Abstract
Loss of the nuclear hormone receptor HNF4α in hepatocytes results in a complex pleiotropic phenotype that includes a block in hepatocyte differentiation and a severe disruption to liver function(1-3). Recent analyses have shown that hepatic gene expression is severely affected by the absence of HNF4α with expression of 567 genes reduced by ≥2.5-fold (P ≤0.05) in Hnf4α-/- fetal livers(4). While many of these genes are direct targets(5), HNF4a has also been shown to regulate expression of other liver transcription factors(6) raising the possibility that the dependency on HNF4α for normal expression of some genes may be indirect. We postulated that the identification of transcription factors whose expression is regulated by HNF4α might reveal roles for HNF4α in controlling hepatic functions that were not previously appreciated. Here we identify CrebH as a transcription factor whose mRNA can be identified in both the embryonic and adult mouse liver and whose expression is dependent on HNF4α. Analyses of genomic DNA revealed an HNF4α binding site upstream of CrebH coding sequence that was occupied by HNF4α in fetal livers and facilitated transcriptional activation of a reporter gene in transient transfection analyses. Although CrebH is highly expressed during hepatogenesis, CrebH-/- mice were viable, healthy, and displayed no overt defects in liver formation. However, upon treatment with tunicamycin, which induces an endoplasmic reticulum (ER)-stress response, CrebH-/- mice displayed reduced expression of acute phase response proteins. These data implicate HNF4α in having a role in controlling the acute phase response of the liver induced by ER-stress through regulating expression of CrebH.
Background
Hepatocyte gene expression is controlled by a complex network of transcription factors that is established during hepatogenesis(5, 6). Recent studies have shown that the nuclear hormone receptor HNF4α, as part of this network, is essential for initiating and maintaining hepatocyte differentiation and liver function(1-4). Loss of HNF4α in the fetal hepatoblasts results in reduced expression of over 500 genes encoding factors that contribute to all aspects of hepatic function (4). HNF4α has also been shown to regulate expression of other transcription factors, including HNF1α (7, 8) and PPARα (9), suggesting that regulation of hepatocyte gene expression by HNF4α may in some cases be indirect. We therefore sought to uncover novel transcription factors that were expressed during hepatic development that could potentially be regulated by HNF4α. We proposed that identification of such factors would implicate HNF4α in regulating aspects of liver function that heretofore were not recognized. In the current study we report the identification of a CREB/ATF transcription factor CrebH (encoded by the gene Creb3l3) as a new target of HNF4α regulation, which is expressed in differentiating hepatocytes throughout liver development and is essential for expression of acute phase response proteins induced by ER-stress.
Results
CrebH mRNA is highly enriched in the fetal liver
We attempted to identify novel transcription factors whose expression was enriched in fetal livers using Affymetrix oligonucleotide array analyses. RNA was isolated from pools of livers, hearts, and heads that were dissected from E10.5 mouse embryos and used to generate probes that were hybridized to Affymetrix oligonucleotide GeneChip Murine Genome U74v2 A and B arrays. A comparison between liver, heart, and head arrays revealed 1208 genes whose expression was predicted to be increased >3.0 fold in the fetal liver samples compared to heart and head. Of these genes, 78 encoded potential fetal liver-enriched transcription factors(10). This list included Hnf4α, CCAAT/enhancer binding protein alpha (c/EBPα), Forkhead box A1 (Foxa1), and Forkhead box A3 (Foxa3) (Table 1 published as supplemental material), all which have previously been shown to be expressed in hepatoblasts during embryonic development; however, other hepatoblast transcription factors, including HNF1α, HNF1ß, and HNF6 were not identified suggesting that this screen was not saturated.
Of the mRNAs identified we chose to focus our analyses on CrebH because it was highly enriched in the fetal liver, was a member of the ATF/CREB family, other members of which are known to have important roles in controlling liver gene expression, and analyses of the human homolog of CrebH (CREBH) had been described as being exclusively expressed in the adult liver and undetectable in other tissues(11). We first confirmed that CrebH mRNA was differentially expressed in the liver compared to heart and head using RT-PCR. Fig. 1a shows that, like Alb1 mRNA, CrebH mRNA was enriched in three independent E10.5 liver samples compared to heart and head samples. Real-time quantitative RT-PCR (not shown) demonstrated that CrebH mRNA levels were 45-fold greater in the liver compared to the heart.
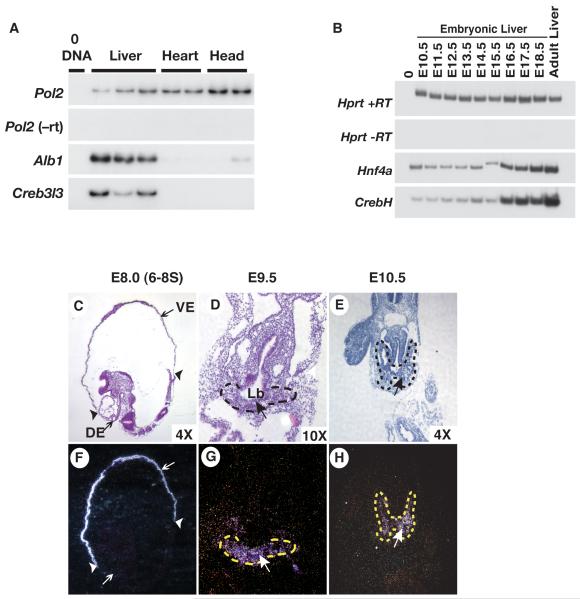
Fig. 1. Fetal expression of CrebH initiates in the primary liver bud and continues throughout hepatogenesis.
A) RT-PCR analyses revealed the presence of CrebH and Albumin (Alb1) mRNAs in livers isolated from E10.5 embryos. Amplification of Rna pol2 (Pol2) was used as a loading control while reactions lacking reverse transcriptase (-RT) and DNA template (0DNA) confirmed the absence of contaminating DNA. B) RT-PCR analyses uncovered CrebH and Hnf4α mRNA in livers isolated mouse embryos at daily intervals ranging from E10.5 through E18.5, as well as in adult livers. Amplification of Hprt was used as loading control while reactions lacking reverse transcriptase (-RT) and DNA template (0DNA) confirmed the absence of contaminating DNA. C-H) Radioactive in situ hybridization analyses revealed the presence of CrebH mRNA (arrows; silver grains) during early development. Sagittal sections through an E8.5 (6-8 somite stage) embryo (C, F) identified CrebH mRNA in the extraembryonic visceral endoderm (VE) but not in the definitive endoderm (DE); extraembryonic/embryonic boundary indicated by arrowheads. CrebH mRNA was also found to be present in the primary liver bud (Lb; outlined with dashes) in transverse sections through an E9.5 embryo (D, G), and in the expanding clusters of hepatoblasts (outlined with dashes) in transverse sections through an E10.5 embryo (E, H). H&E-stained bright field images (C, D, E) and corresponding dark field images (F, G, H) are presented.
Hepatic expression of CrebH initiates during liver bud formation and continues throughout hepatogenesis
As a first step toward determining whether CrebH had the potential to act downstream of HNF4α during liver development and function we compared expression CrebH to that of Hnf4α during hepatic development. Livers were isolated from embryos at E10.5 through E18.5 as well as from adult, and mRNAs encoding CrebH and HNF4α were measured by RT-PCR. Fig. 1b shows that CrebH is expressed in the liver at low levels early in development, begins to increase at around E14.5, and continues increasing until maximal expression is reached in the adult liver. This dynamic pattern of expression is very similar to that of Hnf4α, which was also seen to increase over this developmental time course. In situ hybridization analyses were next performed to assess expression of CrebH mRNA during the onset of hepatic development. Fig. 1c,f show that during specification of the hepatic lineage from the ventral endoderm at E8.0 (6-8 somites), CrebH mRNA is restricted to the extraembryonic visceral endoderm and was not detected in the definitive endoderm, as has previously been described for Hnf4α (12, 13). Approximately a day later in development at E9.5, like Hnf4α (12, 13), CrebH mRNA was detected in the definitive endoderm forming the primary liver bud (Fig. 2d,g) and expression continued in the hepatoblasts as they delaminated from the bud and invaded the surrounding septum transversum at E10.5 (Fig. 1e,h).
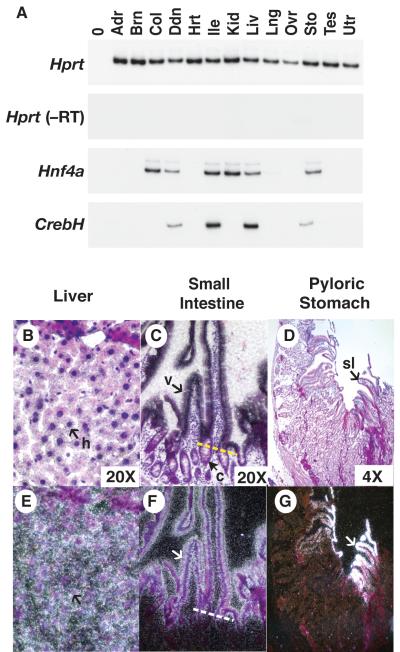
Fig. 2. CrebH is expressed in the adult liver and gastrointestinal tract.
A) RT-PCR analyses of CrebH and Hnf4α was performed on mRNA extracts from the adrenal gland (Adr), brain (Brn), colon (Col), duodenum (Ddn), heart (Hrt) ileum (Ile), kidney (Kid), liver (Liv), lung (Lng), ovary (Ovr), stomach (Sto), testis (Tes) and uterus (Utr). B-G) The distribution of CrebH mRNA in the liver (B, E), small intestine (C, F), and stomach (D, G), was identified (white silver grains) using radioactive in situ hybridization analysis. CrebH mRNA was present in the hepatocytes (h, arrow) of the liver, the epithelial cells of the villi (v,arrow), but not the crypts (c, white arrows; a yellow dashed line demarcates the villi/crypt border) of the small intestine, and in the surface lining cells (sl) of the stomach. H&E-stained bright field images (B, C, D) and corresponding dark field images (E, F, G) are presented.
CrebH is expressed in the epithelial cells of the adult liver, pyloric stomach, and small intestine
The expression profile of CrebH mRNA in adult mouse tissues was next compared to that of Hnf4α. RT-PCR analysis was performed using cDNA derived from a spectrum of adult mouse tissues (Fig. 2a). Hnf4α mRNA was identified in a variety of adult epithelial tissues including the liver, kidney, and gastrointestinal tract as described previously (13-15). Like Hnf4α, CrebH mRNA was identified in the liver, pyloric stomach, duodenum, and ileum; however, in contrast to Hnf4α CrebH mRNA was not identified in the kidney or colon. In sum, all tissues that expressed CrebH also expressed Hnf4α mRNA; although, not all Hnf4α positive tissues expressed CrebH.
We next performed in situ hybridization to identify the specific cell types that expressed CrebH mRNA. Tissues lacking CrebH mRNA, based on RT-PCR analysis, such as the kidney showed no hybridization above background and therefore acted as convenient negative controls (data not shown). CrebH mRNA was detected in the hepatocytes of the adult liver (Fig. 2b,e) as well as in the epithelium of the villi, but not the crypts, of the small intestine (Fig. 2c.f). CrebH transcripts were also identified in the surface epithelial cells of the pyloric stomach (Fig. 2d,g), but not in the glands.
HNF4α regulates transcription through an HNF4α-binding site within the putative transcriptional regulatory region of the CrebH gene
The expression studies described above demonstrate that following specification of the hepatic cell lineage, CrebH is continuously expressed in the hepatic cells throughout liver development in a manner indistinguishable from HNF4α raising the possibility that CrebH is a direct transcriptional target of HNF4α. To identify potential DNA sequences that could facilitate HNF4α-mediated expression of CrebH, we therefore examined 27.5kb of CrebH genomic DNA sequence, including sequences extending 10kb upstream of exon 1, for the presence of any of 215 known HNF4α binding sites using an HNF4α motif finder generated by Sladek and colleagues (http://www.sladeklab.ucr.edu/links.html). This analysis identified a single potential HNF4α binding site (H4.77) lying 3.7kb upstream of CrebH exon 1 (Fig. 3a). The ability of HNF4α to bind the above sequence was confirmed using electrophoretic mobility shift analyses (EMSA) on nuclear extracts from adult liver. Fig. 3b shows that HNF4α protein could be detected in a complex with a well characterized binding site (H4.21;see http://bioinfo.ucr.edu/~ebolotin/h4supp.html) from the Apoc3 gene(14), which could be converted to a slower migrating complex by inclusion of anti-HNF4α antibody. A complex with a similar migration pattern was identified when the same extracts were incubated with the H4.77 HNF4α-binding site from the CrebH gene, but not when extracts were incubated with a FoxA transcription factor binding site from the Ttr gene, which acted as a control for binding specificity. Similar results were obtained when extracts from COS-7 cells expressing exogenous HNF4α, were used (Fig. 3b). Other protein-DNA complexes were also detected in the extracts and likely reflect the binding of additional proteins known to interact with HNF4α binding sites, such as RAR, RXR, and COUP-TF.
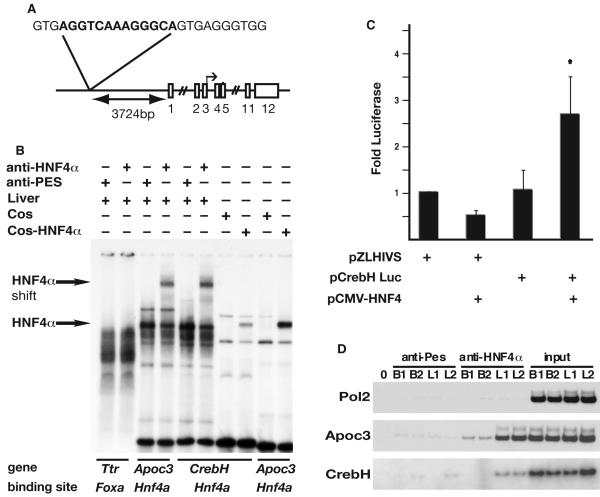
Fig. 3. CrebH is a direct target of HNF4α transcriptional activity.
(A) Schematic showing the genomic location and sequence of the identified HNF4α binding site relative to CrebH (exons shown as boxes). (B) The ability of HNF4α protein to bind the putative HNF4α-binding site was confirmed by EMSA. Radiolabeled oligonucleotides representing binding sites were incubated with liver nuclear extracts in the presence of anti-HNF4α antibody, which resulted in a retarded migration of HNF4α-bound complexes (arrows), or anti-Pes1 antibody (negative control). Alternatively, EMSAs were performed using nuclear extracts from COS-7 cells or COS-7 cells expressing HNF4α. A previously described HNF4α binding site in the Apolipoprotein c3 (Apoc3) promoter served as a positive control and a Foxa (Hnf3) binding site within the Transthyretin (Ttr) promoter served as a negative control. (C) 293T cells were transfected with plasmids in which expression of luciferase was driven by the HIV basal promoter (pZLHIVS) or in addition a 207bp fragment from the CrebH gene that contained the HNF4α binding site (pCrebH-Luc1) in the presence or absence of exogenously expressed HNF4α. Luciferase levels from five independent experiments are presented as fold difference relative to cells transfected with pZLHIVS alone. Significance was determined by Student’s t-test (p<0.05). D) ChIP analyses were performed on chromatin extracted from two independent E18.5 livers (L1, L2) or brains (B1, B2), which acted as a negative control tissue that does not express HNF4α. Chromatin was precipitated using anti-HNF4α or anti-Pes1 (negative control), and specific primers were used to amplify input chromatin or chromatin precipitated from the Pol2 promoter (negative control), Apoc3 promoter (positive control), or CrebH.
The ability of exogenous HNF4α to activate transcription via the HNF4α binding site within the putative CrebH promoter region was studied by transient transfection analyses in 293T cells that do not express endogenous HNF4α. Fig. 3c shows that luciferase levels measured in 293T cells transfected with a reporter plasmid containing only the HIV basal promoter to regulate transcription of the luciferase reporter gene (pHIV-Luc) was not affected by additionally introducing exogenously-expressed HNF4α. Similar results were obtained when 293T cells were transfected with the same reporter plasmid containing an additional 123bp element from CrebH that included the HNF4α-binding site (pCrebH-Luc). However, introduction of exogenously-expressed HNF4α to pCrebH-Luc transfected 293T cells now resulted in an approximate 2.5-fold induction (Student’s t-test, p≤0.05) of luciferase activity compared to controls demonstrating that HNF4α can activate transcription through this element of the CrebH gene.
We next addressed whether HNF4α could occupy the identified binding site within the endogenous CrebH gene in fetal livers using chromatin immunoprecipitation (ChIP) analyses. Fig. 3d shows that, in contrast to sequences from the Pol2 gene that do not contain HNF4α binding sites, a known HNF4α binding site within the Apoc3 promoter (H4.21) as well as the HNF4α binding site within the CrebH gene could be precipitated from chromatin isolated from fetal livers using an antibody that specifically recognizes HNF4α. Importantly, the levels of products identified when precipitations were performed on brain extracts, which lack HNF4α, or when liver extracts were precipitated with an unrelated antibody (anti-Pes) were low to undetectable. Cumulatively, these data demonstrate that the CrebH gene contains an HNF4α recognition element that is occupied by HNF4α in fetal livers.
HNF4α is essential for expression of CrebH in the fetal liver but dispensable for expression in the small intestine
To definitively determine whether CrebH expression is dependent on HNF4α, we generated E18.5 embryos in which HNF4α was specifically deleted in differentiating hepatocytes (Hnf4αloxP/loxPAlfp.cre)(3, 16), colonic epithelial cells (Hnf4αloxP/loxPFoxA3.cre)(17), or small intestinal epithelial cells (Hnf4αloxP/loxPVillin.cre), and measured CrebH levels by RT-PCR. Fig. 4 shows that although CrebH expression was detected in two different control livers, it was not identified in HNF4α-null livers under identical conditions. As expected, CrebH was also undetectable in colons regardless of the presence or absence of HNF4α. Somewhat surprisingly, and in contrast to the liver, CrebH mRNA was detected in both control and HNF4α-null small intestines. We therefore conclude that HNF4α is dispensable for CrebH expression in the gastrointestinal tract but is essential for CrebH expression in the liver.
Fig. 4. HNF4α is essential for expression of CrebH in the liver, but is dispensable for expression in the small intestine.
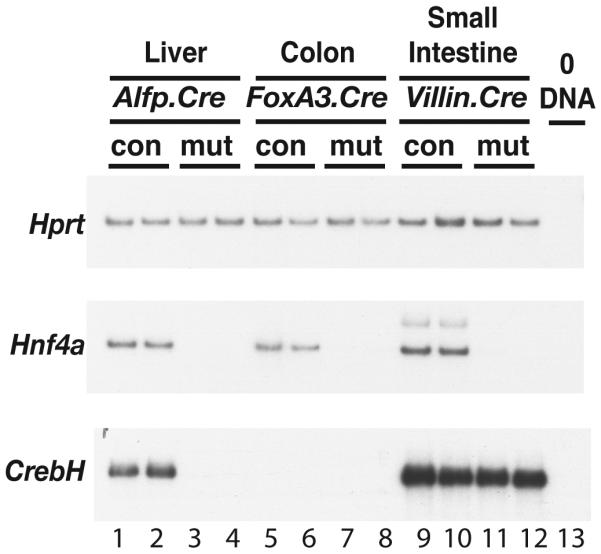
RT-PCR analyses of Hnf4α and CrebH mRNA were performed on RNA isolated from liver (lanes 1-4), colon (lanes 5-8), or small intestine (lanes 9-13), that had been collected from Hnf4αloxP/+ (con) or Hnf4αloxP/loxP (mut) mice that expressed Cre recombinase either in the hepatocytes (Alfp.Cre; lanes 1-4)), colonic epithelial cells (foxa3.Cre; lanes 5-8), or small intestinal epithelial cells (Villin.Cre; lanes 9-13), respectively. Amplification of Hprt was used as a loading control and omitting DNA template from the reaction (0DNA) served as a negative control.
CrebH is dispensable for hepatogenesis and hepatocyte differentiation
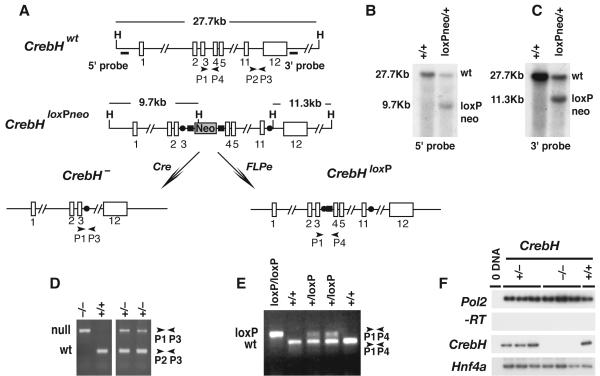
The finding that fetal liver expression of CrebH is dependent on the presence of HNF4α raised the question of whether the absence of CrebH could account for any aspect of the phenotype associated with Hnf4α-/- fetal livers. To test this we generated two strains of mice, one of which harbors a null allele of CrebH (CrebH-/-) and the other a CrebH allele that could be conditionally disrupted by expression of Cre recombinase (CrebHloxP/loxP). Details of the targeting strategy are shown in Fig. 5a. Briefly, ES cells (CrebH+/loxPNeo) were generated that contained a single loxP site between exons 11 and 12 of CrebH and a cassette, which was flanked by FRT sites, containing a loxP site lying 5′ to the neomycin phosphotranferase gene between exons 3 and 4 of CrebH. Correct targeting of the CrebH locus was confirmed by Southern blot analyses of genomic DNA (Fig. 5b and c) and the altered CrebH allele was successfully transmitted through the germline to generate CrebH+/loxPNeo mice. We finally generated mice harboring a null allele of CrebH (CrebH+/-) by mating CrebH+/loxPNeo mice with an EIIa-Cre transgenic mouse (B6.FVB-Tg(EIIa-cre)C5379Lmgd/J), which resulted in Cre-mediated recombination between loxP sites in the germline (18) (Fig. 5 a and d). We also produced mice containing a conditionally null CrebH allele (CrebH+/loxP) by mating CrebH+/loxPNeo mice with mice expressing Flp recombinase from the human ßactin gene promoter (B6;SJL-Tg(ACTFLPe)9205Dym/J) (19) (Fig. 5 a and e).
Fig. 5. Generation of mice harboring conditional and null alleles of CrebH.
A) Diagram showing the targeting strategy used to generate a CrebHloxPneo allele as well as the alleles CrebH-, in response to Cre recombinase activity, and CrebHloxP, in response to Flpe recombinase activity. The position of HindIII restriction endonuclease recognition sequences (H), a neomycin phosphotransferase cassette (Neo), loxP (solid circles) and Frt (solid rectangles) sites, oligonucleotide PCR primers, and Southern blot probes are shown relative to exons (numbered open rectangles). B) Autoradiograph of a Southern blot of HindIII-digested ES cell genomic DNA hybridized to 5′ (left) and 3′ (right) probes, which identify a 27.7kb wild type CrebH (wt) fragment and 9.7kb and 11.3kb CrebHloxPneo (loxPneo) fragments, respectively. D, E) PCR analyses of ear-punch genomic DNA from CrebH+/+, CrebH+/-, CrebH-/-, CrebH+/loxP, and CrebHloxP/loxP mice using primers denoted in A). Sizes of CrebH+, (wt), CrebH- (null), and CrebHloxP (loxP) amplicons are indicated in base pairs. F) RT-PCR analyses of RNA extracted from the individual livers of CrebH+/+(+/+), CrebH+/- (+/-), and CrebH-/- (-/-) mice using primers that identify CrebH, Pol2 (input RNA control), and Hnf4α (hepatocyte control) mRNA. The omission of reverse transcriptase (-RT) and template (0 DNA) ensured the absence of contaminating DNA.
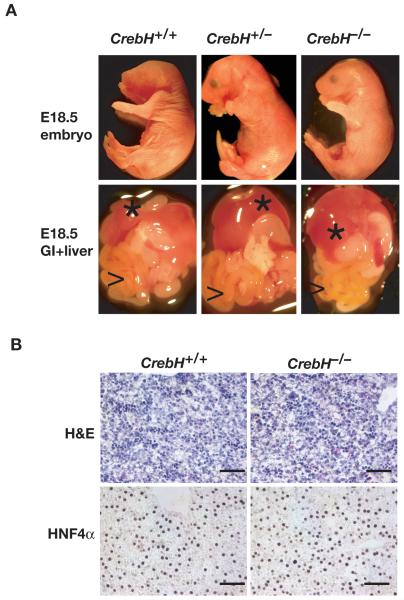
If CrebH were essential for a central aspect of hepatogenesis we predicted that CrebH-/- embryos would die during late gestation stages. However, crosses of CrebH+/- mice yielded CrebH-/- offspring at the expected Mendelian ratio as determined by PCR analyses of genomic DNA isolated from 162 weanlings. RT-PCR analyses identified CrebH mRNA in control livers but not in CrebH-/- livers (Fig. 5f), which is consistent with a loss of CrebH activity in the mutant mice. CrebH-/- mice were found to be long-lived, fecund, and apparently healthy. Examination of E18.5 embryos revealed no obvious difference between control and mutant embryos (Fig. 6a) and the overall anatomy of the liver and gastrointestinal tract of CrebH-/- embryos appeared to be normal. H&E histological staining and HNF4α immunohistochemistry performed on sections through E18.5 CrebH-/- livers found them to be indistinguishable from sections through control livers (Fig. 6b). Finally, oligonucleotide array analyses revealed that mRNA levels were comparable between control and mutant embryos (not shown). Cumulatively, these data demonstrate that CrebH is dispensable for hepatogenesis and hepatocyte differentiation in the mouse.
Fig. 6. CrebH is not essential for development of the liver.
A) Micrographs of viscera (lower panels) showing liver (*) and G.I. tract (>) dissected from E18.5 CrebH+/+, CrebH+/-, and CrebH-/- embryos (upper panels). B) Micrographs of sections through CrebH+/+ and CrebH-/- E18.5 livers stained with hematoxylin and eosin (H&E) or for the presence of HNF4α using immunohistochemistry. Scale bar=100μM.
Loss of CrebH results in reduced expression of acute phase response proteins induced by tunicamycin
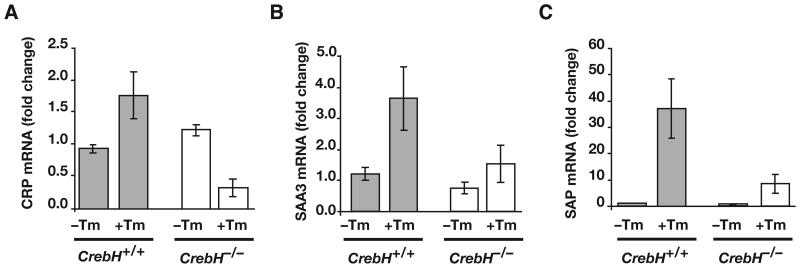
CrebH is a member of the CREB/ATF family of bZip transcription factors that contains a transmembrane domain that directs its localization to the endoplasmic reticulum (11, 20). Recent work has shown that ER-stress results in proteolytic cleavage of CrebH, allowing the N-terminus to translocate to the nucleus where it can activate transcription of target genes including those involved in the acute phase response (20, 21). We therefore examined whether the expression of acute phase genes in response to treatment with tunicamycin, which induces ER stress by blocking glycosylation and protein folding, was affected in CrebH-/- mice. Fig. 7 shows, that expression of mRNAs encoding the acute phase response proteins C-Reactive Protein (CRP), Serum Amyloid P-Component (SAP), Serum Amyloid A3 (SAA3) was robustly induced in CrebH+/+ mice in response to treatment with tunicamycin as expected. However, in contrast to control animals, when CrebH-/- mice were treated with tunicamycin expression of each of these acute phase mRNAs was severely reduced. Similar results were obtained for both liver and serum SAP protein levels (Fig. 8 published as supplemental material). These results confirm that CrebH is an important component of the systemic inflammatory response to ER-stress. Moreover, they imply an indirect role for HNF4α in this response by mediating CrebH expression.
Fig. 7. Response to tunicamycin-induced endoplasmic reticulum stress is reduced by loss of CrebH.
CrebH+/+ (grey bars) and CrebH-/- (white bars) mice were given 2μg/gram body weight tunicamycin by intraperitoneal injection. Livers were isolated at 24 hours and processed for qRT-PCR of mRNAs encoding the acute phase proteins CRP, SAA3, and SAP.
Discussion
In the current analyses we have identified CrebH as a direct target of HNF4α. While our data support the view that CrebH expression is strictly dependent upon HNF4α in the liver we also found that CrebH continues to be robustly expressed in HNF4α null small intestines. Moreover, although HNF4α is dispensable for intestinal CrebH expression, ChIP analyses revealed that, like the liver, HNF4α occupied its binding site within the CrebH promoter in intestinal tissue (data not shown). These data suggest that the regulation of CrebH by HNF4α is tissue dependent and importantly that occupancy of a promoter by HNF4α in vivo does not necessarily correlate with transcriptional regulation (5). It is intriguing why CrebH expression is independent of HNF4α in the small intestine, yet strictly dependent upon HNF4α in the liver. One possible explanation could be that an intestinal regulatory element exists within the CrebH promoter that is controlled specifically by intestinal transcription factors thereby obviating any requirement for HNF4α. Alternatively HNF4g, which in contrast to the liver is robustly expressed in the gut, could potentially compensate for loss of HNF4α in Hnf4αloxP/loxPVillin.cre mice. Efforts to address possible redundancy between these two HNF4 family members through the generation of Hnf4g-/- animals are currently underway.
To examine the role of CrebH in liver function we generated CrebH-/- mice that were viable and healthy. Although at the onset of the project very little was known about this member of the CREB/ATF family of transcription factors a number of reports recently emerged raising a potential role for this factor in controlling hepatic function (11, 20-22). Most relevant was the finding by Zhang et al that proinflammatory cytokines or ER stress could activate CrebH through proteolytic cleavage and that depletion of CrebH using shRNA inhibited expression of acute phase proteins (20). In addition, CrebH was shown to transactivate expression of a luciferase reporter gene through the CRP and SAP promoters suggesting that CrebH directly regulates expression of these acute phase genes. We therefore examined expression of mRNAs encoding acute phase response proteins in CrebH-/- mice following treatment with tunicamycin that induces ER stress. We found that expression of CRP, SAP and SAA3 mRNAs, following treatment was severely diminished in CrebH-/- animals compared to controls. While our data confirm an important role for CrebH in the liver, the contribution of CrebH to gut function, particularly in response to ER-induced stress, remains to be determined. However, with the availability of CrebHloxP/loxP mice it should be possible to address this issue in future studies.
Conclusion
These results confirm that CrebH has an important role in a pathway that controls expression of acute phase proteins in response to ER stress. Moreover, since hepatic expression of CrebH is dependent upon HNF4α, these results demonstrate that HNF4α makes an indirect yet crucial contribution toward the induction of a systemic inflammatory response by ER stress.
Materials and Methods
Oligonucleotide array analysis
RNA was isolated from tissues using RNeasy kit (Qiagen) and cRNA probes were prepared following the directions described in Affymetrix Expression Analysis Technical Manual.
RT-PCR
RT-PCR (1) and quantitative real-time RT-PCR (20) was performed as described previously. Primer sequences are available upon request.
In situ hybridization and immunohistochemistry
In situ hybridization was performed as described elsewhere (23). A probe used to detect CrebH was generated by in vitro transcription of a fragment from a CrebH cDNA, which was generated by PCR using primers atcgaattcccatcagccctttcaactcc, atcggatccaggccagcctggtctacaag and subsequently cloned into the EcoR1/BamH1 sites of pBSII-KS. Immunohistochemistry was performed using previously described procedures(3) with antibodies that detect HNF4α (Santa Cruz sc-6556, 1:500).
Chromatin immunoprecipitation and EMSA
Immunoprecipitation of chromatin was performed using the Upstate ChIP Assay Kit (Upstate #17-295) as described previously(4) and EMSA was performed as described elsewhere (17).
Analysis of Luciferase expression
DNA was introduced into 293T cells using Lipofectamine PLUS™ reagent (Invitrogen). Cell extracts were collected 48hrs after transfection and processed using a Luciferase Assay Dual Reporter System (Promega). Each experiment was performed in triplicate and the entire experiment was repeated on five separate occasions and data were combined. Statistical significance was determined using Student’s t-test with p≤0.05 considered significant.
Animals and ES cells
The Medical College of Wisconsin or the University of Michigan Medical Center IACUC committees approved all animal experiments and procedures. The generation of Hnf4α+/- (Hnf4αtm1Dnl), Hnf4αloxP/loxP (Hnf4αtm1Sad) and Foxa3Cre (Tg(Foxa3-cre)1Khk), AlfpCre (Tg(Alb1-cre)1Khk), and VillinCre (Tg(Vil-cre)997Gum) mice has been described previously(3, 16, 17, 24-26). Noon on the day of the appearance of a vaginal plug was considered as 0.5 d.p.c. (days post coitum) and the genotype of all embryos was determined by PCR analysis of genomic DNA. PMSG used in super ovulation was obtained from A.F. Parlow at the National Hormone and Peptide Program.
Supplementary Material
Acknowledgements
Funding for this project was provided by an American Heart Association fellowship to W.G. and Scientist Development Grant to K.Z., National Institutes of Health grants DK66226 and DK55743 to S.A.D. and DK042394, HL052173, and HL057346 to R.J.K., as well as gifts from the Marcus Family. RJK is an Investigator of the Howard Hughes Medical Institute. The authors also Frances Sladek for providing COS-7 cell extracts and help in identifying HNF4α-binding sites.
References
- 1.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 2.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 4.Battle MA, Konopka G, Parviz F, Gaggl AL, Yang C, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci U S A. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian JM, Schibler U. Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- 8.Kou CJ, Mendel DB, Hansen LP, Crabtree GR. Independent regulation of HNF-1α and HNF-1ß by retinoic acid in F9 teratocarcinoma cells. EMBO J. 1991;10:2231–2236. doi: 10.1002/j.1460-2075.1991.tb07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pineda Torra I, Jamshidi Y, Flavell DM, Fruchart JC, Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol Endocrinol. 2002;16:1013–1028. doi: 10.1210/mend.16.5.0833. [DOI] [PubMed] [Google Scholar]
- 10.Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 11.Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, et al. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29:2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan SA, Manova K, Chen WS, Hoodless P, Weinstein DC, Bachvarova RF, Darnell JE., Jr. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. USA. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taraviras S, Monaghan AP, Schutz G, Kelsey G. Characterization of the mouse HNF-4 gene and its expression during mouse embryogenesis. Mech. Dev. 1994;48:67–79. doi: 10.1016/0925-4773(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 14.Sladek FM, Zhong W, Lai E, Darnell JE., Jr. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 15.Drewes T, Senkel S, Holewa B, Ryffel GU. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol. Cell Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parviz F, Li J, Kaestner KH, Duncan SA. Generation of a conditionally null allele of hnf4alpha. Genesis. 2002;32:130–133. doi: 10.1002/gene.10058. [DOI] [PubMed] [Google Scholar]
- 17.Garrison WD, Battle MA, Yang C, Kaestner KH, Sladek FM, Duncan SA. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 20.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 21.Chin KT, Zhou HJ, Wong CM, Lee JM, Chan CP, Qiang BQ, Yuan JG, et al. The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res. 2005;33:1859–1873. doi: 10.1093/nar/gki332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausa FM, Ye H, Lim L, Duncan SA, Costa RH. In situ hybridization with 33P-labeled RNA probes for determination of cellular expression patterns of liver transcription factors in mouse embryos. Methods. 1998;16:29–41. doi: 10.1006/meth.1998.0642. published erratum appears in Methods 1998 Nov;16(3):359-60. [DOI] [PubMed] [Google Scholar]
- 24.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 25.Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 26.Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.