIV Gag. These findings reinforce the subcellular fractionation experiments (Fig. 2 D), are in keeping with morphological analyses (Fig. 2, A–C), and demonstrate that HIV components and virions intersect with the autophagic pathway with the functional consequence of augmenting Gag processing (Fig. 1 G) and HIV yields (Fig. 1, A–F).
Pharmacological induction of autophagy enhances HIV yields
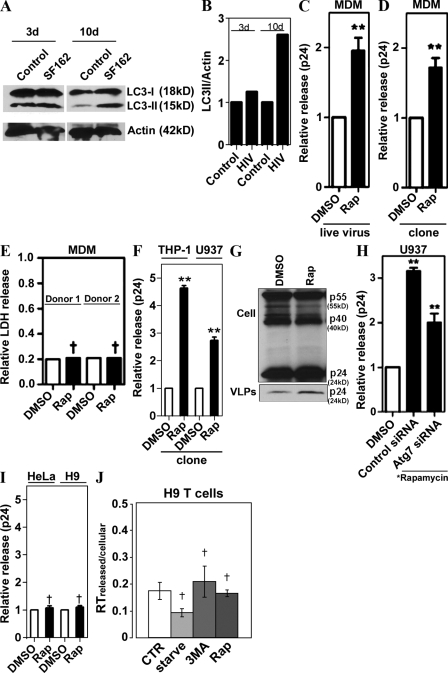
We next reasoned that although the basal autophagy is required for optimal HIV yields, physiologically, pharmacologically, and immunologically induced autophagy might affect HIV yields differently, i.e., by degrading HIV components en route to or at viral assembly sites. Significantly, induction of autophagy occurs during HIV infection of macrophages, as described previously (Delgado et al., 2008). Infection of primary human peripheral blood MDM with HIV-1 strain SF162 virus increased LC3-II levels at 10 d after infection (Fig. 3, A and B), which coincided with the expected (Prasad and Kalpana, 2009) peak HIV production in primary peripheral blood mononuclear cells.
Figure 3.
Induction of autophagy enhances HIV yields. (A) Human peripheral blood MDM were infected with SF162 HIV-1 for 3 or 10 d, and lysates were immunoblotted for LC3 (one of two equal experiments shown). (B) Quantification: LC3-II band intensity relative to actin. Representative data from one of two independent experiments. (C and D) Induction of autophagy promotes viral yields from MDM. (C) MDM were infected with SF162 HIV for 10 d, then washed and incubated with DMSO or 50 µg/ml rapamycin for 4 h. (D) MDM were transfected with a pMSMBA HIV clone for 48 h, then washed and incubated with DMSO or 50 µg/ml rapamycin for 4 h. Core antigen capsid protein (p24) was measured as in Fig. 1. (E) Assessment of nonspecific cytosolic release. Extracellular lactate dehydrogenase (LDH) release assay was performed with a kit from Promega according to manufacturer’s instructions using lysates and media from cells treated with DMSO or rapamycin. Relative LDH was calculated as the ratio between extracellular and total LDH, released from MDM from two different donors. (F) Induction of autophagy increases p24 yields in human monocytic cell lines. THP-1 and U937 cells differentiated into macrophages were transfected with pMSMBA for 48 h, then washed and incubated with DMSO or rapamycin for 4 h. (G) Induction of autophagy increases release of VLPs in human macrophages. U937 cells were infected with VSV-G–pseudotyped pMSMBA-derived virus for 48 h, then washed and treated for 5 h with rapamycin or DMSO alone. VLPs were isolated on a 20% sucrose cushion, and lysates and VLP were immunoblotted for Gag and p24, respectively. (H) U937 cells were cotransfected with siRNA and pMSMBA for 48 h, then washed and incubated with DMSO alone or 50 µg/ml rapamycin for 4 h. (I) HeLa and H9 cells were transfected with pMSMBA for 48 h, then washed and incubated with DMSO or rapamycin for 4 h, and relative viral release (p24) was determined. (J) H9 cells (a T cell line) were infected with T cell tropic HIVLAI for 4 d and washed; relative viral release was found by determining the ratios of extracellular versus intracellular reverse transcription levels. Data indicate means; error bars indicate ±SEM (n ≥ 3). *, P < 0.05; **, P < 0.01; †, P > 0.05 (ANOVA).
To test the effects of induced autophagy, macrophages were treated with the conventional inducer of autophagy rapamycin, and extracellular HIV yields were determined. Primary macrophages treated with rapamycin yielded higher p24 levels than untreated control when infected with the live virus or transfected with a virus molecular clone (Fig. 3, C and D). The increase in p24 was not caused by a nonspecific leakage of cytoplasmic contents, as the cytosolic enzyme lactate dehydrogenase levels found in the medium were not altered by rapamycin treatment (Fig. 3 E). Similar results were obtained when two monocytic cell lines, THP-1 and U937, differentiated into macrophages were tested (Fig. 3 F). Immunoblot analyses of cell-associated viral proteins and p24 in virus-like particles (VLP) in culture medium indicated that the increased p24 yield in macrophages treated with rapamycin was associated with VLP released from macrophages (Fig. 3 G). The autophagy machinery in these experiments was intact, as indicated by response to rapamycin both with LC3-I–to–LC3-II conversion and LC3 puncta formation (Fig. S2, B–D), and responsiveness of the effects to inhibition with the conventional autophagy inhibitor 3MA (Fig. S2, C and D). The enhancement effect of rapamycin on p24 release was counteracted by knocking down the key regulator of autophagy Atg7 (Fig. 3 H). Because Atg7 is a key autophagy factor, this indicates that the rapamycin effects on p24 are autophagy dependent. Thus, pharmacological induction of autophagy, in contrast to our predictions and previous findings of inhibitory effects on viral replication of rapamycin in low concentrations (Heredia et al., 2003; Roy et al., 2002), did not diminish but instead enhanced yields of the virus released from macrophages. The effects of autophagy induction appeared to be specific for macrophages, as we did not observe enhancement using rapamycin in HeLa or H9 T cell lines transfected with an HIV molecular clone (Fig. 3 I) and H9 T cells infected with the CXCR4 coreceptor using (X4, T cell tropic) virus HIVLAI (Fig. 3 J).
HIV protein Nef is required for enhanced HIV yields in response to autophagy induction
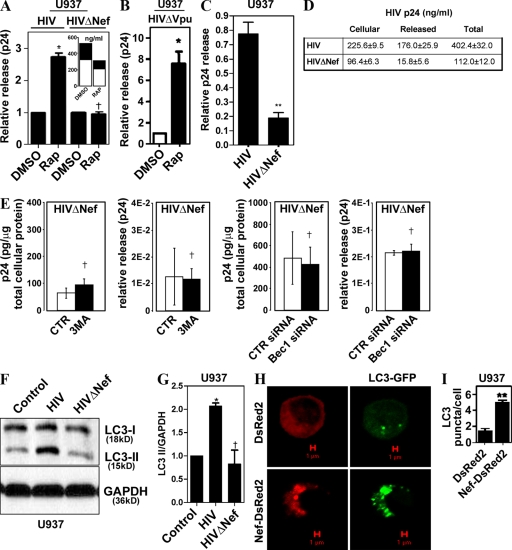
Given the observation that induced autophagy did not harm the virus, but further augmented its yields, we wondered whether the virus, in addition to using basal autophagy to increase its yields, also protected against induced autophagy, which can act as an antimicrobial cell-autonomous defense (Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005; Birmingham et al., 2006, 2008; Singh et al., 2006; Levine and Deretic, 2007; Yano et al., 2008). We investigated whether specific HIV-1 proteins affected autophagic machinery. A release of HIV deleted for nef was not stimulated by rapamycin in cells transfected with pNL4-3ΔNef, as shown in Fig. 4 A, where the data were normalized to represent fold change in relative p24 release. These data show that rapamycin has no additional effect on HIV yields when the virus lacks Nef. The absolute levels of both the released p24 and cell-associated p24 were proportionately reduced with HIVΔNef treated with rapamycin (Fig. 4 A, inset). As a consequence, the ratios remained the same (Fig. 4 A, main graph), although the absolute levels of p24 (both cellular and released) were diminished (Fig. 4 A, inset). In contrast to nef deletion, HIV deleted for vpu still responded to rapamycin stimulation with increased p24 levels (Fig. 4 C). Furthermore, ΔNef virus, although showing reduced relative release of the viral p24 (Fig. 4 C) and cellular p24 levels (Fig. 4 D), showed no further change in yields, release, or cellular p24 when autophagy was inhibited by 3MA (Fig. 4 E, left two panels) or suppressed by Beclin 1 knockdown (Fig. 4 E, right two panels), indicating that Nef is critical for the detectable effects of autophagy on HIV.
Figure 4.
Nef is required for yield-enhancing effects of autophagy on HIV. (A) U937 cells were transfected with pGFP-NL4-3ΔNef (HIVΔNef) for 48 h and incubated with DMSO or 50 µg/ml of rapamycin. (inset) Absolute values of p24 concentrations in cells (open bars) and released into the medium (shaded bars). Note that absolute levels of ΔNef virus are inhibitable by rapamycin but that the ratios of released versus cell-associated virus remain the same, as reflected in the main graph. (B) U937 cells were transfected with pMSMBA-vpu-null (HIVΔVpu) and tested as in A for rapamycin effects. (C) U937 cells were infected with 100 ng/ml each of VSV-G–pseudotyped HIV or HIVΔNef for 48 h, and p24 yields were quantified. (D) Absolute levels of cell-associated, released, and total p24 from samples in B. (E) Absence of basal autophagy inhibition effects on HIVΔNef yields. For experiments with 3MA, U937 cells were infected with NL4-3ΔNef for 48 h, then washed and treated for 4 h. Cells in experiments with Beclin 1 knockdowns were first transfected with siRNA, infected 24 h later, and harvested 48 h after infection. CTR, control. (F and G) Nef increases LC3-II (lapidated form). U937 cells were infected with VSV-G–pseudotyped pMSMBA-derived virus (HIV) or HIVΔNef for 3 d and immunoblotted for LC3. (E) Immunoblot. (F) Quantification (ratio of LC3-II to GAPDH band intensities). (H) U937 macrophages were cotransfected with GFP-LC3 and either DsRed2 or Nef-DsRed2 for 24 h. GFP-LC3 puncta were quantified in three independent experiments. (I) Quantification of LC3 puncta (≥1 µm) per cell. Data indicate means; error bars indicate ±SEM (n ≥ 3). *, P < 0.05; **, P < 0.01; †, P > 0.05 (ANOVA).
HIV causes Nef-dependent accumulation of early autophagic markers
We next examined HIV effects on the execution stages of autophagy. The gold standard for assessment of the early execution phases of autophagy (initiation and elongation) is based on monitoring biochemical and morphological changes that the autophagy protein LC3 undergoes (Kabeya et al., 2000). During the stages when autophagic isolation membranes (phagophores) begin to form and nascent autophagosomes elongate until they are completed by closure, LC3 converts from the nonlipidated cytosolic species (LC3-I) to a predominantly membrane-associated form (LC3-II) covalently modified at the C terminus by phosphatidylethanolamine. During maturation stages, LC3-II is consumed as a portion of it gets degraded in the autolysosomes. The level of LC3 lipidation is monitored by immunoblotting, and detected as a conversion from the unmodified LC3-I band to the lipidated LC3-II form, which shows increased electrophoretic mobility (Kabeya et al., 2000; Mizushima and Yoshimori, 2007). Complete HIV, but not HIV deleted for nef, increased levels of lipidated LC3, as reflected in the increase of LC3-II band on Western blots (Fig. 4 F), and LC3-II/loading control ratios (Mizushima and Yoshimori, 2007) using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the loading reference (Fig. 4 G). We next used another standard assay of autophagy, based on fluorescence microscopy detection of LC3 on autophagic membranes as punctate GFP-LC3 (LC3-II) vis-à-vis diffuse cytosolic GFP-LC3 (LC3-I; Kabeya et al., 2000). U937 cells were transfected with the previously well characterized expression clone of Nef-DsRed2, thoroughly documented in cell biological studies to fully correspond both in distribution and function to the untagged Nef (Roeth et al., 2004). Transfection of U937 cells with Nef-DsRed2 resulted in an increased abundance of GFP-LC3 puncta versus the control DsRed2-transfected cells (Fig. 4, H and I; and Fig. S2, E). Thus, Nef was responsible for accumulation of the early autophagic markers, the lipidated LC3-II form and LC3 puncta.
Nef inhibits autophagic maturation
The observed increase in early autophagic markers associated with Nef action is consistent with: (a) induction of autophagy or (b) a blockage of the maturation stages of autophagy. We first examined whether Nef affected the maturation (degradative) stages of the autophagic pathway. This was performed by testing Nef effects on the marquee autophagic degradative function: proteolysis of long-lived, stable proteins that are normally turned over by autophagy. We tested whether Nef affected autophagic proteolysis using the published assay for stable protein autophagic proteolysis in macrophages, optimized and functional only in the mouse macrophage cell line RAW264.7 (Roberts and Deretic, 2008). Transfection with Nef-DsRed did not induce autophagic proteolysis (Fig. S3 A). Instead, Nef-DsRed inhibited autophagic protein degradation induced by starvation, a gold standard for assessment of autophagy function (Fig. S3 A). Thus, Nef inhibits terminal, degradative stages of autophagy.
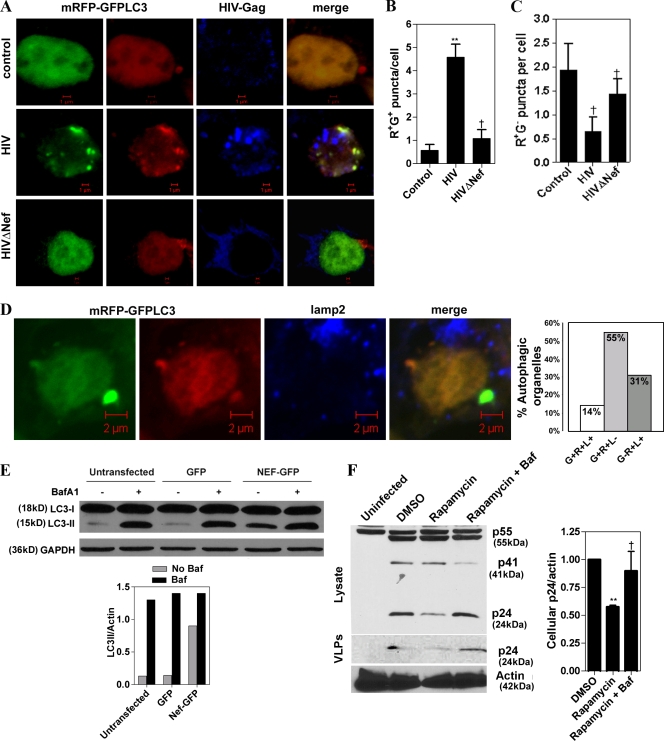
The role of Nef in inhibiting degradative stages of autophagy was further examined in human cells using the RFP-GFP-LC3 probe, a specialized tool for investigation of the autophagic flux, i.e., the maturation of autophagic organelles into degradative autolysosomal compartments (Kimura et al., 2007). Based on the sensitivity of GFP fluorescence to acidic pH and insensitivity of RFP fluorescence to low pH, it is possible to differentiate early, nonacidified autophagosomes (red+green+; yellow in merged images) from acidified, degradative autophagic organelles (red+green−; red in merged images; Kimura et al., 2007). In cells infected with Nef+ HIV, there was a pronounced accumulation of red+green+ (yellow) puncta, compared with uninfected cells or cells infected with ΔNef HIV (Fig. 5, A–C). This is in keeping with the conclusion that Nef blocks maturation of early autophagic organelles into acidified, degradative autolysosomes. Of the Nef-dependent red+green+ puncta, 85% were negative for the lysosomal protein Lamp2 (Fig. 5 D). All red+green− puncta (representing 31% of the total mRFP-GFP-LC3 puncta) were Lamp2 positive (Fig. 5 D). Expression of Nef-GFP resulted in an increase of LC3-II (Fig. 5 E). This was not or only slightly enhanced in the presence of bafilomycin A1 (Fig. 5 E, graph), an inhibitor of autophagosomal/autolysosomal acidification used to differentiate between effects on autophagy induction versus maturation (Mizushima and Yoshimori, 2007), which suggests that the bulk of Nef effects on autophagy were based on blocking autophagic flux.
Figure 5.
Nef inhibits autophagic maturation. (A) Nef blocks maturation of early autophagic organelles into autolysosomes. 293T cells were infected with VSV-G–pseudotyped HIV or HIVΔNef and transfected with mRFP-GFP-LC3 for 48 h, then immunostained for Gag and analyzed by confocal microscopy. Based on differential pH sensitivity of RFP and GFP, the mRFP-GFP-LC3 probe differentiates between early, nonacidified autophagosomes (red+green+; yellow in merged images) from acidified, degradative autolysosomes (red+green−; red in merged images). (B and C) Quantification of (red+green+) R+G+ and R+G− puncta per cell, respectively. (D) Analysis of Lamp2 association with RGP-GFP-LC3 profiles in HIV-infected 293T cells (HIV infection of >90% determined by staining with antibody to Gag). L+, percentage of Lamp2-positive profiles; L−, percentage of Lamp2-negative profiles. Data are from 42 cells from three slides. (E) LC3-II levels in Nef-transfected cells in the presence or absence of bafilomycin A1. 293T cells were transfected with GFP alone or Nef-GFP for 48 h. Cells were then incubated with or without bafilomycin A1 (Baf A1 or Baf) for 4 h and immunoblotted for LC3 and GAPDH. (E, bottom) Quantification: LC3/GAPDH ratios, representative of one of two experiments. (F) Inhibition of autophagic flux/maturation protects Nef-null virus from degradation. U937 cells were infected with VSV-G–pseudotyped HIVΔNef for 72 h, then washed and treated with rapamycin or rapamycin plus bafilomycin A1 (100 nM) for 5 h. VLP and cell lysates were subjected to immunoblot analysis. (F, right) Quantification of cellular p24 (n = 3). Data indicate means; error bars indicate ±SEM. *, P < 0.05; **, P < 0.01; †, P ≥ 0.05 (ANOVA).
Nef blocks autophagic degradation of HIV
We next tested whether Nef blocks HIV-specific autophagic degradation by monitoring the yields of HIV p24. U937 cells were infected with VSV-G–pseudotyped Nef-null HIV and treated with rapamycin. This led to a marked decrease in intracellular p24 levels and in lower p24 levels in VLP preparations (Fig. 5 F). The decrease in p24 was abrogated with bafilomycin A1, which blocks autophagic degradation (Fig. 5 F). Similar results were observed with cellular p24 levels (Fig. 5 F). These findings strongly indicate that Nef inhibits autophagic degradation of HIV biosynthetic intermediates or virions, and that this in turn enhances HIV yields.
HIV Nef colocalizes with autophagy regulators and is found in Beclin 1 protein complexes
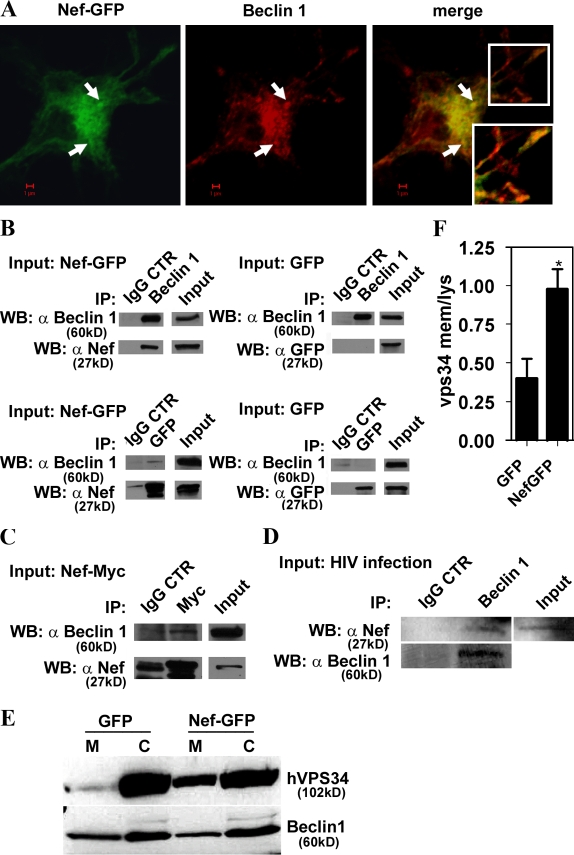
We next investigated intracellular distribution of Nef in relationship to autophagy regulators. Nef did not colocalize with mTOR (Fig. S3 B), so it is unlikely that it affects Tor directly. Nef showed a partial colocalization with 2xFYVE-GFP (Fig. S3 C), a probe binding to membranes containing phosphatidylinositol 3-phosphate (PI3P), the enzymatic product of type III PI3K hVPS34 that plays a critical role in autophagy when complexed with Beclin 1 (Kihara et al., 2001; Furuya et al., 2005; Pattingre et al., 2005; Zeng et al., 2006). Nef showed colocalization with autophagy factors Atg7 and Atg12 (Fig. S3, D and E), and colocalized (Figs. 6 A and S3 F) with the autophagic protein Beclin 1, which is the central regulator of autophagy at multiple stages (Liang et al., 1999; Pattingre et al., 2005). Immunoprecipitation of Beclin 1 in extracts from cells transfected with Nef-GFP resulted in the presence of Nef-GFP in the precipitated protein complexes (Fig. 6 B, top left). GFP was absent from the control samples when Beclin 1 was immunoprecipitated from cells transfected with GFP alone (Fig. 6 B, top right). A converse experiment using immunoprecipitation of GFP revealed the presence of Beclin 1 in immune complexes in cells transfected with Nef-GFP (Fig. 6 B, bottom left) but not in extracts from cells transfected with GFP alone (Fig. 6 B, bottom right). In a different configuration, using cells transfected with C-terminally myc epitope–tagged Nef, Beclin 1 was found in immunoprecipitates generated with myc antibodies (Fig. 6 C). In all immunoprecipitation experiments, IgG control showed negative results for the specific proteins analyzed (Fig. 6). The blots shown with the IgG control were developed until a very faint band (representing background in any type of immunoprecipitation experiments) was revealed when possible; shorter development times left IgG controls completely blank, whereas the specifically coimmunoprecipitated bands were still detected. Importantly, HIV Nef also coimmunoprecipitated with Beclin 1 in extracts from cells infected with HIV virus (Fig. 6 D), demonstrating that Nef–Beclin 1 complexes form during viral infection. Thus, Beclin 1 and Nef colocalize (Fig. 6 A) and are present in a shared protein complex (Fig. 6, B–D), associating directly or indirectly via an intermediate partner. Furthermore, Nef affected hVPS34 distribution (Fig. 6, E and F), as a consequence of its association with Beclin 1, resulting in an increased presence of hVPS34 on membranes.
Figure 6.
Nef is in protein complexes with autophagy regulator Beclin 1. (A) Macrophages were transfected with Nef-GFP and immunostained for Beclin 1. Arrows, perinuclear profiles exemplifying Nef and Beclin 1 colocalization. (inset) Peripheral colocalization. (B) 293T cells were transfected with Nef-GFP or GFP alone for 48 h. Lysates were immunoprecipitated either with Beclin 1 antibody and immunoblotted with Nef antibody (top) or with GFP and immunoblotted for Beclin 1 (bottom). Specific protein levels in cell lysates (input) and immunoprecipitates with IgG controls are shown. (C) Coimmunoprecipitation of Beclin 1 with Nef-Myc. 293T cells were transfected with Nef-Myc for 48 h, then cells were lysed and immunoprecipitation was performed using monoclonal Myc antibody or IgG control. Western blots were probed with Beclin 1 or Nef antibodies. The blots shown represent one of four independent experiments. (D) U937 cells were infected for 48 h with VSV-G–pseudotyped HIV. Cells were lysed and immunoprecipitated with Beclin 1–specific antibody or control IgG. Western blots of immunoprecipitated material were probed with Nef antibody. (E and F) 293T cells were cotransfected with Nef-GFP or GFP and Flag-hVPS34 for 48 h. Cells were fractionated into membranes (M) and cytosol (C) and immunoblotted with anti-Flag and Beclin 1 antibodies. (E) Immunoblots. (F) Quantification (ratios of membrane-associated hVPS34 to cytosolic hVPS34. Data indicate means; error bars indicate ±SEM. *, P < 0.05 (n = 3).
Mutational analysis of HIV Nef–Beclin 1 interactions and Nef effects on autophagy
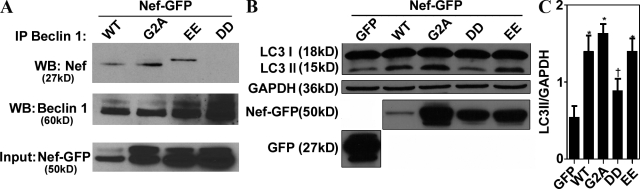
We next used a panel of Nef mutants to test whether any of the known motifs were necessary for Nef interactions with Beclin 1 and Nef effects on autophagy (Figs. 7 A and S3 G). In an identical coimmunoprecipitation approach as in Fig. 6, the previously characterized Nef mutant construct (Olivetta and Federico, 2006) with changes in the diacidic motif (174DD175 → 174AA175), responsible for interactions with the V1 domain of vacuolar H+ ATPase and required for CD4 down-regulation (Roeth and Collins, 2006), lost the capacity to coimmunoprecipitate Beclin 1 (Fig. 7 A). In contrast, the mutation 154EE155 → 154QQ155, in another region of Nef, i.e., the diacidic motif required for β-COP interactions (Piguet et al., 1999; Roeth and Collins, 2006), did not significantly diminish the capacity of Nef to coimmunoprecipitate with Beclin 1 (Fig. 7 A). Another mutation 2G → 2A, abrogating the ability of Nef to be N-terminally myristoylated, a posttranslational modification assisting Nef in membrane localization and required for many Nef functions (Roeth and Collins, 2006), did not affect the capacity of Nef to coimmunoprecipitate with Beclin 1 (Fig. 7 A). Myristoylation of Nef is often considered a sine qua non posttranslational modification required for nearly all previously known functions of Nef (Roeth and Collins, 2006), with the exception of Hck activation by Nef (Briggs et al., 2001), and thus it may appear surprising that this did not nullify Nef’s action in our assays. However, it has been shown (Bentham et al., 2006) that membrane association of NefG2A is not fully abrogated despite the loss of myristoylation, but that instead it may be shifted from plasma membrane to endomembranes, which is compatible with the action of Nef within the autophagic pathway.
Figure 7.
Nef 174DD175 motif is required for interaction with Beclin 1 and inhibition of autophagic maturation. (A) 293T cells were transfected with the wild-type HIV-1 Nef fusion with GFP or the indicated mutants: G2ANefGFP, 154EE-QQ155NefGFP, and 174DD-AA175NefGFP for 48 h. Beclin 1 immunoprecipitates were analyzed by immunoblotting. (B and C) 293T cells were transfected with the indicated constructs and immunoblotted for LC3. (B) Immunoblot. (C) Quantification (ratio of LC3-II to GAPDH band intensities). Data indicate means; error bars indicate ±SEM. *, P < 0.01; †, P ≥ 0.05 (ANOVA; n = 3).
The same set of Nef mutants was tested for their capacity to increase the LC3-II form (Fig. 7, B and C). The 174DD175 → 174AA175 mutant was again the only Nef variant tested that resulted in reduced increase in the autophagic marker LC3-II. Identical results were obtained when expression levels of Nef mutants were adjusted (Fig. S3 G). Thus, based on interaction assays with Beclin 1, and functional analysis with LC3-I–to–LC3-II conversion, the diacidic motif at the positions 174 and 175 of Nef is critical for the ability of Nef to control autophagic flux.
Discussion
This study shows that autophagy factors interact with HIV components, that basal autophagy augments Gag processing and HIV yields in macrophages, and that HIV inhibits degradative stages of autophagy. Our data show that HIV proteins Gag and Nef interact with two different autophagy factors and that this controls the autophagic pathway in infected cells with the net result of enhancing HIV yields. The rationale for our study was based on the growing recognition of autophagy as an antimicrobial cell-autonomous defense mechanism endowing eukaryotic cells with the ability to eliminate invading microbes (Deretic and Levine, 2009). However, we found that autophagy, even when pharmacologically induced, could not decrease HIV yields in macrophages, and that instead it enhanced HIV production. HIV protein Nef binds to the protein complexes containing the mammalian autophagy protein Beclin 1. Nef inhibits the degradative stages of the autophagy pathway, thus protecting HIV or its biosynthetic intermediates from autophagic destruction. The net effect of Nef may at first appear akin to that of herpes simplex virus type 1 protein ICP34.5, which binds to Beclin 1 and inhibits its function in autophagy (Orvedahl et al., 2007). However, the action of Nef is preferentially related to the maturation stages of autophagosomal pathway, as show in our work. Recent studies have indicated that Beclin 1–hVPS34 complexes in mammalian cells include potential equivalents of yeast Atg14 (Itakura et al., 2008), which appears to be autophagy initiation specific, and VPS38 (UVRAG), which acts at maturation and possibly initiation stages (Liang et al., 2008). Thus, different parts of the autophagic pathway may be targeted by viral factors affecting Beclin 1: autophagy initiation or maturation as in the case of ICP34.5 (Orvedahl et al., 2007), and Nef (this paper). Our findings also provide a functional and molecular basis for the results of a recent comprehensive siRNA screen in HeLa cells identifying among >250 host genes several Atg factors as playing a role in the HIV life cycle (Brass et al., 2008).
It has been previously reported (Roy et al., 2002; Heredia et al., 2003) that chronic treatment (for 3–7 d) of cells with low concentrations of rapamycin, which do not induce autophagy, may inhibit viral replication. The use of rapamycin in our study was limited to acute doses inducing autophagy, as we used rapamycin only as one of the agents to study how autophagy affects viral yields, irrespective of any long term effects that rapamycin may have on viral replication. Furthermore, induction of autophagy either with rapamycin or by starvation (unpublished data) both increased HIV yields, provided that the virus encoded Nef. Our work nevertheless indicates that, when unopposed by Nef, autophagy can act as a cell-autonomous anti-HIV defense. This is likely to be of importance, as autophagy is induced during HIV infection, as shown here and as recently noted in the context of TLR7 and TLR8 signaling (Delgado et al., 2008). In terms of the mechanism of the antiautophagic degradation action of Nef, we found that the diacidic 174DD175 motif, responsible for interactions with the V1 domain of vacuolar H+ ATPase and needed for CD4 down-regulation (Roeth and Collins, 2006), is required for effects of Nef on autophagy. Hence, the simplest explanation would be that Nef influences H+ ATPase assembly or activity, precluding autophagosomal acidification and maturation into autolysosomes. However, although HIV inhibits acidification of compartments with newly budded virions, the pH effect has been reported to be independent of Nef (Jouve et al., 2007). Thus, the protein complex containing Nef and Beclin 1 may act through a mechanism other than acidification. The effect of Nef on redistribution of hVPS34 (Fig. 6 E) to membranes may be related to inhibition of autophagic maturation.
Within the portfolio of Nef effects, which includes down-regulation of MHC class I and CD4 cell surface expression, altered T cell activation, and augmented viral infectivity (Peterlin, 2006; Roeth and Collins, 2006; Schindler et al., 2006, 2008), a less understood effect is the Nef-induced accumulation of MVB-like organelles (Stumptner-Cuvelette et al., 2003; Sandrin and Cosset, 2006) and emergence of large vacuoles (Sanfridson et al., 1997). This phenomenon can now be explained at least in part by the inhibition of MVB or amphisome consumption due to a Nef-dependent blockage of autophagic degradation.
In primary human macrophages, the virus transits through the intracellular compartments that intersect with autophagy factors such as LC3, as illustrated in Fig. 2. The intersection between HIV and the autophagic pathway is not limited to conditions when autophagy is induced to high levels. For example, the HIV precursor protein Gag is found in complexes with LC3 (Fig. 2 E) even when macrophages are not pharmacologically stimulated for autophagy, which indicates engagement of the basal autophagy in biosynthesis, processing, or assembly of HIV intermediates. In other cell types, such as 293T cells used to dissect mechanistically molecular aspects of Nef action in the context of the autophagic pathway, the viral Gag did not show colocalization with the RFP-GFP-LC3 probe (Fig. 5 A), reflecting the likely differences in intracellular trafficking of viral precursors in these cells versus macrophages (Gendelman et al., 1988; Raposo et al., 2002; Pelchen-Matthews et al., 2003; Morita and Sundquist, 2004; Jouvenet et al., 2006; Deneka et al., 2007; Jouve et al., 2007; Welsch et al., 2007). Nevertheless, viral Nef did inhibit autophagic maturation even in 293T cells, indicating that this activity does not necessarily coincide with the location of the viral particles or Gag in relation to LC3.
This is further underscored by the effects of Atg7 and Beclin 1 knockdowns on total p24 yields in resting macrophages infected with HIV. This effect has been independently observed in HeLa cells upon knockdown of other Atg factors (Brass et al., 2008). The enhancement by autophagy of HIV yields coincides with the association of Gag with LC3 uncovered in our work. Furthermore, our findings of enhanced Gag processing associated with autophagy indicate that this process plays a role in promoting certain steps in HIV biogenesis. Although autophagy is commonly viewed as a catabolic, degradative pathway primarily engaged in turning over macromolecules and removing toxic protein aggregates, or whole or parts of intracellular organelles and pathogens, it can also play a biosynthetic, anabolic role; this is clearly seen in the Cvt pathway in yeast, where Atg proteins are needed for completion of a functional vacuole (Scott et al., 1996; Xie and Klionsky, 2007)
Nef also inhibits apoptosis and cell death in macrophages (Olivetta and Federico, 2006). It has been shown that HIV Env induces death in bystander CD4+ CXCR4+ cells via a temporal succession of autophagy followed by apoptosis, and that completion of autophagy was a prerequisite for the execution of the subsequent apoptotic cell death (Espert et al., 2006). This effect was recently narrowed down to gp41 (Denizot et al., 2008). Based on Nef’s ability to inhibit terminal stages of autophagy, it follows that Nef may protect infected macrophages against cell death, in addition to guarding virions from autophagic elimination. Extending the life span of macrophages (Olivetta and Federico, 2006) and protecting virions from degradation may lead to higher HIV yields that are important for progression to AIDS (Daniel et al., 1992; Kirchhoff et al., 1995). Pharmacological intervention to modulate autophagy in HIV-infected macrophages may help delay or prevent development of clinical AIDS.
Materials and methods
Cells
For macrophages, human monocytes were prepared from HIV-negative donors by density gradient centrifugation (400 g for 30 min) through a Ficoll-Hypaque gradient (GE Healthcare). Adherent monocytes were matured into macrophages for 7 d before infection. THP-1 and U937 cells were maintained in RPMI supplemented with glucose, glutamine, Hepes, and pyruvate. HeLa and 293T cells were maintained in DME supplemented with glutamine and FBS. MAGI cells were obtained from the National Institutes of Health AIDS reagent program.
Antibodies and chemicals
Atg7 and Beclin 1 antibodies were obtained from Santa Cruz Biotechnology, Inc.; monoclonal p24 antibody was obtained from Novus Biologicals; LC3 antibody was obtained from T. Ueno (Juntendo University School of Medicine, Tokyo, Japan) or from Sigma-Aldrich, and Atg12 antibody was obtained from N. Mizushima (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan); and the GAPDH antibody was obtained from Abcam. Gag monoclonal p17 and actin antibodies were obtained from Abcam. Secondary Alexa Fluor 488– and 568–conjugated antibodies were obtained from Invitrogen. Gag rabbit polyclonal antibody was obtained from the National Institutes of Health AIDS reagents program. Rapamycin, 3MA, bafilomycin, and lipopolysccharide were obtained from Sigma-Aldrich.
Autophagy methods
Autophagy was triggered by treatment with 25–50 ng/µl rapamycin for 5 h in full nutrient medium. Alternatively, autophagy was induced by amino acid and serum starvation. Cells were washed three times with PBS and incubated in 1 ml Earle’s balanced salts solution (starvation medium) at 37°C for 5 h. Autophagy was quantified by the GFP-LC3 puncta, LC3-I–to–LC3-II conversion, and proteolysis assays. Autophagy was inhibited with 10 mM 3MA. Where used, bafilomycin A1 was at a concentration of 100 nm. Cells were transfected or cotransfected with GFP-LC3, RFP-GFP-LC3, DsRed2, Nef-DsRed2 (Nef-DsRed2 was provided by K. Collins, University of Michigan, Ann Arbor, MI) and other constructs for 24 h. The total number of puncta (≥1 µm) per cell was counted.
HIV extracellular yield
Methods to monitor HIV p24 yields are described in the legend to Table S2. For experiments with H9 T cells infected with live virus, the RT-PCR–based assay EnzChek (Invitrogen) was used to measure reverse transcription activity. Assays were performed according to the manufacturer’s instructions. Relative viral release was calculated as the ratio of extracellular-to-intracellular reverse transcription activity (Peden et al., 1991; Fan and Peden, 1992; Conti et al., 1998; Prasad and Kalpana, 2009).
Transfections and infections
Cells were transfected using the nucleoporation protocol (Amaxa) as described previously (Chua and Deretic, 2004), with 10 µg of DNA or 1.5 µg of siRNA, as required. Atg7 and Beclin 1 knockdown protein was achieved using siGENOME SMART pool (Thermo Fisher Scientific). All effects of siRNA were compared with siCONTROL nontargeting siRNA pool (Thermo Fischer Scientific). For VSV-G–pseudotyped HIV infections, U937 cells were differentiated overnight with 50 ng/ml of phorbol 12-myristate 13-acetate (Sigma-Aldrich), and viral infections were performed as described previously (Olivetta and Federico, 2006). MDM were infected with 105 tissue culture infections dose of SF162. Infections were allowed to go for 10 d with replacement of media every other day. Supernatants from these cells and cell lysates were frozen at −70°C until used for p24 ELISA or MAGI assays. The MAGI assay for HIV infectivity was performed as described previously (Chackerian et al., 1997). HIVLAI (Barre-Sinoussi et al., 1983; Nguyen et al., 1994) was expanded for 8 d in H9 T cells as described previously (Peden et al., 1991). Supernatants from these cells were used to infect H9 T cells for experiments using titers as described in Prasad and Kalpana, (2009).
Fluorescence microscopy and image acquisition
Cells were fixed for 10 min with 1% paraformaldehyde, washed with PBS, and, when immunostained, permeabilized with 0.1% Triton X-100. Secondary antibody controls were routinely performed and showed no similarity to the pattern obtained when the primary antibody was included. Images were taken and processed on a confocal microscope system (META; Carl Zeiss, Inc.), equipped with 63× 1.4 NA oil differential interference contrast Plan-Apochromat objective, using Immersol (n = 1.518) at room temperature. Acquisition software was LSM 510, Expert mode (Carl Zeiss, Inc.). Images were processed by Photoshop (Adobe) using proportional adjustments.
Electron and immunoelectron microscopy
Electron microscopy of viral budding in HIV-infected cells was performed as follows: control or U937 cells infected with VSV-G–pseudotyped pMSMBA were fixed with 3% formaldehyde (from paraformaldehyde) + 2% glutaraldehyde in 0.1 M cacodylate, pH 7.4. Cells were then washed and postfixed in 1% osmium tetroxide, 100 mM cacodylate buffer; dehydrated with increasing concentrations of ethanol; and gradually infiltrated with epon resin, embedded in straight resin, and examined using a transmission electron microscope (EM 900; Carl Zeiss, Inc.). Immunoelectron microscopy was performed using rabbit polyclonal LC3 antibody (Tanida et al., 2008), applying the preembedding gold enhancement method as described previously (Luo et al., 2006). U937 cells cultured on plastic coverslips (LF; Sumitomo Bakelite) were fixed with 4% paraformaldehyde (Nacalai Tesque) in 0.1 M sodium PBS, pH 7.4, for 30 min. After washing with the same buffer three times for 5 min, the fixed cells were permeabilized using 0.25% saponin in PBS. The cells were washed with PBS, blocked by incubating for 30 min in PBS containing 0.1% saponin, 10% BSA, 10% normal goat serum, and 0.1% cold water fish skin gelatin, then exposed overnight to 0.01 mg/ml of anti-LC3 rabbit polyclonal antibody or to 0.01 mg/ml of nonimmunized rabbit IgG in the blocking solution. After washing with PBS containing 0.005% saponin, the cells were incubated with colloidal gold (1.4-nm diameter; Nanoprobes)-conjugated goat anti–rabbit IgG in the blocking solution for 2 h. The cells were then washed with PBS and fixed with 1% glutaraldehyde in PBS for 10 min. After washing with 50 mM glycine in PBS, 1% BSA in PBS, and finally with milliQ water (Millipore), gold labeling was intensified with a gold enhancement kit (GoldEnhance EM; Nanoprobes) for 3 min at room temperature according to the manufacturer’s instructions. After washing with distilled water, the cells were postfixed in 1% OsO4 containing 1.5% potassium ferrocyanide in PBS for 60 min at room temperature, and washed with distilled water. The cells were dehydrated in a series of graded ethanol solutions and embedded in epoxy resin. After the epoxy resin hardened, the plastic coverslip was removed from it. Ultrathin sections were cut horizontally to the cell layer and double stained with uranyl acetate and lead citrate. Samples were analyzed with an electron microscope (H7600; Hitachi).
Western blots and immunoprecipitations
Cells were washed in PBS and lysed with buffer containing 10 mM Tris HCl, pH 8.0, 150 mM NaCl, 0.5% deoxycholate, 2 mM EDTA, 2% NP-40, 1 mM PMSF, and protease inhibitor cocktail (Roche). 50 µm of protein was loaded and separated on a 12.5% SDS-polyacrylamide gel and transferred to nitrocellulose. The membrane was blocked overnight at 4°C in 5% milk in PBS/Tween 20 (0.1%) and probed with primary antibodies for 1 h at room temperature. After washing with PBS/Tween, the blot was probed with appropriate HRP-conjugated secondary antibody for 1 h at room temperature and stained with SuperSignal West Dura chemiluminescent substrate from Thermo Fisher Scientific. GAPDH was used as a loading control. For immunoprecipitations, transfected 293T cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 1 mM EDTA, with protease and phosphatase inhibitors) for 1 h, followed by centrifugation to remove cell debris. Supernatants were precleared and incubated for 2 h with rabbit anti–Beclin 1 (Novus Biologicals), anti-LC3B, or rabbit anti-GFP (Abcam) at 4°C. The immune complexes were captured with protein G–agarose beads (EMD) overnight at 4°C. Immunoprecipitates were washed four times with PBS, eluted with Laemmli SDS-PAGE sample buffer for 5 min at 100°C, and subjected to immunoblot analysis with mouse anti-NEF (United States Biological), goat anti–Beclin 1 (Santa Cruz Biotechnology, Inc.), and mouse anti-GFP (Abcam). Note, in immunoprecipitation experiments with the 154EE156 → 154QQ155 Nef mutant, this Nef variant consistently showed anomalous electrophoretic mobility.
Subcellular fractionation and cytosol preparation
U937 cells were infected with VSV-G–pseudotyped HIV for 2 d. Cells were lysed by passage through a tubing-interconnected two-syringe apparatus, and nuclei were cleared by low-speed centrifugation. The postnuclear supernatant was put on 20, 30, 40, 45, 50, 55, and 60% sucrose gradients, then centrifuged overnight at 100,000 g. 12 fractions collected from the top were pelleted at 100,000 g for 1 h and immunoblotted for the indicated organellar markers.
Online supplemental material
Fig. S1 shows that Atg7 and Beclin 1 knockdowns inhibit autophagy in U937 cells, as well as transmission and immunoelectron micrograph HIV profiles in macrophages. Fig. S2 shows a comparison of LC3 forms in whole cell lysate versus LC3 forms associated with membranes, that autophagy induction is operational in cells used to detect HIV yield-enhancing effects of acute rapamycin treatment, and that Nef causes accumulation of LC3 puncta. Fig. S3 shows that Nef inhibits autophagic proteolysis; intracellular localization of Nef relative to mTor, 2xFYVE-GFP, and autophagy markers Atg7, Atg12, and Beclin 1; and that Nef motif 174DD175 is but G2A motif is not required for Nef-dependent increase in LC3II levels. Table S1 shows HIV molecular clones, viruses, and viral preparations and use. Table S2 shows ELISA (p24) and quantification of viral release. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903070/DC1.
Acknowledgments
We thank N. Mizushima for Atg12 antibody, K. Collins for Nef clones, T. Howard and the electron microscopy facility for assistance, and M. Peterlin for discussion.
This work was supported by National Institute of Allergy and Infectious Diseases grants AI069345, AI45148, AI42999 to V. Deretic, AI06849 to L. Wu, and by amfAR grant 107160-44-RGRL and a Bill and Melinda Gates Foundation Grand Challenges Explorations grant G#52068 to V. Deretic. C. Dinkins was supported by National Institutes of Health Biology of Infectious Diseases and Inflammation training grant T32AI007538.
Footnotes
Abbreviations used in this paper: 3MA, 3-methyl adenine; AIDS, acquired immune deficiency syndrome; ANOVA, analysis of variance; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HIV, human immunodeficiency virus; MDM, monocyte-derived macrophages; MVB, multivesicular body; PI3K, phosphatidylinositol 3-kinase; Tor, target of rapamycin; VLP, virus-like particle; VSV-G, vesicular stomatitis virus G.
References
- Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS).Science. 220:868–871 [DOI] [PubMed] [Google Scholar]
- Bentham M., Mazaleyrat S., Harris M. 2006. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein.J. Gen. Virol. 87:563–571 [DOI] [PubMed] [Google Scholar]
- Birmingham C.L., Smith A.C., Bakowski M.A., Yoshimori T., Brumell J.H. 2006. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole.J. Biol. Chem. 281:11374–11383 [DOI] [PubMed] [Google Scholar]
- Birmingham C.L., Canadien V., Kaniuk N.A., Steinberg B.E., Higgins D.E., Brumell J.H. 2008. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles.Nature. 451:350–354 [DOI] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. 2008. Identification of host proteins required for HIV infection through a functional genomic screen.Science. 319:921–926 [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Scholtz B., Jacque J.M., Swingler S., Stevenson M., Smithgall T.E. 2001. HIV-1 Nef promotes survival of myeloid cells by a Stat3-dependent pathway.J. Biol. Chem. 276:25605–25611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chackerian B., Long E.M., Luciw P.A., Overbaugh J. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection.J. Virol. 71:3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua J., Deretic V. 2004. Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles.J. Biol. Chem. 279:36982–36992 [DOI] [PubMed] [Google Scholar]
- Conti L., Rainaldi G., Matarrese P., Varano B., Rivabene R., Columba S., Sato A., Belardelli F., Malorni W., Gessani S. 1998. The HIV-1 vpr protein acts as a negative regulator of apoptosis in a human lymphoblastoid T cell line: possible implications for the pathogenesis of AIDS.J. Exp. Med. 187:403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M.D., Kirchhoff F., Czajak S.C., Sehgal P.K., Desrosiers R.C. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene.Science. 258:1938–1941 [DOI] [PubMed] [Google Scholar]
- Deacon N.J., Tsykin A., Solomon A., Smith K., Ludford-Menting M., Hooker D.J., McPhee D.A., Greenway A.L., Ellett A., Chatfield C., et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients.Science. 270:988–991 [DOI] [PubMed] [Google Scholar]
- Delgado M.A., Elmaoued R.A., Davis A.S., Kyei G., Deretic V. 2008. Toll-like receptors control autophagy.EMBO J. 27:1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneka M., Pelchen-Matthews A., Byland R., Ruiz-Mateos E., Marsh M. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53.J. Cell Biol. 177:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizot M., Varbanov M., Espert L., Robert-Hebmann V., Sagnier S., Garcia E., Curriu M., Mamoun R., Blanco J., Biard-Piechaczyk M. 2008. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells.Autophagy. 4:998–1008 [DOI] [PubMed] [Google Scholar]
- Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations.Cell Host and Microbe. 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espert L., Denizot M., Grimaldi M., Robert-Hebmann V., Gay B., Varbanov M., Codogno P., Biard-Piechaczyk M. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4.J. Clin. Invest. 116:2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Peden K. 1992. Cell-free transmission of Vif mutants of HIV-1.Virology. 190:19–29 [DOI] [PubMed] [Google Scholar]
- Furuya N., Yu J., Byfield M., Pattingre S., Levine B. 2005. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function.Autophagy. 1:46–52 [DOI] [PubMed] [Google Scholar]
- Geleziunas R., Xu W., Takeda K., Ichijo H., Greene W.C. 2001. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell.Nature. 410:834–838 [DOI] [PubMed] [Google Scholar]
- Gendelman H.E., Orenstein J.M., Martin M.A., Ferrua C., Mitra R., Phipps T., Wahl L.A., Lane H.C., Fauci A.S., Burke D.S., Skillman D., Meltzer M.S. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes.J. Exp. Med. 167:1428–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. 2004. The biogenesis of multivesicular endosomes.Nat. Rev. Mol. Cell Biol. 5:317–323 [DOI] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages.Cell. 119:753–766 [DOI] [PubMed] [Google Scholar]
- Harris J., De Haro S.A., Master S.S., Keane J., Roberts E.A., Delgado M., Deretic V. 2007. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis.Immunity. 27:505–517 [DOI] [PubMed] [Google Scholar]
- Heredia A., Amoroso A., Davis C., Le N., Reardon E., Dominique J.K., Klingebiel E., Gallo R.C., Redfield R.R. 2003. Rapamycin causes down-regulation of CCR5 and accumulation of anti-HIV beta-chemokines: an approach to suppress R5 strains of HIV-1.Proc. Natl. Acad. Sci. USA. 100:10411–10416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.Mol. Biol. Cell. 19:5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Giddings T.H., Jr., Taylor M.P., Mulinyawe S., Rabinovitch M., Kopito R.R., Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses.PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve M., Sol-Foulon N., Watson S., Schwartz O., Benaroch P. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages.Cell Host Microbe. 2:85–95 [DOI] [PubMed] [Google Scholar]
- Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. 2006. Plasma membrane is the site of productive HIV-1 particle assembly.PLoS Biol. 4:e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing.EMBO J. 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T. 2001. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network.EMBO Rep. 2:330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3.Autophagy. 3:452–460 [DOI] [PubMed] [Google Scholar]
- Kirchhoff F., Greenough T.C., Brettler D.B., Sullivan J.L., Desrosiers R.C. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection.N. Engl. J. Med. 332:228–232 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. 2007. Autophagy: from phenomenology to molecular understanding in less than a decade.Nat. Rev. Mol. Cell Biol. 8:931–937 [DOI] [PubMed] [Google Scholar]
- Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells.Science. 315:1398–1401 [DOI] [PubMed] [Google Scholar]
- Levine B., Deretic V. 2007. Unveiling the roles of autophagy in innate and adaptive immunity.Nat. Rev. Immunol. 7:767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Klionsky D.J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy.Dev. Cell. 6:463–477 [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease.Cell. 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking.Nat. Cell Biol. 10:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1.Nature. 402:672–676 [DOI] [PubMed] [Google Scholar]
- Luo H., Nakatsu F., Furuno A., Kato H., Yamamoto A., Ohno H. 2006. Visualization of the post-Golgi trafficking of multiphoton photoactivated transferrin receptors.Cell Struct. Funct. 31:63–75 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T. 2007. How to interpret LC3 immunoblotting.Autophagy. 3:542–545 [DOI] [PubMed] [Google Scholar]
- Morita E., Sundquist W.I. 2004. Retrovirus budding.Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., et al. 2004. Autophagy defends cells against invading group A Streptococcus.Science. 306:1037–1040 [DOI] [PubMed] [Google Scholar]
- Nguyen M.H., Schinazi R.F., Shi C., Goudgaon N.M., McKenna P.M., Mellors J.W. 1994. Resistance of human immunodeficiency virus type 1 to acyclic 6-phenylselenenyl- and 6-phenylthiopyrimidines.Antimicrob. Agents Chemother. 38:2409–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. 2005. Escape of intracellular Shigella from autophagy.Science. 307:727–731 [DOI] [PubMed] [Google Scholar]
- Olivetta E., Federico M. 2006. HIV-1 Nef protects human-monocyte-derived macrophages from HIV-1-induced apoptosis.Exp. Cell Res. 312:890–900 [DOI] [PubMed] [Google Scholar]
- Ono A., Freed E.O. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body.J. Virol. 78:1552–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Talloczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein.Cell Host Microbe. 1:23–35 [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy.Cell. 122:927–939 [DOI] [PubMed] [Google Scholar]
- Peden K., Emerman M., Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI.Virology. 185:661–672 [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A., Kramer B., Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages.J. Cell Biol. 162:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B.M. 2006. Nef: out and in? Nat. Immunol. 7:229–230 [DOI] [PubMed] [Google Scholar]
- Piguet V., Gu F., Foti M., Demaurex N., Gruenberg J., Carpentier J.L., Trono D. 1999. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes.Cell. 97:63–73 [DOI] [PubMed] [Google Scholar]
- Prasad V.R., Kalpana G.V. 2009. HIV protocols. Humana Press, New York: 457 pp [Google Scholar]
- Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments.Traffic. 3:718–729 [DOI] [PubMed] [Google Scholar]
- Roberts E.A., Deretic V. 2008. Autophagic proteolysis of long-lived proteins in nonliver cells.Methods Mol. Biol. 445:111–117 [DOI] [PubMed] [Google Scholar]
- Roeth J.F., Collins K.L. 2006. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways.Microbiol. Mol. Biol. Rev. 70:548–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeth J.F., Williams M., Kasper M.R., Filzen T.M., Collins K.L. 2004. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail.J. Cell Biol. 167:903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J., Paquette J.S., Fortin J.F., Tremblay M.J. 2002. The immunosuppressant rapamycin represses human immunodeficiency virus type 1 replication.Antimicrob. Agents Chemother. 46:3447–3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrin V., Cosset F.L. 2006. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein.J. Biol. Chem. 281:528–542 [DOI] [PubMed] [Google Scholar]
- Sanfridson A., Hester S., Doyle C. 1997. Nef proteins encoded by human and simian immunodeficiency viruses induce the accumulation of endosomes and lysosomes in human T cells.Proc. Natl. Acad. Sci. USA. 94:873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis.Nature. 450:1253–1257 [DOI] [PubMed] [Google Scholar]
- Schindler M., Munch J., Kutsch O., Li H., Santiago M.L., Bibollet-Ruche F., Muller-Trutwin M.C., Novembre F.J., Peeters M., Courgnaud V., et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1.Cell. 125:1055–1067 [DOI] [PubMed] [Google Scholar]
- Schindler M., Schmokel J., Specht A., Li H., Munch J., Khalid M., Sodora D.L., Hahn B.H., Silvestri G., Kirchhoff F. 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+T cells in natural SIV infection.PLoS Pathog. 4:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Munz C. 2007. Innate and adaptive immunity through autophagy.Immunity. 27:11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Munz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes.Immunity. 26:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Hefner-Gravink A., Morano K.A., Noda T., Ohsumi Y., Klionsky D.J. 1996. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole.Proc. Natl. Acad. Sci. USA. 93:12304–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. 2004. Autophagy in health and disease: a double-edged sword.Science. 306:990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.B., Davis A.S., Taylor G.A., Deretic V. 2006. Human IRGM induces autophagy to eliminate intracellular mycobacteria.Science. 313:1438–1441 [DOI] [PubMed] [Google Scholar]
- Stumptner-Cuvelette P., Jouve M., Helft J., Dugast M., Glouzman A.S., Jooss K., Raposo G., Benaroch P. 2003. Human immunodeficiency virus-1 Nef expression induces intracellular accumulation of multivesicular bodies and major histocompatibility complex class II complexes: potential role of phosphatidylinositol 3-kinase.Mol. Biol. Cell. 14:4857–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingler S., Brichacek B., Jacque J.M., Ulich C., Zhou J., Stevenson M. 2003. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection.Nature. 424:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I., Ueno T., Kominami E. 2008. LC3 and Autophagy.Methods Mol. Biol. 445:77–88 [DOI] [PubMed] [Google Scholar]
- Welsch S., Keppler O.T., Habermann A., Allespach I., Krijnse-Locker J., Krausslich H.G. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages.PLoS Pathog. 3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations.Nat. Cell Biol. 9:1102–1109 [DOI] [PubMed] [Google Scholar]
- Xu Y., Jagannath C., Liu X.D., Sharafkhaneh A., Kolodziejska K.E., Eissa N.T. 2007. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity.Immunity. 27:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T., Mita S., Ohmori H., Oshima Y., Fujimoto Y., Ueda R., Takada H., Goldman W.E., Fukase K., Silverman N., et al. 2008. Autophagic control of listeria through intracellular innate immune recognition in drosophila.Nat. Immunol. 9:908–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Overmeyer J.H., Maltese W.A. 2006. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking.J. Cell Sci. 119:259–270 [DOI] [PubMed] [Google Scholar]