Abstract
The outbreak of human infection due to the novel swine-origin influenza A (H1N1) virus began in Mexico in March 2009. As of July 6, 2009, more than 94 000 laboratory-confirmed cases were reported in over 100 countries, including 7983 cases in Canada. In this report, we describe the epidemiologic and clinical characteristics of the first cluster of reported cases of human-to-human transmission of the new influenza virus in Canada.
In March and April 2009, an outbreak of severe respiratory infection began in Mexico that has subsequently been attributed to a novel human swine-origin influenza A (H1N1) virus.1 Since then, the virus has spread to more than 100 countries, with over 94 000 laboratory-confirmed cases reported as of July 6, including 7983 cases in Canada.2,3
On Apr. 23, 2009, a cluster of cases of acute respiratory illness in a private school was reported to public health officials in Nova Scotia. Through routine surveillance, investigation of the outbreak revealed that a group of 25 students and 3 chaperones had travelled to various parts of the Yucatan Peninsula in Mexico from Apr. 1–8, 2009. Only 3 days before notification of this outbreak, an alert describing severe respiratory illness in Mexico (later identified as human infection with the new swine-origin influenza A [H1N1] virus) was posted by the Canadian Integrated Outbreak Surveillance Centre, Public Health Agency of Canada.
Nasal specimens from 5 symptomatic individuals in Nova Scotia were sent to the Capital District Health Authority Microbiology Laboratory. On Apr. 24, 2009, reverse transcriptase polymerase chain reaction revealed that 3 of the individuals had influenza A that could not be typed as human H1 or H3 in origin. These samples were forwarded to the National Microbiology Laboratory in Winnipeg, Manitoba. On Apr. 25, 2009, the National Microbiology Laboratory confirmed that 4 of the 5 individuals had infection with the new influenza A virus.
In this article, we summarize the epidemiologic and clinical characteristics of this cluster of cases. For the purposes of this report, we include only cases identified up to May 8, 2009.
Case definitions
After reviewing preliminary data, we determined that only 55% of the confirmed and probable cases met the Public Health Agency of Canada’s case definition for influenza-like illness.4 Therefore, we adapted the agency’s case definition to meet the needs of this investigation (Table 1).
Table 1.
Case definitions for the investigation of a cluster of severe respiratory illnesses at a private school in Nova Scotia, Apr. 30, 2009
| Case status | Definition |
|---|---|
| Confirmed | A person with:
|
| Probable | A person with:
|
| Suspect | A person with:
|
| Acute respiratory illness | Cough or fever and one or more of sore throat, headache, eye pain or myalgia |
| Close contact | Household contact or other individual who has had sustained contact (several consecutive hours) in close proximity to the case (< 2 m) |
Epidemiologic characteristics
As of May 8, 2009, 99 cases of human infection with the new influenza A (H1N1) virus were identified as being associated with the school cluster in Nova Scotia; 43 were laboratory-confirmed cases, 1 was probable and 55 were suspect cases.
Data from the 99 cases were collected and analyzed without review from an institutional review board because this activity was part of a public health outbreak investigation. However, for the purpose of publication of this article, each case was asked to provide written consent to have their data included in this report. We describe only the 87 cases (41 laboratory confirmed, 1 probable and 45 suspect) who provided consent. We identified no statistically significant difference between the results of the analysis of all 99 cases and the analysis of the 87 cases from whom consent was obtained.
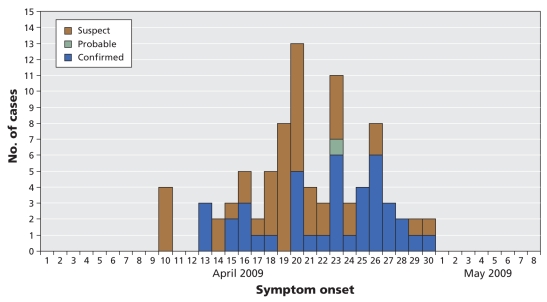
The dates of symptom onset of the 87 cases ranged from Apr. 9 to 30, 2009 (Figure 1). The first cases reported to the public health officials involved 4 students who had not travelled to Mexico. During the investigation it was learned that 6 of the students who had travelled to Mexico on the school trip became ill within a week of their return. A seventh student travelled independently to Mexico and also became ill during the same period.
Figure 1.
Epidemic curve of 87 confirmed, probable and suspect cases of infection with the novel human swine-origin influenza A (H1N1) virus in Nova Scotia, Canada, Apr. 1 to May 8, 2009, by date of symptom onset.
The attack rate, including both suspect and confirmed cases, was 21% among boarding students (n = 207) and 17% among day students (n = 136). Characteristics of the 87 cases are outlined in Table 2.
Table 2.
Characteristics of 87 reported cases of the novel human swine-origin influenza A (H1N1) virus as of May 8, 2009, in Nova Scotia, Canada
| Type of case; no. (%) of individuals* |
||||
|---|---|---|---|---|
| Characteristic | Confirmed n = 41 | Probable n = 1 | Suspect n = 45 | Total n = 87 |
| Sex | ||||
| Male | 21 (51) | 1 (100) | 23 (51) | 45 (52) |
| Female | 20 (49) | 0 | 22 (49) | 42 (48) |
| Age, yr | ||||
| Mean | 18.2 | 16.0 | 23.7 | 20.1 |
| Median (range) | 17.0 (12–46) | 16.0 (16) | 17.0 (0–60) | 17.0 (0–60) |
| School association | ||||
| Campus resident | 21 (51) | 1 (100) | 21 (47) | 43 (49) |
| Day student | 15 (37) | 0 | 8 (18) | 23 (26) |
| Staff | 2 (5) | 0 | 7 (16) | 9 (10) |
| Close contact | 3 (7) | 0 | 9 (23) | 12 (14) |
| Seasonal flu vaccine | ||||
| Yes | 23 (56) | 1 (100) | 25 (56) | 49 (56) |
| No | 14 (34) | 0 | 17 (38) | 31 (36) |
| Unknown | 4 (10) | 0 | 3 (7) | 7 (8) |
| Travel to Mexico on school trip | ||||
| Yes | 4 (10) | 0 | 8 (18) | 12 (14) |
| No | 37 (90) | 1 (100) | 37 (82) | 75 (86) |
Unless stated otherwise.
Cough was reported in 90% and fever in 59% of confirmed and probable cases. Other common symptoms included headache (83%), sore throat (76%) and nasal congestion (76%). Among suspect cases, cough was reported in 89%, nasal congestion in 78%, sore throat in 64% and headache in 64%. Eighty percent of cases sought medical attention, usually presenting to the campus infirmary for assessment. None of the cases was admitted to hospital. No deaths were associated with the cluster.
Diagnostic testing
As of May 8, 2009, the Capital District Health Authority Microbiology Laboratory had tested about 1075 specimens for suspected human swine-origin influenza virus. Initial testing was done using a multiplexed in-house protocol for reverse transcriptase polymerase chain reaction that targeted influenza A, influenza B and respiratory syncytial virus. The primers, specific for influenza A, were directed against conserved regions of the matrix gene that detects all subtypes of influenza A of human, avian and swine origin.5 Specimens positive for influenza A were further tested using reverse transcriptase polymerase chain reaction specific for human H1 and H3 hemagglutinin genes.6 This subtyping method is specifically designed to detect human strains and not the novel swine-origin virus. As such, patients with the novel swine-origin influenza A (H1N1) virus are considered to have “nontypeable” influenza A. Confirmation of the novel virus requires nucleic acid detection designed to identify the H1 gene of the virus. Alternatively, the matrix gene of the virus can be sequenced and compared with known swine-origin influenza sequences. Both methods were used by the National Microbiology Laboratory in Winnipeg, Manitoba, to confirm the initial 18 cases in Nova Scotia.
The conventional protocol for reverse transcriptase polymerase chain reaction for confirmation of swine H1 was developed by the National Microbiology Laboratory on the basis of data on the hemagglutinin (EPI176470) and neur-aminidase (EPI176472) gene sequences for A/California/04/2009 obtained from the Global Initiative on Sharing Avian Influenza Data website (methodology available from the corresponding author upon request). As part of a rapid response to this global outbreak, and in accordance to annex C of the Canadian Pandemic Influenza Plan,7 this method was distributed to all Canadian provincial public health laboratories or designates, including the Capital District Health Authority Microbiology Laboratory. Since that time, the Capital District Health Authority Microbiology Laboratory has used this swine-specific assay to confirm an additional 25 cases. As of May 8, 2009, there was 1 case identified as “nontypeable” influenza A awaiting confirmation testing at the National Microbiology Laboratory.
In addition, other respiratory viruses, including both human strains of H1N1 and H3N2 and respiratory syncytial virus, continue to circulate in the community. During this outbreak investigation, 19 human influenza strains (10 H1N1 and 9 H3N2) were identified.
Measures to control the outbreak
To date, all cases associated with the cluster in Nova Scotia either travelled to Mexico or were close contacts of a case. As the international situation evolved during the early stages of this investigation, it became clear that a containment strategy was unlikely to succeed and was not recommended. Efforts have been focused on slowing the transmission of the virus in the school and surrounding community by means of antiviral treatment and isolation of symptomatic individuals as well as quarantine of close contacts and postexposure prophylaxis with oseltamivir.
Cases were isolated for 7 days after symptom onset, with females on one floor of the girls’ residence and males on one floor of the boys’ residence. All cases were given oseltamivir therapy. Although the illness in this cluster has been mild, it was thought that the use of antiviral medication could potentially reduce the infectious period and, thereby, viral transmission.
Public health measures to manage this cluster were modified as the outbreak evolved: The resources required for contact tracing soon exceeded public health capacity; quarantine measures were difficult to enforce and were of questionable effectiveness, and there was no evidence that the virus had spread from this cluster to the surrounding community. Therefore, the strategy was changed to consider this private school as a closed residential facility. Analogous to the standard approach to the management of an influenza outbreak in a long-term care facility, students and staff were given prophylaxis with oseltamivir in an attempt to decrease transmission both within the school and to the community. This is consistent with the measures outlined in the Nova Scotia pandemic plan and with the overall goal of mitigation of spread, with the clear understanding that this strategy would be reevaluated if evidence of spread outside this cluster developed.
Summary
This was the first identified and, to date, one of the largest clusters of human infection with the novel swine-origin influenza A (H1N1) virus in Canada. The findings of our investigation of this cluster may not accurately represent the transmissibility or pathogenicity of the virus in the general population, given the institutional setting, the age of the cluster population and potential case-ascertainment bias.
In at least 7 cases, the virus was likely acquired in Mexico. This was followed by person-to-person transmission in the school setting. However, because the virus was introduced into the school by several individuals at the same time, it has proven difficult to determine a chain of transmission in order to calculate an incubation period. The epidemic curve, however, suggests an interval of about 3 days between successive generations of cases. Further analysis of transmission within dormitories and households may allow calculation of the reproductive number.
All students were offered the seasonal influenza vaccine in the fall of 2008. Vaccine uptake was estimated to be close to 100% among boarding students and 15% among day students. Despite the differences in vaccine uptake, we observed similar attack rates in the 2 groups of students. Our findings suggest that the vaccine does not offer substantial cross- protection against the novel human swine-origin influenza A (H1N1) virus. This is consistent with studies that showed that use of seasonal influenza vaccine from recent seasons did not produce cross-reactive antibodies against the novel influenza virus.8
Seasonal human influenza continues to circulate at low levels in the community. Because clinical symptoms of infection with the novel influenza virus do not differ from those of seasonal human influenza, there is continued need for subtyping and laboratory confirmation. Whether the novel virus will supplant seasonal influenza viruses is unclear. If future management decisions require differentiation of the viruses, persistent concurrent circulation of the novel virus and the seasonal influenza viruses may strongly influence the amount of diagnostic testing required.
The ideal specimen type for diagnosis of the novel influenza virus has not been defined. However, patients suspected of having this virus strain should have a nasopharyngeal swab collected and sent for testing by means of nucleic acid detection. Even though some of the cases in the United States were initially identified with the use of rapid antigen point-of-care detection kits, the performance characteristics of the kits for detecting the novel swine-origin virus are un -known. These point-of-care kits are clearly less sensitive than nucleic acid detection;9 therefore, a negative result from a suspect case must be forwarded to the provincial public health laboratory or designated laboratory for confirmation. Because point-of-care kits currently available in Canada cannot subtype influenza A, a positive result for influenza A using such a kit must be forwarded to a public health laboratory for subtyping. Therefore, because specimens that yield either positive or negative results with a point-of-care kit require further testing, we do not currently recommend the use of these kits.
It was always assumed that the emergence of a novel influenza strain would result in severe disease. Currently, the number of reported cases of infection with the novel influenza A (H1N1) virus and the number of countries continues to rise daily, which indicates that the virus is able to transmit efficiently from person to person. Although some individuals have required hospital care and deaths have occurred, the majority of cases, including the cluster we have described, have had a mild, self-limiting illness.10,11 This suggests that the virus may continue to spread and cause mild illness, similar to the pattern of illness during an aggressive influenza season. However, it remains prudent to be vigilant and to monitor for severe disease.
As the outbreak evolves, public health officials have to rise to the challenge of providing the best recommendations based on data that are constantly changing, while trying to provide a consistent message to front-line clinicians and the public. Updated clinical information and guidelines can be found on the Public Health Agency of Canada’s website (www.phac-aspc.gc.ca/alert-alerte/swine-porcine/hp-index-eng.php).
Supplementary Material
Footnotes
This article has been peer reviewed.
Competing interests: None declared.
Contributors: Jennifer Cutler, Emily Schleihauf and Bev Billard were responsible for data entry and analysis. Todd Hatchette and Ross Davidson oversaw the diagnostic testing at the Capital District Health Authority Microbiology Laboratory, and the collection and analysis of data. Gaynor Watson-Creed coordinated the outbreak response and facilitated the data collection. Shelly Sarwal coordinated the epidemiologic investigation and analysis. Yan Li was responsible for the accuracy of the data analysis at the National Microbiology Laboratory, where confirmatory testing of all positive samples occurred. Yan Li and Nathalie Bastien were responsible for genotyping of the novel influenza A (H1N1) virus in patient samples and the design of the molecular diagnostic assay for the novel virus used at both the National Microbiology Laboratory and the Capital District Health Authority Microbiology Laboratory. Jennifer Cutler, Emily Schleihauf, Bev Billard, Todd Hatchette and Shelly Sarwal developed the first draft of the manuscript. All of the authors contributed to the revising of the manuscript and approved the final version submitted for publication.
The Nova Scotia Human Swine Influenza Investigation Team: Maureen Baikie, Nova Scotia Department of Health Promotion and Protection, Halifax, NS; Kimberlee Barro, Capital District Health Authority, Halifax, NS; Terri-Lyn Bennett, Nathalie Blanchet, Nova Scotia Department of Health Promotion and Protection; Lisa Boucher, Capital District Health Authority; Teri Cole, Nova Scotia Department of Health Promotion and Protection; Ann Coombs, Nova Scotia Department of Health Promotion and Protection; Sarah Fleming, First Nations and Inuit Health, Health Canada; Kevin Forward, Division of Microbiology, Department of Pathology and Laboratory Medicine, Capital District Health Authority; Rosanne Gillis, Capital District Health Authority; Richard Gould, Nova Scotia Department of Health Promotion and Protection; Morag Graham, National Microbiology Laboratory, Public Health Agency of Canada; David Haldane, Division of Microbiology, Department of Pathology and Laboratory Medicine, Capital District Health Authority; Laura Hermann, Department of Medicine, Dalhousie University; Cara-Leah Hmidan, Capital District Health Authority; Elaine Holmes, Nova Scotia Department of Health Promotion and Protection; Lynn Johnston, Department of Medicine, Dalhousie University; Joanne Langley, Canadian Centre for Vaccinology, IWK Health Centre, Halifax, NS; Jason LeBlanc, Division of Microbiology, Department of Pathology and Laboratory Medicine, Capital District Health Authority; Mark Kazimirski, Capital District Health Authority; Shelly McNeil, Department of Medicine, Dalhousie University; Kim McGill, Dee Mombourquette, Gertrude Oliver, Gary O’Toole, Nova Scotia Department of Health Promotion and Protection; Kathy Penny, Capital District Health Authority; Janice Pettipas, Division of Microbiology, Department of Pathology and Laboratory Medicine, Capital District Health Authority; Carol Lynn Raithby, Capital District Health Authority; Patsy Rawding, Nova Scotia Department of Health, Halifax, NS; Robert Strang, Nova Scotia Department of Health Promotion and Protection; Gerry Styles, Caryll Tawse, Capital District Health Authority; Shaun Tyler, National Microbiology Laboratory, Public Health Agency of Canada; Noella Whelan, Capital District Health Authority; and the laboratory technologists at the Division of Microbiology, Department of Pathology and Laboratory Medicine, Capital District Health Authority, Halifax, NS
REFERENCES
- 1.General Directorate of Epidemiology, Ministry of Health, Mexico; Pan American Health Organization; World Health Organization; Public Health Agency of Canada; US Centers for Disease Control and Prevention. Outbreak of swine-origin influenza A (H1N1) virus infection — Mexico, March–April 2009. [(accessed 2009 June 24)];MMWR Morb Mortal Weekly Rep. 2009 58:1–3. Available: www.wpro.who.int/NR/rdonlyres/5D3B17F6-8F19-4269-A547-C5F61C1AC8E2/0/MMWRswineoriginfluMexico_mm58d0430a2.pdf. [Google Scholar]
- 2.Public Health Agency of Canada. Cases of H1N1 flu virus in Canada. Ottawa (ON): the agency; 2009. [(accessed 2009 June 24)]. Available: www.phac-aspc.gc.ca/alert-alerte/swine-porcine/surveillance-eng.php. [Google Scholar]
- 3.World Health Organization. Influenza A (H1N1) — update 58. Geneva (Switzerland): the organization; 2009. Jul 6, [(accessed 2009 July 8)]. Available: www.who.int/csr/don/2009_07_06/en/index.html. [Google Scholar]
- 4.Public Health Agency of Canada. FluWatch. Ottawa (ON): the agency; 2009. [(accessed 2009 June 24)]. Definitions for the 2008–2009 season. Available: wwwphac-aspcgcca/fluwatch/08-09/def08-09-engphp. [Google Scholar]
- 5.Foucher RA, Bestebroer TM, Herfst S, et al. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Chen S, Evans DH. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J Clin Microbiol. 2001;39:696–704. doi: 10.1128/JCM.39.2.696-704.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Public Health Agency of Canada. The Canadian pandemic influenza plan for the health sector. Ottawa (ON): the agency; 2009. [(accessed 2009 June 24)]. Annex C: pandemic influenza laboratory guidelines. Available: wwwphac-aspcgcca/cpip-pclcpi/ann-c-engphp. [Google Scholar]
- 8.US Centers for Disease Control and Prevention. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Weekly Rep. 2009;58:521–4. [PubMed] [Google Scholar]
- 9.Espy MJ, Uhl JR, Sloan LM, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Centers for Disease Control and Prevention. Swine-origin influenza A (H1N1) virus infections in a school — New York City, April 2009. MMWR. 2009;58:470–2. [PubMed] [Google Scholar]
- 11.Dawood FS, Jain S, Finelli L, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.