Abstract
Retrovirus assembly and maturation involve folding and transport of viral proteins to the virus assembly site followed by subsequent proteolytic cleavage of the Gag polyprotein within the nascent virion. We report that inhibiting proteasomes severely decreases the budding, maturation, and infectivity of HIV. Although processing of the Env glycoproteins is not changed, proteasome inhibitors inhibit processing of Gag polyprotein by the viral protease without affecting the activity of the HIV-1 viral protease itself, as demonstrated by in vitro processing of HIV-1 Gag polyprotein Pr55. Furthermore, this effect occurs independently of the virus release function of the HIV-1 accessory protein Vpu and is not limited to HIV-1, as proteasome inhibitors also reduce virus release and Gag processing of HIV-2. Electron microscopy analysis revealed ultrastructural changes in budding virions similar to mutants in the late assembly domain of p6gag, a C-terminal domain of Pr55 required for efficient virus maturation and release. Proteasome inhibition reduced the level of free ubiquitin in HIV-1-infected cells and prevented monoubiquitination of p6gag. Consistent with this, viruses with mutations in PR or p6gag were resistant to detrimental effects mediated by proteasome inhibitors. These results indicate the requirement for an active proteasome/ubiquitin system in release and maturation of infectious HIV particles and provide a potential pharmaceutical strategy for interfering with retrovirus replication.
Proteasomes are multicatalytic multisubunit proteases that comprise the major proteolytic system in the cytosol and nuclei of eukaryotic cells for disposing of damaged, misfolded, or unwanted proteins. Most proteasome substrates are covalently attached to ubiquitin (Ub), a 76-aa highly conserved polypeptide. Ub is linked to proteins via an isopeptide bond between its C terminus and ɛ-NH2 groups of Lys residues present either on the target protein itself or on Ub already attached to the target protein. The latter results in the chain formation of poly-Ub. Oligomers of four (or more) Ub molecules target the protein for proteasomal destruction (reviewed in ref. 1), whereas linkage of mono-Ub is used to regulate protein functions, e.g., the internalization of cell-surface proteins (2).
Given its central role in cellular metabolism, it is expected that the proteasome/Ub system is involved in viral replication, and numerous examples have been reported for a variety of viruses. Work has begun to unravel the involvement of the proteasome/Ub system in the replication of HIV. The system is used for the degradation of the primary virus receptor CD4 induced by the HIV-1 protein Vpu (3). Additionally, unconjugated Ub was found to be incorporated into virus particles of HIV-1, simian immunodeficiency virus (SIV), avian leukosis virus, and Moloney murine leukemia virus (Mo-MuLV), and a single Ub was detected covalently attached to the C-terminal domains of HIV-1 and SIV Gag proteins and to the p12 domain of Mo-MuLV Gag (4, 5). Finally, proteasomes may degrade structural proteins of incoming HIV particles, decreasing viral infectivity (6).
The main structural components of retrovirus particles are synthesized as three polyproteins that produce either the inner virion core (Gag), the viral enzymes (Pol), or the glycoproteins of the virion envelope (Env). The processing of the HIV-1 Gag polyprotein Pr55 by the viral protease (PR) generates the matrix (MA), capsid (CA), nucleocapsid (NC), and p6gag proteins. HIV particles bud from the plasma membrane as immature noninfectious viruses, consisting predominantly of uncleaved polyproteins. Subsequently, and in concert with PR activation, processing of Gag polyproteins and condensation of the inner core structure occur, resulting in the formation of mature infectious virus (reviewed in ref. 7). Besides PR (8), at least two other viral factors are known to promote efficient budding and release of virus particles: the HIV-1 specific accessory protein Vpu (9) and the p6gag domain (10). Although Vpu supports virus release by ion channel activity, the C-terminal Gag domain, p6gag, contains the late assembly (L) domain that is required for efficient separation of assembled virions from the cell surface (10, 11). However, the mechanism of L-domain function in virus release has not yet been solved.
In the present study, we demonstrate that proteasomal blockade profoundly interferes with the processing of Gag polyproteins and decreases release and infectivity of secreted virions. This phenomenon occurred independently of the virus release function of Vpu but depended on the integrity of PR and p6gag, two viral factors that govern both processing of Gag polyproteins and release of budding virions and for which a mutual interaction has been previously suggested (11). In addition, we found that proteasome inhibition reduced the level of free Ub in HIV-1- infected cells and prevented monoubiquitination of p6gag. The findings presented in this study, together with accompanying papers by Strack et al. (12) and Patnaick et al. (13), point to a hitherto unappreciated function of the Ub–proteasome pathway in late steps of retrovirus replication.
Materials and Methods
Cell Culture.
H9, A3.01, MT-4, and C8166 were cultured in RPMI medium 1640, and HeLa cells (ATCC CCL2) were propagated in Dulbecco's modified Eagle's medium. Pulse–chase experiments and immunoprecipitation were performed as described (3, 14). The following plasmic DNAs were used for transfection: p6ILterm and p6PTAP (11), pNL4-3 (15), pROD10 (16), vpuDEI-1 (17), and pD25A, which carries an A-to-C nucleotide exchange in position 2326.
In Vitro Processing of Gag Polyproteins.
HIV-1 PR was preincubated with zLLL/LC for 5 min, and 1 μM purified Pr55 was incubated with 40 nM recombinant HIV-1 PR for 30 min. PR was expressed and purified as described in ref. 18.
Detection of p6gag-Ub Conjugates.
H9 cells acutely infected with HIV-1NL4-3 were treated at peak virus production with 25 μM zLLL and LC. Virions released during a period of 36 h were collected by centrifugation through a 20% sucrose cushion and analyzed by reversed-phase micro-HPLC as described in ref. 4.
Other Methods.
Procedures for infectivity assay, virus step gradient, and electron microscopy are published as supplemental material on the PNAS web site, www.pnas.org.
Results
Inhibition of Proteasome Activity Reduces HIV-1 Gag Processing and Virus Particle Release.
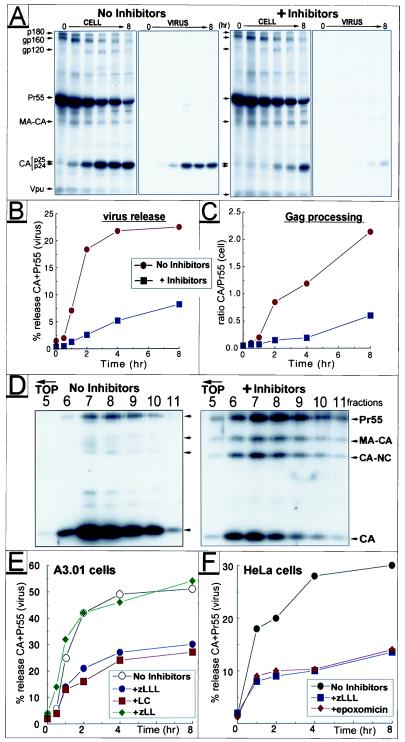
To study whether proteasome activity is important for assembly and release of HIV-1, parallel cultures of HeLa cells transfected with the molecular clone HIV-1NL4–3 were either left untreated or were treated with two proteasome inhibitors: the peptide aldehyde N-carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (zLLL, also known as MG132), which reversibly blocks all activities of the 26S proteasome (19), and lactacystin (LC), a highly specific and irreversible proteasome inhibitor (20). Inhibitors were added 45 min before pulse labeling with [35S]methionine and were maintained throughout the chase period. Comparable amounts of radiolabeled cellular and viral proteins were recovered from treated and control cultures at the end of pulse labeling (not shown), confirming that short zLLL/LC treatment does not affect protein synthesis in HIV-1-expressing HeLa cells, similar to results previously reported for uninfected cells (14). At various times after radiolabeling detergent lysates of cells, the viral pellet and the clarified supernatant fractions were subjected to immunoprecipitation with HIV-1-specific antisera and analyzed by SDS/PAGE and fluorography (Fig. 1A). Relative amounts of the Gag polyprotein Pr55 and its major processing product CA were quantitated by using a PhosphorImager (Molecular Dynamics), and percentages of particle-associated Gag proteins were plotted as a function of time (Fig. 1B).
Figure 1.
Effect of proteasome inhibitors on HIV-1 release and Gag processing. HeLa cells transfected with HIV-1NL4–3 were treated with 10 μM each of zLLL and LC or were left untreated during a pulse–chase experiment. Viral proteins were immunoprecipitated from the cell lysates, pelleted virions, and clarified supernatant, separated by SDS/PAGE, and analyzed by fluorography (A). Positions of the two major CA products, p24 and p25, are indicated by double arrows. The time course of virus release was calculated as the percentage of Gag (Pr55 and CA) present in the virus pellet relative to the total amount of Gag detected intra- and extracellularly (B). The rate of Pr55 processing was estimated by calculating the ratio of CA vs. Pr55 detected intracellularly at different time points (C). D shows a sucrose density gradient analysis of virus particles produced from HIV-1-infected A3.01 cells in the presence or absence of zLLL/LC. Individual fractions of the gradient were analyzed by Western blot by using HIV-1-specific antiserum. For studies on proteasome specificity, infected A3.01 (E) or transfected HeLa cells (F) were treated individually with proteasome inhibitors zLLL, LC, and epoxomicine or the control inhibitor zLL, respectively (final concentration 10 μM). After pulse–chase, virus release was calculated as above.
We observed a significant delay and reduction of HIV-1 release in the presence of proteasome inhibitors (Fig. 1 A and B). This effect was most dramatic early during the chase period, reaching ≈6-fold difference in extracellular particulate Gag after 2 h. By the end of the 8-h chase period, ≈3-fold less Gag was still recovered from the virus fractions (Fig. 1 A and B). In addition, zLLL/LC treatment inhibited the processing of Pr55 into CA as evidenced by a 3- to 5-fold lower ratio of intracellular CA over Pr55 in the treated culture throughout the chase period (Fig. 1C). This effect was specific for Gag, because neither the cleavage nor the stability of the HIV-1 envelope glycoproteins was significantly altered (Fig. 1A). Similar findings were made in additional experiments by using a variety of HIV-1 isolates, including macrophage tropic viruses (not shown), indicating that the effects of proteasome inhibitors are not restricted to a particular HIV-1 isolate or strain.
To analyze the sedimentation characteristic of released virions and the profile of virus-associated Gag proteins, we characterized HIV-1 particles released over a 24-h period of treatment with zLLL/LC. Virions produced by an acutely HIV-1-infected T cell line, A3.01, cultured in the absence or presence of inhibitors, were subjected to centrifugation on a sucrose density gradient followed by immunoblot analysis (Fig. 1D). The sedimentation of virus particles was comparable, although peak-of-virus particles produced by inhibitor-treated cells were slightly shifted (one fraction) toward the top of the gradient compared with virions from control cells (Fig. 1D). This subtle difference might reflect variations in Gag processing and virus composition. Most important, and consistent with short-term release kinetics (Fig. 1 A–C), this steady-state analysis (Fig. 1D) revealed ≈3-fold reduction of virus release from inhibitor-treated cells. Furthermore, Gag processing was significantly impaired in virions generated in the presence of inhibitors resulting in an up to 10-fold lower ratio of CA/Pr55 compared with virions produced from untreated cultures. Complete cleavage of Pr55 leads in general to formation of matrix (MA), CA, NC, and p6gag, as well as small spacer peptides separating individual domains. Several of these sequential cleavage steps were impaired in the presence of inhibitors, and intermediate cleavage products probably corresponding to MA-CA (p41), CA-NC (p39) (Fig. 1D), and a CA protein still containing the C-terminally adjacent 14-aa spacer peptide 1 (p25) were detected (Fig. 1A).
Inhibition of HIV-1 Polyprotein Processing and Release Is Specific for Compounds That Block Proteasome Activity.
Besides blocking proteasomes at higher concentrations, peptide aldehydes like zLLL also inhibit cellular cysteine proteases, including calpains (reviewed in ref. 19). To control for proteasome specificity, we included N-carbobenzoxy-l-leucyl-l-leucinal (zLL), a dipeptide analogue that inhibits a similar spectrum of thiol proteases but does not affect the proteasome at the concentrations used (21). Acutely infected A3.01 cells were treated individually with 20 μM zLLL, zLL, or the most specific proteasome inhibitor LC, and pulse–chase experiments were conducted as above. Both LC and zLLL reduced Pr55 processing (not shown) and virus release with equal efficiency, whereas the control inhibitor zLL had no effect (Fig. 1E).
In additional experiments (not shown), we found that the maximal effect was observed when LC and zLLL were used in a combination of 10 μM each or when either inhibitor was applied individually at 20–30 μM. At these inhibitor concentrations, cell viability, as determined by trypan blue staining, was not affected in the time frame of the pulse–chase experiments reported here, nor was the secretion of free nonparticulate viral proteins enhanced, as demonstrated by immunoprecipitation of soluble Gag and Env gp120 proteins from the clarified supernatants after pelleting of virus particles (not shown). Therefore, it is concluded that drug treatment had not caused nonspecific release of viral proteins by cell leakage and that suppression of virus release and Gag processing is specific for the inactivation of proteasomes.
We also tested a novel proteasome inhibitor, epoxomicin, a natural epoxy–ketone containing peptoid produced by Actinomycetes that possesses antitumor and antiinflammatory activities in vivo. Epoxomicin is a highly selective, irreversible, and potent inhibitor of the chymotrypsin-like activity of the 20 S proteasome (22). In HIV-1-expressing HeLa cells, epoxomicin reduced Gag processing (not shown) and virus release (Fig. 1F) to a similar extent as zLLL tested in parallel at the same relatively low concentration of 10 μM.
Proteasome Inhibitor Treatment Decreases HIV-1 Infectivity.
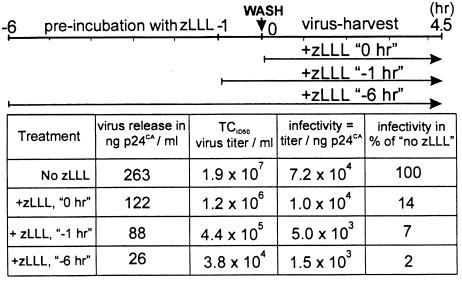
Considering the reduction of virus release and Gag processing, proteasome inhibitors should also affect virus infectivity. A3.01 cells acutely infected with HIV-1NL4–3 were treated with zLLL for different time periods, and the infectious titer of virus-containing supernatants collected during a 4.5-h harvest period was determined by endpoint titration (Table 1). HIV-1 from an untreated culture had a titer of 1.9 × 107 infectious units ml−1, whereas treatment of virus-producing cells with zLLL starting at the beginning of the harvest period led to ≈15-fold reduction in virus titer, and longer pretreatment caused a further decline in infectivity. Loss of infectivity resulted from decrease in virus release and, additionally, an up to 50-fold reduction in specific infectivity of released particles, most likely because of defects in Gag processing (Table 1).
Table 1.
Proteasome inhibitors decrease virus infectivity
Parallel cultures of HIV-1NL4–3-infected A3.01 cells either were left untreated or were incubated with 40 μM zLLL for 1 or 6 h. Cells were washed, and incubation in the presence or absence of zLLL was continued for another 4.5 h as indicated in the time scheme on top. Virus-containing medium was collected, and CA antigen was quantitated by ELISA. Virus titer was determined by endpoint dilution on C8166 cells. The specific infectivity was calculated as infectious titer per nanogram of CA and expressed in percent of the untreated sample.
Proteasome Inhibitors Interfere with Gag Processing and Release of HIV-1 and HIV-2 in a Vpu-Independent Manner.
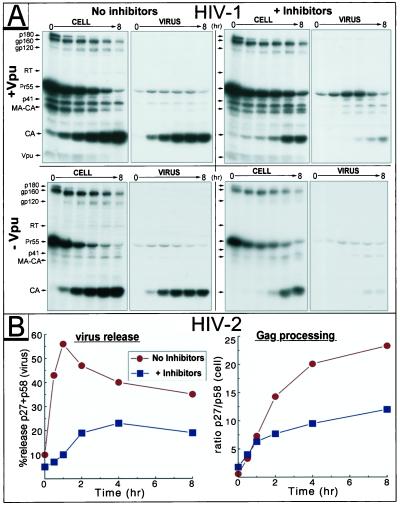
The HIV-1 accessory protein Vpu regulates virus release (9, 17), and proteasome activity is required for one of Vpu's distinct functions, the degradation of the primary virus receptor CD4 (3). To investigate the potential involvement of Vpu in the proteasome inhibitor-mediated impairment of virus release, A3.01 cells were infected with wild-type HIV-1NL4–3 or with a vpu-deficient mutant. Standard pulse–chase experiments revealed a similar reduction in processing of intracellular and virus-associated Pr55 accompanied by lower virus release after zLLL/LC application for wild-type and mutant viruses (Fig. 2A). The Vpu-dependent increase in virus production was clearly detectable even in cultures with inactive proteasomes. In addition, because of the high level of Gag expression in acutely infected T-cells when compared with transfected HeLa cells (Fig. 1A), pulse–chase data presented in Fig. 2 demonstrate impaired Gag processing even in the virus particle fractions after treatment with zLLL/LC. In similar experiments in A3.01 cells infected with variants of HIV-1NL4–3, we found that proteasome inhibitors blocked the release of a phosphorylation mutant of Vpu (3), which is unable to induce CD4 degradation, or an ion channel inactive mutant of Vpu, which is unable to promote virus release (data not shown). Taken together, these findings clearly indicate that proteasome inhibitors act independently of Vpu in blocking virus release and do not negate the virus-release function of Vpu.
Figure 2.
Proteasome inhibitors interfere with Gag processing and release of HIV-1 and HIV-2 in a Vpu-independent manner. A3.01 cells infected with HIV-1NL4–3 (+Vpu) or the Vpu-mutant vpuDEL-1 (−Vpu) (A), and HeLa cells transfected with the HIV-2 proviral plasmid pROD10 (B) were incubated in the presence or absence of inhibitors (10 μM each of zLLL and LC) and subjected to pulse–chase studies. In B, the HIV-2 Gag precursor Pr58 and the major processing product p27CA collected by immunoprecipitation were quantitated, and the time course of particle release and efficiency of intracellular Gag processing were calculated as described for Fig. 1.
We next examined the effects of proteasome inhibitors on HeLa cells transfected with the molecular clone HIV-2ROD10 (16). HIV-2 is a closely related lentivirus that does not encode Vpu. Consistent with the effects observed for HIV-1, processing of the Pr58 Gag polyprotein and release of HIV-2 particles were significantly impaired in cells with inactivated proteasomes (Fig. 2B), suggesting that these phenomena apply in general to human immunodeficiency viruses.
Proteasome Inhibitors Do Not Inhibit PR Activity.
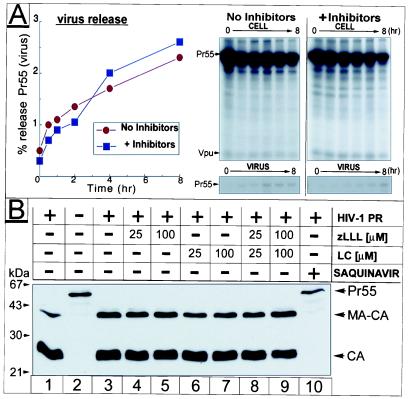
To determine whether the deleterious effects of proteasome inhibitors on HIV release and maturation depend on PR activity, we studied the effect of zLLL/LC on a variant of HIV-1NL4–3 (pD25A) that carries a point mutation in the PR active site. PR activity is not essential for release of immature noninfectious particles but appears to promote virus production, at least in the case of HIV-1 (8). Accordingly, particle production of pD25A was ≈8-fold lower (Fig. 3A) compared with wild-type virus (Fig. 1 A and B). In contrast to findings with Rous sarcoma virus (RSV) and SIV Gag particles (12, 13), we observed under various experimental conditions in context of vpu+ full length HIV-1NL4–3 a significant decline in virus release after PR inactivation. However, no additional effect on virus release was observed on treatment of PR-deficient HIV-1 with proteasome inhibitors, suggesting that the inhibitors directly target Gag processing (Fig. 3A).
Figure 3.
Effect of proteasome inhibitors on PR activity. In A, a pulse–chase similar to Fig. 1 was conducted in HeLa cells transfected with the PR mutant pD25A. Relevant parts of the fluorograms depicting cell and virus fractions are shown (Right), and calculation of release kinetics is depicted (Left). (B) Recombinant Pr55 and PR were incubated in the presence or absence of inhibitors at the concentrations indicated. Cleavage reactions were analyzed by Western blot by using HIV-1 CA-specific antiserum.
The simplest explanation for our observations would be a direct inhibition of HIV-1 PR by proteasome inhibitors. Although the known activities and catalytic mechanisms of the proteasome are quite distinct from that of HIV-1 PR, crossinhibition is conceivable, particularly because the HIV-1 PR inhibitor ritonavir has been reported to inhibit the chymotrypsin-like activity of the 20S proteasome (23, 24). We therefore examined the effect of inhibitors on in vitro processing of HIV-1 Gag. Pr55 was incubated with HIV-1 PR at an enzyme-to-substrate ratio of 1:25, where cleavage was only partial and inhibitory effects could be detected with maximal sensitivity (Fig. 3B, lane 1). No effect on PR activity was observed for LC or zLLL or a combination of both at concentrations of up to 100 μM (Fig. 3B, lanes 4–9), whereas the HIV PR-specific inhibitor saquinavir completely blocked processing (Fig. 3B, lane 10). Thus, under these in vitro assay conditions, proteasome inhibitors do not affect HIV-1 PR activity and are likely to block HIV maturation through another mechanism.
Electron Microscopy Analysis of HIV-1-Infected Cells Treated with Proteasome Inhibitor.
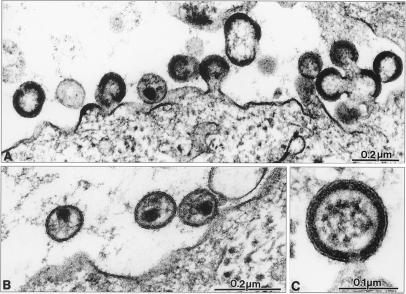
We next examined the effects of proteasome inhibitors on assembly and release and on virion structure by thin-section electron microscopy (Fig. 4). MT-4 cells acutely infected with HIV-1NL4–3 were either left untreated or were treated with 50 μM zLLL for 5 h. During the treatment period, cells were drawn into cellulose capillary tubes, leading to retention of secreted virions, therefore obviating the need for centrifugation. In agreement with our biochemical data on virus release and Gag processing, zLLL caused two obvious alterations: (i) a major reduction in the number of extracellular particles and budding structures, and (ii) a relative increase in the number of immature virions (Fig. 4A). However, some mature particles of regular size and morphology were also detected in the zLLL-treated sample (Fig. 4B).
Figure 4.
Electron microscopy analysis of proteasome inhibitor-treated HIV-1-infected cells. HIV-1-infected MT-4 cells were treated with 50 μM zLLL for 5 h and fixed for thin-section electron microscopy. A shows an overview of infected cells with budding structures and immature virus. B shows mature extracellular HIV-1 particles, and C shows a higher-magnification view of an immature particle still connected to the cellular membrane.
Electron microscopy analysis usually reveals various assembly structures at different stages, ranging from early accretions at the membrane to very late-budding structures resembling immature virions but still tethered to the plasma membrane. A representative analysis of zLLL-treated cells revealed a relative increase in the number of these very late-budding structures compared with untreated control cells (Fig. 4C). Budding arrest at a very late stage has previously been observed for HIV-1 isolates that carry mutations within the L-domain of the C-terminal p6gag region of Pr55 (10, 11). These variants exhibited reduced virus release accompanied by an increase in relative and absolute numbers of late-budding structures (10). Although causing budding arrest, zLLL also reduced the total number of budding structures, an effect that is probably related to the decrease in protein synthesis associated with zLLL treatment for up to 5 h (14).
Proteasome Inhibitors Interfere with Ubiquitination of p6gag.
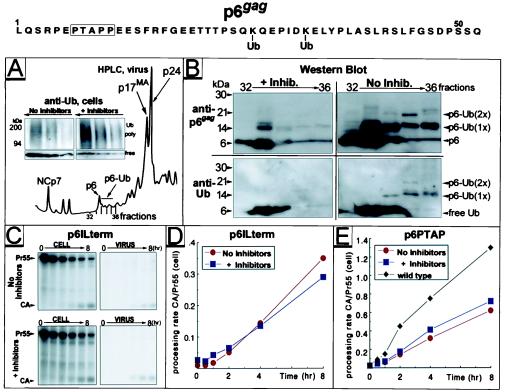
A direct connection between the ubiquitin/proteasome pathway and the function of p6gag is implied by the recent finding that single molecules of Ub are conjugated to HIV-1 and SIV p6gag proteins and to the functionally analogous p12gag protein of Moloney murine leukemia virus (4). A major consequence of proteasome shutdown is changes in the dynamic of Ub expression and turnover resulting in an accumulation of polyubiquitinated proteins and reduced levels of free Ub available for conjugation (25). This was readily detected by Western blot analysis of HIV-1NL4–3-infected H9 cells after treatment with zLLL/LC: whereas proteasome shutdown induced accumulation of polyubiquitinated proteins, the level of free Ub declined (Fig. 5A Inset).
Figure 5.
Variants of HIV-1 with alterations in p6gag are insensitive to proteasome inhibitors that prevent monoubiquitination in wild-type p6gag. Proteins from purified HIV-1NL4–3 particles produced in H9 cells in the presence or absence of 25 μM zLLL/LC were separated by HPLC. The HPLC A206 profile is shown in A. Increasing aliquots, 5–30 μl of lysates of cells used for virus production, were analyzed by Western blot by using monoclonal antibody detecting both poly- and mono-Ub (A Inset). Selected HPLC fractions of separated virions were analyzed by Western blot (B) by using antibodies specific for Ub, followed by stripping and reprobing with anti-p6gag. Positions of free Ub and p6gag as well as those of mono- [p6-Ub (1×)] and di- [p6-Ub (2×)] ubiquitinated conjugates are indicated (Right). HeLa cells transfected with p6ILterm (C and D) or p6PTAP (E) were subjected to treatment with 25 μM zLLL/LC, pulse–chase experiments were conducted as described for Figs. 1–3, and rates of Gag processing were calculated (D and E). Only relevant parts of fluorograms depicting cell and virus fractions for p6ILterm-transfected cells are shown in C. In E, rate of Gag processing established for wild-type HIV-1NL4–3 in cells not treated with inhibitors was included for comparison. Amino acid sequence of the p6gag domain derived from HIV-1NL4–3 is indicated (Top).
To analyze the impact of proteasome inhibition on monoubiquitination of p6gag, components of HIV-1 particles produced in the presence or absence of inhibitors were fractionated by micro-HPLC (Fig. 5A), and fractions containing p6gag, free Ub, and Ub–p6gag conjugates were analyzed by Western blot (Fig. 5B). Virions produced from untreated cells contained mono- (17 kDa, fractions 33–36) and diubiquitinated (24 kDa, fraction 35) p6gag conjugates, in addition to free p6gag (fractions 32 and 33 with trailing to 36) and Ub. In contrast, virions produced in the presence of inhibitors contained little, if any, p6gag–Ub conjugates, whereas free p6gag and Ub molecules were readily detectable (Fig. 5B). The ≈14-kDa proteins detected in fraction 33 of treated and control samples reacted with anti-p6gag but not with anti-Ub antibody (Fig. 5B) or antisera directed against NC or the Gag-spacer peptide SP2 (not shown) and therefore represent uncharacterized forms of p6gag.
L-Domain Mutants of HIV-1 Are Not Affected by Proteasome Inhibitors.
Proteasome inhibition induces ultrastructural changes similar to L-domain mutants in p6gag (Fig. 4) and, at the same time, forestalls monoubiquitination of two conserved Lys residues in p6gag (Fig. 5 A and B). This led us to examine whether variants containing mutations in this domain respond to proteasome inhibitors. Two previously described variants were analyzed: p6lLterm contains a stop codon immediately following the NC-SP2-p6gag cleavage site, leading to a complete loss of p6gag expression, whereas p6PTAP contains several point mutations in the PTAP motif (11) essential for the late-budding function (10). In agreement with previously published results (11), pulse–chase analysis of p6gag variants revealed reduced virus release and Pr55 processing in the absence of inhibitor treatment when compared with wild-type HIV-1 (Fig. 5 C–E). However, no additional decrease in Gag processing and virus release was observed for either of the two p6gag variants on treatment with zLLL/LC (Fig. 5 C–E). This finding is in contrast to the observations for vpu-deficient mutants of HIV-1 (Fig. 2A), which were also reduced in virus release but remained responsive to inhibitor treatment. Taken together, these results indicate that the detrimental effect of proteasome inhibitors depends on the integrity of two functional domains, p6gag (Fig. 5A) and PR (Fig. 3A), which are in general required for efficient virus maturation and release.
Discussion
There is only limited knowledge regarding the mechanisms involved in the highly regulated process of retrovirus assembly such as activation of PR and transport, folding, and processing of Gag polyproteins. We found that virus budding, maturation, and infectivity of released virions are dramatically reduced when host cells are exposed to proteasome inhibitors. Our findings suggest that this deficiency coincides with a defect in the proteolytic processing of Gag by PR. These phenomena may result from one or a combination of the following models:
Model 1: Proteasome inhibitors interfere with virus budding and, because proteolytic processing is a late event, there is less cleavage of Gag polyproteins.
Model 2: Proteasome inhibitors interfere with Gag processing and, because PR activity promotes efficient HIV-1 release (8), there is less virus budding.
There are several possibilities that may apply to either model 1 or 2 or both. First would be a mechanism that involves the interaction of functional late domain within p6gag with the Ub machinery. Proteasome inhibition in general causes depletion of pools of Ub available for protein conjugation resulting in rapid deubiquitination of nucleosomal histones (23). Indeed, we found that proteasome inhibitors significantly reduce levels of free Ub in HIV-1-infected cells that may account for the absence of mono-Ub residues normally present in p6gag. This finding together with our observation that variants of HIV-1 that either do not express p6gag or possess mutations within the critical PTAP region of p6gag are insensitive to proteasome inhibitors suggests that virus release and proteolytic maturation require monoubiquitination of p6gag. This hypothesis is consistent with recent findings by Patnaik et al. (see accompanying paper, ref. 13) demonstrating an important function of Ub in assembly and release of RSV. Similar to our findings on HIV and those of others on SIV Gag (12), proteasome inhibition caused defects in virus budding, although in the case of RSV, Gag processing was only slightly diminished (13). The effects of proteasome inhibitors were overcome by expressing Ub from a transfected gene or by fusing Ub directly to the C terminus of RSV Gag (13).
The mechanism by which Ub promotes retrovirus release is rather obscure at the moment, as are the Ub-attachment sites in Gag and the number of Ub molecules added to each site. In an additional study, we found that exchanging Arg for the Lys residues in positions 27 and 33 in p6gag does not affect maturation, release, infectivity, and consequently replication characteristic of HIV-1 in T cell cultures. Furthermore, removal of Ub-attachment sites in p6gag does not influence the level of free Ub detected in secreted HIV-1 virions (24). However, it is well known that ubiquitination can occur at multiple Lys residues, and removal of one acceptor can shift ubiquitination machinery to adjacent lysine within the targeted protein (25). Thus, further investigations are necessary to study the importance of various potential Ub sites in HIV-1 Gag for virus release and maturation.
The findings of Strack et al. (12) and Patnaik et al. (13), in conjunction with our own, are consistent with two possibilities: first, ubiquitination of Gag is required for its optimal function in virus release and maturation. Second, the L-domain of Gag functions to recruit Ub ligases to the site of viral assembly where they act on cellular factors that contribute to viral maturation, with Gag ubiquitination occurring incidentally. Distinguishing between these mechanisms will require additional experiments.
At the present time, we cannot absolutely distinguish the relative contributions of either Gag processing or virus release to the detrimental effect of proteasome inhibitors on HIV assembly. In favor of the effect on virus budding is our observation that proteasome inhibitors do not affect PR activity in vitro. Four observations, however, suggest it may be more important that proteasome inhibitors interfere with Gag polyprotein processing: first, proteasome inhibitors have no significant effect on release of immature Pr55 particles produced by a HIV-1 PR deficient mutant. Second, release of these PR- inactive virions is significantly reduced compared with wild-type HIV-1 in the absence of proteasome inhibitors. Third, the virus-release factor Vpu does not counteract the effect of proteasome inhibitors on HIV-1 release, nor is the effect modulated by the differing kinetics of release of various HIV-1 and HIV-2 isolates. Fourth, proteasome inhibitors block the processing of Gag when expressed by recombinant vaccinia virus, despite the very low efficiency of particle production in this system (U.S., unpublished observations).
Although more studies are required to decipher the mechanism(s) by which proteasome inhibitors interfere with retrovirus release and maturation, the involvement of the proteasome/Ub system in retrovirus replication provides potential targets for antiviral drug development. Given the cytotoxic properties of proteasome inhibitors in cultured cells, it would be expected that they would be highly toxic in vivo at concentrations necessary to efficiently block virus replication. Surprisingly, the proteasome inhibitor epoxomicin effectively blocked HIV-1 maturation in cell culture at concentrations previously demonstrated to be tolerated in vivo where it exhibited antitumor and antiinflammatory effects in mice (22). It is therefore of interest to test highly specific proteasome inhibitors like epoxomicin for their capacity to block HIV replication in vivo. Alternatively, it is also possible that the antiviral activity of the proteasome inhibitors is based on downstream effects on cellular metabolism, which may target a specific aspect of the viral maturation process. A thorough understanding of the phenomenon, therefore, may lead to novel and highly specific strategies for blocking retroviral replication.
Additional discussion is published as supplemental material on the PNAS web site, www.pnas.org.
Supplementary Material
Acknowledgments
We are grateful to H. Hohenberg, G. Rutter, C. Huckhagel, and I. Ellhof for performing electron microscopy analysis; to Lori Coren for Western blot analysis; to Eric Freed, Luis Anton, and James Gibbs for many helpful discussions; to Eric Freed for the generous gift of p6gag mutants, and to Jan Konvalinka (Technical University, Prague, Czech Republic) for a gift of Saquinavir. This work was supported by federal funds from the National Cancer Institute, the National Institutes of Health, under Contract No. NO1-CO-56000 (to D.O.) and in part by grants from the German Ministry for Education and Research and the Deutsche Forschungsgemeinschaft (DFG) (to H.G.K.). U.S. was supported by Grant Schu11/2–1 and a Heisenberg Grant from the DFG.
Abbreviations
- Ub
ubiquitin
- SIV
simian immunodeficiency virus
- PR
viral protease
- CA
capsid
- NC
nucleocapsid
- L
late assembly
- zLLL
N-carbobenzoxy-l-leucyl-l-leucyl-l-leucinal
- zLL
N-carbobenzoxy-l-leucyl-l-leucinal
- LC
lactacystin
- RSV
Rous sarcoma virus
- DRiPs
defective ribosomal products
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12945.
References
- 1.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Shih S C, Sloper-Mould K E, Hicke L. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubert U, Antón L C, Bacik I, Cox J H, Bour S, Bennink J R, Orlowski M, Strebel K, Yewdell J W. J Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder C, 2nd, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putterman D, Pepinsky R B, Vogt V M. Virology. 1990;176:633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard J-M. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanstrom R, Wills J. In: Retroviruses. Coffin J, Hughes S, Varmus H, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 263–334. [Google Scholar]
- 8.Kaplan A H, Manchester M, Swanstrom R. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strebel K, Klimkait T, Martin M A. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- 10.Göttlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strack B, Calistri A, Accola M A, Palù G, Göttlinger H G. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patnaik A, Chau V, Wills J W. Proc Natl Acad Sci USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schubert U, Antón L C, Gibbs J, Norbury C, Yewdell J W, Bennink J R. Nature (London) 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 15.Adachi A, Gentleman H E, König S, Folks T, Willey R, Rabson A, Martin M A. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bour S, Schubert U, Strebel K. J Virol. 1995;69:1510–1520. doi: 10.1128/jvi.69.3.1510-1520.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klimkait T, Strebel K, Hoggan M D, Martin M A, Orenstein J M. J Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konvalinka J, Litterst M A, Welker R, Kottler H, Rippmann F, Heuser A M, Kräusslich H G. J Virol. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 20.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 21.Tsubuki S, Saito Y, Tomioka M, Hisashi I, Kawashima S. J Biochem. 1996;11:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- 22.Meng L, Mohan R, Kwok B H, Elofsson M, Sin N, Crews C M. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimnaugh E G, Chen H Y, Davie J R, Celis J E, Neckers L. Biochemistry. 1997;36:14418–14429. doi: 10.1021/bi970998j. [DOI] [PubMed] [Google Scholar]
- 24.Ott, D. E., Coren, L. V., Chertova, E. N., Gagliardi, T. D. & Schubert, U. (2000) Virology, in press. [DOI] [PubMed]
- 25.Hou D, Cenciarelli C, Jensen J P, Nguygen H B, Weissman A M. J Biol Chem. 1994;269:14244–14247. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.