Abstract
Purpose
Currently, we lack biomarkers to predict whether high-risk women with mammary atypia will respond to tamoxifen chemoprevention.
Experimental Design
Thirty-four women with cytologic mammary atypia from the Duke University High-Risk clinic were offered tamoxifen chemoprevention. We tested whether ESR1 promoter hypermethylation and/or estrogen receptor (ER) protein expression by immunohistochemistry predicted persistent atypia in 18 women who were treated with tamoxifen for 12 months and in 16 untreated controls.
Results
We observed a statistically significant decrease in the Masood score of women on tamoxifen chemoprevention for 12 months compared with control women. This was a significant interaction effect of time (0, 6, and 12 months) and treatment group (tamoxifen versus control) P = 0.0007. However, neither ESR1 promoter hypermethylation nor low ER expression predicted persistent atypia in Random Periareolar Fine Needle Aspiration after 12 months tamoxifen prevention.
Conclusions
Results from this single institution pilot study provide evidence that, unlike for invasive breast cancer, ESR1 promoter hypermethylation and/or low ER expression is not a reliable marker of tamoxifen-resistant atypia.
Introduction
In cancer, epigenetic silencing through promoter hypermethylation of tumor suppressor and other genes has been discovered to occur at least as frequently as mutations or deletions (1). Numerous studies support the idea that promoter hypermethylation plays a causal role in the earliest stages of carcinogenesis (2-4). Evidence also suggests that hypermethylation of DNA repair genes may affect overall and disease-free survival in patients with malignancy (5).
The estrogen receptor-α (ESR1) promoter and first exon contain a CpG island of which aberrant hypermethylation occurs in breast, endometrial, prostate, and lung cancer, as well as adult acute myeloid leukemia and hepatocellular carcinoma (6-11). ESR1 promoter hypermethylation is observed in both estrogen receptor ER(+) and ER(−) breast cancer specimens at widely varying rates—from 0% to 100%—although these studies often stratify specimens according to multiple characteristics such as hormone receptor status (12-18). Some reports have found that in breast cancer cell lines, ESR1 promoter hypermethylation exhibits a tight inverse relationship with ER expression (15). The suppression of estrogen receptor in these ER(−) cell lines is lifted after treatment with the demethylating agent 5-azacytidine (19), and previous findings suggest a correlation between ESR1 promoter hypermethylation and suppression of message and protein levels in breast cancer (15, 17). However, other reports find no or only a weak association, suggesting it may not be a clear inverse relationship (13, 14, 20). Recent studies show that that ESR1 promoter hypermethylation outdid hormone receptor status as a predictor of clinical response to the selective estrogen receptor modulator tamoxifen in women with invasive breast cancer (20).
Random Periareolar Fine Needle Aspiration (RPFNA) is a research technique developed to repeatedly sample mammary cells from the whole breast of asymptomatic high-risk women to assess both breast cancer risk and response to chemoprevention (21-23). The presence of atypia in RPFNA has been prospectively validated to confer a 5.6-fold increase in breast cancer risk in high-risk women (23). The presence of persistent atypia in RPFNA has been recently used as a surrogate marker to track cytologic response to chemoprevention agents (21, 22). These studies underscore the utility of RPFNA as a translational research tool, and as in our previous work, RPFNA can be used to couple cytologic analysis, methylation studies, and chemoprevention (21, 24, 25).
Although ER expression is routinely used to predict tamoxifen sensitivity in invasive breast cancers and ductal carcinoma in situ, ER expression is not clinically used to predict the tamoxifen sensitivity of atypia. This is because normal breast tissue expresses low levels of ER (26-28). Over 90% of epithelial cells in normal breast tissue express low ER and only 5% to 10% cells express moderate levels of ER (26-28). Importantly though, unlike ER(−) estrogen-resistant invasive breast cancer, normal breast tissue is estrogen responsive. Furthermore, in normal breast tissue, only epithelial cells expressing low ER (and not ER+ epithelial cells) proliferate in response to estrogen (27, 28). It is hypothesized that this inverse relationship is disturbed during early mammary carcinogenesis (27).
In our single institution pilot study, we tested for the ability of ESR1 promoter hypermethylation and low ER expression to predict persistent atypia in RPFNA in 18 high-risk women who took tamoxifen chemoprevention for 12 months. Here, we provide evidence that although tamoxifen reduced the incidence of mammary atypia in high-risk women, neither ESR1 promoter hypermethylation nor low ER expression predicted persistent atypia after 12 months tamoxifen chemoprevention, suggesting they may not be suitable markers for tamoxifen-resistant atypia. Larger multi-institutional studies are needed to confirm these results.
Materials and Methods
Informed Consent
The study was approved by the Human Subjects Committee and Institutional Review Board at Duke University Medical Center, in accordance with assurances filed with and approved by the Department of Health and Human Services. Women were offered tamoxifen chemoprevention as part of standard-of-care for high-risk women.
RPFNA Eligibility
To be eligible for RPFNA, women were required to have at least one of the following major risk factors for breast cancer: (a) 5-y Gail risk calculation of ≥1.7%; (b) prior biopsy exhibiting atypical hyperplasia, lobular carcinoma in situ, and ductal carcinoma in situ; (c) known BRCA1/2 mutation carrier; or (d) contralateral breast cancer (23).
Tamoxifen Chemoprevention
To be eligible this study, women were required to have (a) a prior excisional biopsy exhibiting atypical hyperplasia, lobular carcinoma in situ, or contralateral ductal carcinoma in situ and (b) a minimum of one prior RPFNA demonstrating a Masood Cytology Index of 14 to 16 (atypia). Women with a history of clotting disorder, stroke, or abnormal uterine bleeding were not eligible to participate. Thirty-four women were offered tamoxifen chemoprevention; 18 women elected to take tamoxifen and 16 declined. This is consistent with our previously published studies demonstrating that approximately half of high-risk women in our cohort with atypia in RPFNA elect to take tamoxifen chemoprevention and half decline (29). Eighteen high-risk women with atypia (Masood 14−16) in a minimum of one prior RPFNA received 20 mg/d tamoxifen chemoprevention. Sixteen high-risk women with atypia in RPFNA (Masood 14−16) who declined tamoxifen chemoprevention served as controls. RPFNA was done at the time of starting tamoxifen chemoprevention (0 mo) and at 6 and 12 mo after initiation of tamoxifen. Persistent atypia was defined by the continued presence of aspirates with Masood Cytology Indices of 14 to 16 after 12 mo tamoxifen treatment.
RPFNA
RPFNA was done as previously published (25). A minimum of one epithelial cell cluster with at least 10 epithelial cells was required to sufficiently determine pathology; the most atypical cell cluster was examined and scored (22, 23). Cells were classified qualitatively as nonproliferative, hyperplasia, or hyperplasia with atypia (30). Cytology preparations were also given a semiquantitative index score through evaluation by the Masood Cytology Index (31). As previously described, cells were given a score of 1 to 4 points for each of six morphologic characteristics that include cell arrangement, pleomorphism, number of myoepithelial cells, anisonucleosis, nucleoli, and chromatin clumping; the sum of these points computed the Masood score: ≤10 nonproliferative (normal); 11 to 13 hyperplasia; 14 to 17 atypia; >17 suspicious cytology (23, 31).
Materials and Cell Culture Lines
Sodium bisulfite (Sigma; A.C.S.) and hydroquinone (Sigma; 99+%) were used under reduced lighting and stored in a dessicator. The T47D breast cancer cell line was obtained from the American Type Culture Collection and grown in supplemented αMEM (Life Technologies).
Methylation-Specific PCR
DNA was extracted from breast cancer cell lines and RPFNA as previously published; bisulfite treatment was as previously published (25). Previous work has elucidated appropriate methylation-specific PCR primers within exon 1 of the ESR1 promoter, from nt +376 to nt +494 relative to the transcription start site (18). The primer sequences used were as follows:Methylated (M) forward 5′-GTG TAT TTG GAT AGT AGT AAG TTC GTC-3′; M reverse 5′-CGT AAA AAA AAC CGA TCT AAC CG-3′; Unmethylated (U) forward 5′-GGT GTA TTT GGA TAG TAG TAA GTT TGT-3′; U reverse 5′-CCA TAA AAA AAA CCA ATC TAA CCA-3′ (18). All PCR reactions consisted of 50 ng bisulfite-treated DNA, 1× PCR buffer, 250 μmol/L of each deoxynucleotide triphosphate, 200 nmol/L of each primer, and 2.5 U of HotStar Taq polymerase (Qiagen) in 30 μL total volume.
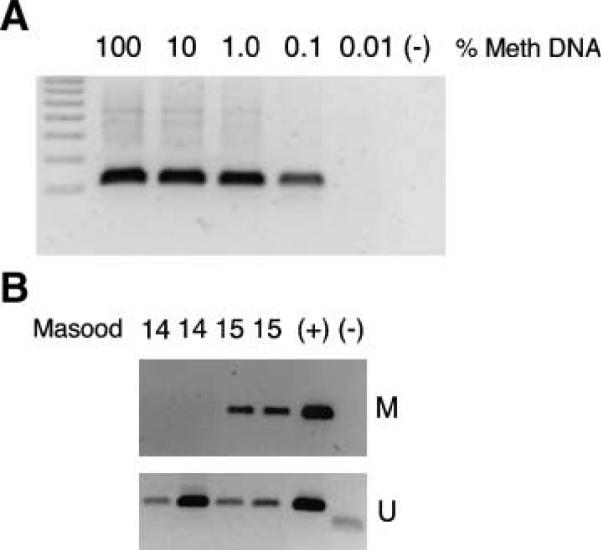
PCR buffers were individually optimized for the methylated and unmethylated programs. The 1× M buffer consisted of 15 mmol/L (NH4)2SO4, 60 mmol/L Tris (pH 8.0), 4.0 mmol/L MgCl2, and 100 mmol/L 2-pyrrolidinone (Fluka; 99+%); the 1× U buffer consisted of 15 mmol/L (NH4)2SO4, 60 mmol/L Tris (pH 8.5), and 4.5 mmol/L MgCl2. The methylated PCR program consisted of 95°C for 5 min followed by 40 amplification cycles (94°C for 1 min, 56°C for 1 min, and 72°C for 1 min) and a final extension of 72°C for 4 min; the unmethylated PCR program was identical except a 52°C annealing temperature was used. A GeneAmp PCR System 9700 (Applied Biosystems) was used for all amplifications. PCR products were visualized on 1.5% ethidium bromide agarose gels using an Image Station 440 (Kodak). Optimization with methylated primers was achieved using minute amounts (∼50 pg) of CpGenome Universal Methylated DNA (Chemicon) to model RPFNA samples. To estimate PCR sensitivity, titrated experiments were done using known amounts of methylated genomic positive control DNA (1 μg-100 pg) spiked in unmethylated T47D genomic DNA for a total of 1 μg (Fig. 1A; ref. 18).
Figure 1.
ESR1 promoter hypermethylation. A. Methylation-specific PCR control assays detected 0.1% methylated sequence (1 ng of positive control supplemented with unmethylated cell line for a total of 1 μg genomic DNA). Titration experiments were as described in Materials and Methods; ESR1 M, hypermethylation of the ESR1 promoter. B. Hypermethylation of the ESR1 promoter in RPFNA obtained from representative high-risk women with atypical RPFNA. M and U, the use of methylation-specific PCR primers to identify methylated and unmethylated ESR1 promoter, respectively. (+), a hypermethylated positive control in the M gels and the T47D breast cancer cell line unmethylated positive control in the U gels. (−), negative PCR control.
ER Immunohistochemistry
ERα protein expression was tested in RPFNA cytology specimens by immunohistochemistry (IHC) by a modification of published methods (32). Antigen retrieval was at 90°C with 0.2× nuclear decloker (pH 9.5) for 2 min. Slides were then placed in glucose oxidase blocker at 37°C for 30 min. Cells were incubated with anti-ERα antibody (DAKO, 1D5). A minimum of 100 cells was assessed on the sample judged to be most abnormal by 2 readers. The proportion of cells stained at 5 intensity levels (0+ to 4+) were assessed to provide a weighted intensity score, calculated by multiplying each intensity level by the proportion of cells at that level and then summing over all levels. For example, if 80% of cells exhibited 0+ staining and 20% exhibited 1+ staining, the weighted intensity score would be 0.0 + 0.2 = 0.2. The slide was judged to by ER(+) if the intensity score was ≥0.2.
Statistical Methods
The Wilcoxon Rank-Sums test was used to test for the association between ESR1 promoter hypermethylation and ER expression as well as the potential association between ESR1 promoter hypermethylation and/or ER expression and persistent atypia after 12 months of tamoxifen chemoprevention. A SAS Mixed model analysis for repeated measures was used to test for differences in Masood scores over time and between groups.
Results
Study Demographics
Eighteen high-risk women received 20 mg/day tamoxifen as part of standard-of-care for high-risk women. Sixteen high-risk women who declined tamoxifen chemoprevention served as controls. All 34 women had a minimum of one prior RPFNA that scored a Masood Cytology Index of 14 to 16 (atypia). Women underwent RPFNA aspiration at 0, 6, and 12 months. A Masood Cytology Index of 14 to 16 was confirmed on the 0 month RPFNA. The average age of women receiving tamoxifen prevention was 43.5 years (range, 36−50 years); average age for controls was 45.5 years (range, 39−55 years). The median RPFNA Masood score for women receiving tamoxifen prevention at 0 month was 15 and for controls subjects was 14 (Table 1). Seventeen of 18 high-risk women completed 12 months of tamoxifen prevention; 1 woman elected to undergo prophylactic surgery after 6 months of tamoxifen prevention and did not complete 12 months of tamoxifen.
Table 1.
Summary of Masood scores, ER methylation, and expression
| Age in y (range) | T = 0 | T = 6 mo | T = 12 mo | ESR1 Methylation | ER low (IHC) | |
|---|---|---|---|---|---|---|
| Control group | 45.5 (39−55) | 14 | 14 | 15 | 5/16 (31%) | 12/16 (75%) |
| Tamoxifen group | 43.5 (36−50) | 15 | 14 | 13 | 2/18 (11%) | 13/18 (72.2%) |
NOTE: Summary of Masood scores, ER methylation, and expression. This table provides information on 18 women who received tamoxifen chemoprevention (tamoxifen group) and 16 women who did not (control group). Median age, ESR1 methylation, and ER expression by IHC were determined at the T = 0 time point of the study. Numbers in T = 0, T = 6 mo, and T = 12 mo columns are the median Masood scores at that time point.
The median RPFNA Masood score for women receiving tamoxifen prevention at 6 and 12 months was 14 and 13.5, respectively (Table 1). The median RPFNA Masood score for controls subjects at 6 and 12 months was 14 and 15, respectively (Table 1). There was a statistically significant difference in the average RPFNA Masood score at 12 months for women taking tamoxifen relative to control subjects (P = 0.028; Table 3) but not at 6 months (P = 0.32; Table 3). Furthermore, in a SAS mixed model analysis for repeated measures of Masood score, there is a significant interaction effect of time (0, 6, and 12 months) and treatment group (tamoxifen versus no tamoxifen; P = 0.0007; Table 3).
Table 3.
Summary of variables tested for statistical significance
| Variable(s) tested | Groups compared | P |
|---|---|---|
| Masood score | Control vs tamoxifen group at 6 mo | 0.32 |
| Masood score | Control vs tamoxifen group at 12 mo | 0.028 |
| Masood score | Effect of time (0, 6, 12 mo) and treatment group (control vs tamoxifen) | 0.0007 |
| Masood score | Tamoxifen group persistent vs loss of atypia at 12 mo | 0.001 |
| ESR1 promoter hypermethylation vs ER expression (IHC) | Whole cohort | 0.15 |
| ESR1 promoter hypermethylation | Tamoxifen group persistent vs loss of atypia | 0.32 |
| ER expression (IHC) | Tamoxifen group persistent vs loss of atypia | 0.5 |
NOTE: Summary of variables tested for statistical significance. This table is a summary of all statistical data presented in the article. Significant statistical effects are in bold.
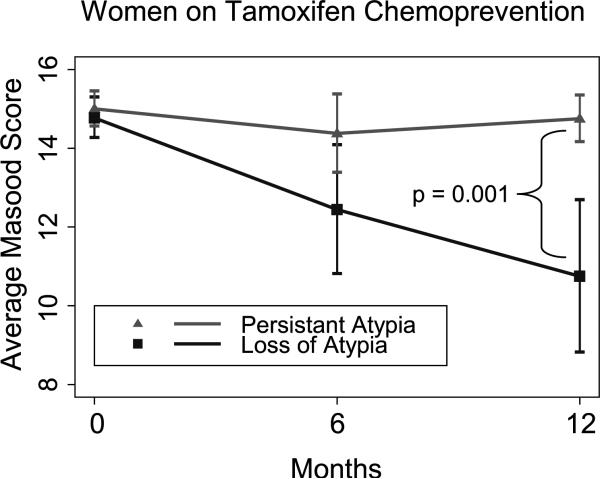
Atypia in RPFNA was defined by an initial (0 month) RPFNA with a Masood Cytology Index of 14 to 16. Persistent atypia was defined by the continued presence of RPFNA cells with a Masood score of 14 to 16 after 12 months of tamoxifen treatment. In the control group, 15 of 16 women had persistent atypia 12 months after their initial RPFNA. After 12 months of tamoxifen treatment, 8 of 17 (47%) women had persistent atypia, and 9 of 17 (53%) women had a loss of atypia. The median Masood score at 12 months for the group of tamoxifien-treated women with persistant atypia was 14.75, whereas the median Masood score at 12 months for the group of tamoxifen-treated women with loss of atypia was 10.75 (Fig. 1). These medians are statistically significantly different (P = 0.001; Table 3).
ESR1 Promoter Hypermethylation in Atypical RPFNA Samples Does Not Predict Low ER Expression
Hypermethylation from nt +367 to nt +494 of the ESR1 promoter was tested using methylation-specific PCR in all RPFNA specimens obtained. Four representative samples are shown in Fig. 1B. Strong unmethylated bands were detected in all included samples, confirming both the presence of DNA and the promoter sequence itself (Fig. 1B). ER protein expression was tested by IHC using a modification of published methods. The frequency of ESR1 promoter hypermethylation and low ER in atypical RPFNA was 7 of 34 (21%) and 25 of 34 (74%), respectively (Table 1). ESR1 promoter hypermethylation was observed in 2 of 18 atypical RPFNA samples obtained from high-risk women who received tamoxifen prevention and in 5 of 16 high-risk control subjects who did not receive tamoxifen (Table 1). Low ER expression was observed in 13 of 18 (72%) atypical RPFNA samples obtained from women receiving tamoxifen prevention and in 12 of 16 (75%) control subjects (Table 1). No association was found between ESR1 promoter hypermethylation and ER expression by IHC in the combined group of subjects (P = 0.15; Table 3).
Neither ESR1 Promoter Hypermethylation nor Low ER Expression Predicts Persistent Atypia after 12 Months of Tamoxifen Prevention
Although there was no relationship between ESR1 promoter hypermethylation and ER protein levels in our cohort, we sought to test whether these variables may be potenital biomarkers for predicting persistent atypia after tamoxifen chemoprevention. From the overall group of 17 women who received 12 months of tamoxifen prevention, 12 women had low ER expression (Fig. 2). Of these 12 women, 7 with low ER expression had persistent atypia, whereas the other 5 women with low ER expression had a loss of atypia at 12 months (Table 2). Only one woman who received tamoxifen chemoprevention had ESR1 promoter hypermethylation (Table 2). Of the 16 women without ESR1 promoter hypermethylation who received tamoxifen, 8 of 16 (50%) had persistent atypia. Neither ESR1 promoter hypermethylation nor low ER expression was statistically associated with the presence of persistent atypia in high-risk women who received tamoxifen prevention (P = 0.32 and P = 0.5, respectively; Table 3).
Figure 2.
Median Masood score from tamoxifen-treated women with persistent atypia or loss of atypia. The 17 women that completed 12 mo of tamoxifen treatment were divided into 2 groups: persistent aytpia (▲, Masood score of ≥14) or loss of atypia at 12 mo (■, Masood score of <14). The median Masood value at 0, 6, and 12 mo for these two groups was plotted over time. There was a statistically significant difference in the median Masood score at 12 mo (P = 0.001). Errors bars, 95% confidence interval.
Table 2.
Summary of ER methylation and expression in Control and tamoxifen groups divided into women with persistent atypia or loss of atypia
| ESR1 promoter methylation | Low ER expression (IHC) | |
|---|---|---|
| Women on tamoxifen (n = 17) | ||
| Persistent atypia (n = 8) | 0 | 7 |
| Loss of atypia (n = 9) | 1 | 5 |
| Control women (n = 16) | ||
| Persistent atypia (n = 15) | 4 | 11 |
| Loss of atypia (n = 1) | 1 | 1 |
| Whole cohort (n = 33) | 6 | 24 |
| 21% | 74% |
NOTE: Summary of ER methylation and expression in control and tamoxifen groups divided into women with persistent atypia or loss of atypia. This table provides information for ESR1 promoter methylation and ER expression on 17 women who completed 12 mo of tamoxifen chemoprevention and 16 women who served as controls. Both the tamoxifen and control groups were divided into either persistent atypia (Masood ≥ 14) or loss of atypia (Masood < 14) at 12 mo. The data for the two groups combined is also represented (whole cohort).
Discussion
The National Surgical Adjuvant Breast and Bowel Project Trial Breast Cancer Prevention Trial (P1) showed a 50% reduction in estrogen-sensitive breast cancer in premenopausal high-risk women who took tamoxifen chemoprevention (33). Importantly, tamoxifen decreased risk of invasive breast cancer in women with lobular carcinoma in situ and atypical hyperplasia by 56% and 86%, respectively (33). These findings provide evidence that women with lobular carcinoma in situ and atypia benefit most from tamoxifen (33). As a result, tamoxifen chemoprevention is an option for premenopausal women with mammary atypia. Although tamoxifen has been shown to have significant benefit in many women, not all women benefit from tamoxifen prevention and there are significant side effects associated with tamoxifen treatment. Progress in breast cancer prevention is currently limited by our lack of biological markers to identify which women will respond to prevention therapies such as tamoxifen, as well as to identify women with tamoxifen-resistant atypia. Here, we tested whether ESR1 promoter hypermethylation predicted persistent atypia in high-risk women who were treated with tamoxifen chemoprevention. Recent reports show that ESR1 promoter hypermethylation outdid hormone receptor status as a predictor of clinical response to tamoxifen hormonal therapy in women with invasive breast cancer (20). In contrast to these prior studies in invasive breast cancer, neither ESR1 promoter hypermethylation nor low ER expression predicted persistent atypia at 12 months in this study. These studies highlight potential differences between ER+ breast cancer and mammary atypia.
Although the presence of atypia has been shown to increase breast cancer risk, it is currently unknown if the presence of persistent atypia in RPFNA can be used as a surrogate marker of breast cancer risk or as a marker of resistance to tamoxifen chemoprevention. We are unable to make conclusions relative to women who have a loss of atypia as this may be due to sampling error; further studies are necessary. However, in women with persistent atypia, it is clear that tamoxifen did not result in the elimination of atypia; yet the long term implications of these observations must be interpreted with caution. It is possible that women who have persistent atypia on RPFNA will still have a risk reduction benefit. Currently we are unable to identify whether an individual woman with either atypia on excisional biopsy or cytologic atypia on RPFNA will progress to develop breast cancer. Further longitudinal studies are ongoing to determine whether women with persistent atypia on RPFNA are at increased short-term breast cancer risk relative to women who have had disappearance of atypia. It is important to recognize that this study is limited by being a small, single institution study. Although these results are interesting, further validation in a larger multi-institution study is required before our findings can be generalized to a larger, high-risk population.
Although the molecular mechanism of tamoxifen action in ER+ breast cancer is well-studied, there is little information on how tamoxifen may act in noncancerous human breast tissue that normally expresses low levels of ER (ER “poor”). Prevention models for tamoxifen action in ER-poor, noncancerous breast tissue have been primarily based on observations in ER+ breast cancer cells. Normal mammary tissue is composed of a heterogeneous population of cells. Greater than 90% of normal mammary epithelial cells express low ER, and only 5% to 10% of normal mammary epithelial cells express moderate ER (26). If normal breast tissue is evaluated solely by ER expression, it would be classified as ER(−). This classification system, however, may not be adequate for describing normal mammary tissue. Although normal mammary tissue exhibits low ER, it responds to estrogen and, therefore, is not equivalent to ER(−), estrogen-resistant invasive breast cancer.
There are a number of possible explanations why ER-poor mammary atypia may respond to tamoxifen chemoprevention therapy. First, in vivo atypical mammary epithelial cells may be more sensitive to extragenomic effects of tamoxifen. In our in vitro cellular models of early mammary carcinogenesis, we observed that immediately after the acute loss of p53 function, primary human mammary epithelial cells exhibit a narrow window of tamoxifen-induced apoptosis sensitivity (34-37). It is possible that loss of p53 or other tumor suppressors early in mammary carcinogenesis (38, 39) sensitizes cells to tamoxifen chemoprevention, leading to elimination of these atyplical cells in high-risk women.
Although inhibition of ER transcriptional activity and signaling is the predominate effect of tamoxifen in the breast, not all of the effects of tamoxifen can be directly attributed to competitive interactions with ER. Tamoxifen induces apoptosis in cholangiocarcinoma cells and inhibits angiogenesis in fibrosarcomas (40, 41). Tamoxifen also has a wide variety of other pharmacologic activities including stimulation of transforming growth factor-β, blockade of various chloride channels (42), inhibition of protein kinase C (43), and antagonism of calmodulin activity (44). Furthermore, tamoxifen-binding sites, independent of ER, have been identified. For example, Sutherland, et al. (45) reported a high-affinity antiestrogen binding site in human and rat uterine cells, as well as in other tissues. Tamoxifen also directly inhibits calmodulin in a calcium-dependent manner (44). Because therapeutic concentrations of tamoxifen are several orders of magnitude higher than required to saturate ER (46), these “extragenomic” effects of tamoxifen may play an important role in ER-poor normal breast tissue.
The molecular mechanisms for tamoxifen resistance in ER(+) invasive breast cancer is an area of intense investigation. It is possible that similar mechanisms for tamoxifen resistance in invasive breast cancer may exist during early mammary carcinogenesis. For example, ER/progesterone receptor–positive invasive breast cancer is more likely to respond to antiestrogen therapy than ER(+)/progesterone receptor(−) breast cancer. The present study did not examine progesterone receptor status. Growth factor signaling pathways downstream of ErbB family members, such as the Akt/mammalian target of rapamycin pathway, are often up-regulated in tamoxifen-resistant ER(+) breast cancers. Therefore, if these pathways are up-regulated in high-risk women with mammary atypia, it is possible that these women would be resistant to tamoxifen chemoprevention. Furthermore, the ER coactivators AIB1/SRC-3 and MNAR/PELP1 have both been shown to promote tamoxifen resistance in models of ER(+) breast cancer, both dependent and independent of ER signaling (47, 48). Finally, alterations in tamoxifen metabolism may also predict resistance. Production of tamoxifen metabolites occurs in the liver via cytochrome P450 CYP2D6 (49, 50). Several polymorphisms in CYP2D6 that result in a decrease in tamoxifen metabolism have been identified (51, 52). Although the effect of these polymorphisms on the efficacy of tamoxifen is still controversial (53, 54), it has been reported that specific mutations in CYP2D6 results in a higher risk of disease relapse but a decrease in the incidence of tamoxifen side effects (55). Although there is no clinical data associated with tamoxifen chemoprevention, it is reasonable to hypothesize that women with CYP2D6 polymorphisms may be resistant to tamoxifen chemoprevention.
In this study, we found that ESR1 hypermethylation or low ER expression was not able to predict persistent atypia in response to tamoxifen treatment in our high-risk cohort. Additionally, our results indicate that, similar to invasive breast cancer, not all mammary atypia is responsive to tamoxifen; 53% of women in this cohort showed a loss of atypia at 12 months. Although the implications of persistant atypia relative to breast cancer risk are still unclear, further analysis of RPFNA samples from ayptia that is tamoxifen responsive versus tamoxifen resistant may be useful to determine biomarkers of tamoxifen sensitivity, as well as to increase our understanding of the early events underlying mammary carcinogenesis and the different tumor types that arise in the breast.
Acknowledgments
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Grant support: NIH/National Cancer Institute grants P50CA68438 (AVON/National Cancer Institute Partners in Progress), P30CA14236-26, R01CA88799 (V.L. Seewaldt), R01CA98441 (V.L. Seewaldt), and R01CA114068 (V.L. Seewaldt); Susan G. Komen Breast Cancer Award BCTR061314 (V.L. Seewaldt), and a V-Foundation Award (V.L. Seewaldt).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holst CR, Nuovo GJ, Esteller M, et al. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–601. [PubMed] [Google Scholar]
- 4.Veigl ML, Kasturi L, Olechnowicz J, et al. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopelovich L, Crowell JA, Fay JR. The epigenome as a target for cancer chemoprevention. J Natl Cancer Inst. 2003;95:1747–57. doi: 10.1093/jnci/dig109. [DOI] [PubMed] [Google Scholar]
- 6.Hori M, Iwasaki M, Shimazaki J, Inagawa S, Itabashi M. Assessment of hypermethylated DNA in two promoter regions of the estrogen receptor α gene in human endometrial diseases. Gynecol Oncol. 2000;76:89–96. doi: 10.1006/gyno.1999.5662. [DOI] [PubMed] [Google Scholar]
- 7.Lai JC, Cheng YW, Chiou HL, Wu MF, Chen CY, Lee H. Gender difference in estrogen receptor α promoter hypermethylation and its prognostic value in non-small cell lung cancer. Int J Cancer. 2005;117:974–80. doi: 10.1002/ijc.21278. [DOI] [PubMed] [Google Scholar]
- 8.Li LC, Chui R, Nakajima K, Oh BR, Au HC, Dahiya R. Frequent methylation of estrogen receptor in prostate cancer:correlation with tumor progression. Cancer Res. 2000;60:702–6. [PubMed] [Google Scholar]
- 9.Li Q, Kopecky KJ, Mohan A, et al. Estrogen receptor methylation is associated with improved survival in adult acute myeloid leukemia. Clin Cancer Res. 1999;5:1077–84. [PubMed] [Google Scholar]
- 10.Ottaviano YL, Issa JP, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–5. [PubMed] [Google Scholar]
- 11.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–61. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 12.Archey WB, McEachern KA, Robson M, et al. Increased CpG methylation of the estrogen receptor gene in BRCA1-linked estrogen receptor-negative breast cancers. Oncogene. 2002;21:7034–41. doi: 10.1038/sj.onc.1205844. [DOI] [PubMed] [Google Scholar]
- 13.Falette NS, Fuqua SA, Chamness GC, Cheah MS, Greene GL, McGuire WL. Estrogen receptor gene methylation in human breast tumors. Cancer Res. 1990;50:3974–8. [PubMed] [Google Scholar]
- 14.Hori M, Iwasaki M, Yoshimi F, Asato Y, Itabashi M. Hypermethylation of the Estrogen Receptor α Gene Is Not Related to Lack of Receptor Protein in Human Breast Cancer. Breast Cancer. 1999;6:79–86. doi: 10.1007/BF02966912. [DOI] [PubMed] [Google Scholar]
- 15.Lapidus RG, Ferguson AT, Ottaviano YL, et al. Methylation of estrogen and progesterone receptor gene 5′ CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin Cancer Res. 1996;2:805–10. [PubMed] [Google Scholar]
- 16.Nass SJ, Herman JG, Gabrielson E, et al. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–8. [PubMed] [Google Scholar]
- 17.Piva R, Rimondi AP, Hanau S, et al. Different methylation of oestrogen receptor DNA in human breast carcinomas with and without oestrogen receptor. Br J Cancer. 1990;61:270–5. doi: 10.1038/bjc.1990.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapidus RG, Nass SJ, Butash KA, et al. Mapping of ER gene CpG island methylation-specific polymerase chain reaction. Cancer Res. 1998;58:2515–9. [PubMed] [Google Scholar]
- 19.Ferguson AT, Lapidus RG, Baylin SB, Davidson NE. Demethylation of the estrogen receptor gene in estrogen receptor-negative breast cancer cells can reactivate estrogen receptor gene expression. Cancer Res. 1995;55:2279–83. [PubMed] [Google Scholar]
- 20.Widschwendter M, Siegmund KD, Muller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64:3807–13. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 21.Bean GR, Kimler BF, Seewaldt VL. Long-term raloxifene in a woman at high risk for breast cancer. N Engl J Med. 2006;355:1620–2. doi: 10.1056/NEJMc061954. [DOI] [PubMed] [Google Scholar]
- 22.Fabian CJ, Kimler BF, Brady DA, et al. phase II breast cancer chemoprevention trial of oral α-difluoromethylornithine: breast tissue, imaging, and serum and urine biomarkers. Clin Cancer Res. 2002;8:3105–17. [PubMed] [Google Scholar]
- 23.Fabian CJ, Kimler BF, Zalles, et al. Short-term breast cancer prediction by random periareolar fine-needle aspiration cytology and the Gail risk model. J Natl Cancer Inst. 2000;92:1217–27. doi: 10.1093/jnci/92.15.1217. [DOI] [PubMed] [Google Scholar]
- 24.Bean GR, Ibarra Drendall C, Goldenberg VK, et al. Hypermethylation of the breast cancer-associated gene 1 promoter does not predict cytologic atypia or correlate with surrogate end points of breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2007;16:50–6. doi: 10.1158/1055-9965.EPI-06-0598. [DOI] [PubMed] [Google Scholar]
- 25.Bean GR, Scott V, Yee L, et al. Retinoic acid receptor-β2 promoter methylation in random periareolar fine needle aspiration. Cancer Epidemiol Biomarkers Prev. 2005;14:790–8. doi: 10.1158/1055-9965.EPI-04-0580. [DOI] [PubMed] [Google Scholar]
- 26.Anderson E, Clarke RB. Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia. 2004;9:3–13. doi: 10.1023/B:JOMG.0000023584.01750.16. [DOI] [PubMed] [Google Scholar]
- 27.Anderson E, Clarke RB, Howell A. Estrogen responsiveness and control of normal human breast proliferation. J Mammary Gland Biol Neoplasia. 1998;3:23–35. doi: 10.1023/a:1018718117113. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Mohsin SK, Mao S, Hilsenbeck SG, Medina D, Allred DC. Hormones, receptors, and growth in hyperplastic enlarged lobular units:early potential precursors of breast cancer. Breast Cancer Res. 2006;8:R6. doi: 10.1186/bcr1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldenberg VK, Seewaldt VL, Scott V, et al. Atypia in random periareolar fine-needle aspiration affects the decision of women at high risk to take tamoxifen for breast cancer chemoprevention. Cancer Epidemiol Biomarkers Prev. 2007;16:1032–4. doi: 10.1158/1055-9965.EPI-06-0910. [DOI] [PubMed] [Google Scholar]
- 30.Zalles CM, Kimler BF, Kamel S, McKittrick R, Fabian CJ. Cytology patterns in random aspirates from women at high and low risk for breast cancer. Breast J. 1995;1:343–9. [Google Scholar]
- 31.Masood S, Frykberg ER, McLellan GL, Scalapino MC, Mitchum DG, Bullard JB. Prospective evaluation of radiologically directed fine-needle aspiration biopsy of nonpalpable breast lesions. Cancer. 1990;66:1480–7. doi: 10.1002/1097-0142(19901001)66:7<1480::aid-cncr2820660708>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P, Kimler BF, Warner C, et al. Estrogen receptor expression in benign breast ductal cells obtained from random periareolar fine needle aspiration correlates with menopausal status and cytomorphology index score. Breast Cancer Res Treat. 2006;100:71–6. doi: 10.1007/s10549-006-9234-8. [DOI] [PubMed] [Google Scholar]
- 33.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 34.Bowie ML, Dietze EC, Delrow J, et al. Interferon-regulatory factor-1 is critical for tamoxifen-mediated apoptosis in human mammary epithelial cells. Oncogene. 2004;23:8743–55. doi: 10.1038/sj.onc.1208120. [DOI] [PubMed] [Google Scholar]
- 35.Bowie ML, Troch MM, Delrow J, et al. Interferon regulatory factor-1 regulates reconstituted extracellular matrix (rECM)-mediated apoptosis in human mammary epithelial cells. Oncogene. 2007;26:2017–26. doi: 10.1038/sj.onc.1210013. [DOI] [PubMed] [Google Scholar]
- 36.Dietze EC, Caldwell LE, Grupin SL, Mancini M, Seewaldt VL. Tamoxifen but not 4-hydroxytamoxifen initiates apoptosis in p53(-) normal human mammary epithelial cells by inducing mitochondrial depolarization. J Biol Chem. 2001;276:5384–94. doi: 10.1074/jbc.M007915200. [DOI] [PubMed] [Google Scholar]
- 37.Dietze EC, Troch MM, Bean GR, et al. Tamoxifen and tamoxifen ethyl bromide induce apoptosis in acutely damaged mammary epithelial cells through modulation of AKT activity. Oncogene. 2004;23:3851–62. doi: 10.1038/sj.onc.1207480. [DOI] [PubMed] [Google Scholar]
- 38.Aldaz CM, Hu Y, Daniel R, Gaddis S, Kittrell F, Medina D. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene. 2002;21:6366–76. doi: 10.1038/sj.onc.1205816. [DOI] [PubMed] [Google Scholar]
- 39.Rohan TE, Hartwick W, Miller AB, Kandel RA. Immunohistochemical detection of c-erbB-2 and p53 in benign breast disease and breast cancer risk. J Natl Cancer Inst. 1998;90:1262–9. doi: 10.1093/jnci/90.17.1262. [DOI] [PubMed] [Google Scholar]
- 40.Blackwell KL, Haroon ZA, Shan S, et al. Tamoxifen inhibits angiogenesis in estrogen receptor-negative animal models. Clin Cancer Res. 2000;6:4359–64. [PubMed] [Google Scholar]
- 41.Pan G, Vickers SM, Pickens A, et al. Apoptosis and tumorigenesis in human cholangiocarcinoma cells. Involvement of Fas/APO-1 (CD95) and calmodulin. Am J Pathol. 1999;155:193–203. doi: 10.1016/S0002-9440(10)65113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson JR, Wakefield LM, Baum M, Colletta AA. Synthesis and secretion of transforming growth factor β isoforms by primary cultures of human breast tumour fibroblasts in vitro and their modulation by tamoxifen. Br J Cancer. 1996;74:352–8. doi: 10.1038/bjc.1996.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Brian CA, Liskamp RM, Solomon DH, Weinstein IB. Inhibition of protein kinase C by tamoxifen. Cancer Res. 1985;45:2462–5. [PubMed] [Google Scholar]
- 44.Lopes MC, Vale MG, Carvalho AP. Ca2(+)-dependent binding of tamoxifen to calmodulin isolated from bovine brain. Cancer Res. 1990;50:2753–8. [PubMed] [Google Scholar]
- 45.Murphy LC, Sutherland RL. A high-affinity binding site for the antioestrogens, tamoxifen and CI 628, in immature rat uterine cytosol which is distinct from the oestrogen receptor. J Endocrinol. 1981;91:155–61. doi: 10.1677/joe.0.0910155. [DOI] [PubMed] [Google Scholar]
- 46.Friedman ZY. Recent advances in understanding the molecular mechanisms of tamoxifen action. Cancer Invest. 1998;16:391–6. doi: 10.3109/07357909809115779. [DOI] [PubMed] [Google Scholar]
- 47.Osborne CK, Bardou V, Hopp TA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–61. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 48.Vadlamudi RK, Manavathi B, Balasenthil S, et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005;65:7724–32. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan VC. New insights into the metabolism of tamoxifen and its role in the treatment and prevention of breast cancer. Steroids. 2007;72:829–42. doi: 10.1016/j.steroids.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 51.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3:229–43. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 52.Raimundo S, Toscano C, Klein K, et al. A novel intronic mutation, 2988G>A, with high predictivity for impaired function of cytochrome P450 2D6 in white subjects. Clin Pharmacol Ther. 2004;76:128–38. doi: 10.1016/j.clpt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wegman PP, Wingren S. CYP2D6 variants and the prediction of tamoxifen response in randomized patients:author response. Breast Cancer Res. 2005;7:E7. doi: 10.1186/bcr1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]