Abstract
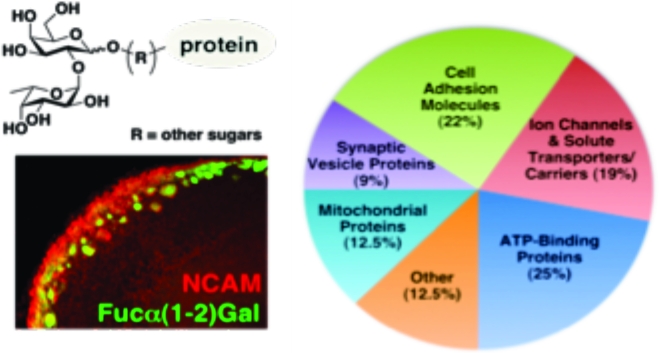
Fucose-α(1−2)-galactose [Fucα(1−2)Gal] sugars have been implicated in the molecular mechanisms that underlie neuronal development, learning, and memory. However, an understanding of their precise roles has been hampered by a lack of information regarding Fucα(1−2)Gal glycoproteins. Here, we report the first proteomic studies of this plasticity-relevant epitope. We identify five classes of putative Fucα(1−2)Gal glycoproteins: cell adhesion molecules, ion channels and solute carriers/transporters, ATP-binding proteins, synaptic vesicle-associated proteins, and mitochondrial proteins. In addition, we show that Fucα(1−2)Gal glycoproteins are enriched in the developing mouse olfactory bulb (OB) and exhibit a distinct spatiotemporal expression that is consistent with the presence of a “glycocode” to help direct olfactory sensory neuron (OSN) axonal pathfinding. We find that expression of Fucα(1−2)Gal sugars in the OB is regulated by the α(1−2)fucosyltransferase FUT1. FUT1-deficient mice exhibit developmental defects, including fewer and smaller glomeruli and a thinner olfactory nerve layer, suggesting that fucosylation contributes to OB development. Our findings significantly expand the number of Fucα(1−2)Gal glycoproteins and provide new insights into the molecular mechanisms by which fucosyl sugars contribute to neuronal processes.
Fucose-α(1−2)-galactose [Fucα(1−2)Gal], a terminal sugar found on N- and O-linked glycoproteins, has been implicated in cognitive processes such as learning and memory. Both task-specific learning and long-term potentiation (LTP), an electrophysiological model of learning and memory, have been shown to induce protein fucosylation in hippocampal neurons (1). Moreover, injection of exogenous l-fucose (l-Fuc) or 2′-fucosyllactose enhanced LTP in hippocampal slices and in vivo (2,3). Conversely, inhibition of Fucα(1−2)Gal linkages on proteins by incorporation of 2-deoxy-d-galactose (2-dGal) into glycans caused reversible amnesia in animals and interfered with the maintenance of LTP (4−7).
In addition to modulating synaptic plasticity, Fucα(1−2)Gal may play important roles in regulating neurite outgrowth and neuronal morphology. Delayed synapse formation and stunted neurite outgrowth were observed in hippocampal cultures treated with 2-dGal (8,9). Fucα(1−2)Gal sugars displayed on multivalent polyacrylamide polymers were found to stimulate the outgrowth of hippocampal neurons, whereas other sugars such as l-Fuc, d-galactose (d-Gal), and fucose-α(1−3)-N-acetylglucosamine [Fucα(1−3)GlcNAc] had no effect (8). Furthermore, addition of exogenous lectin Ulex europaeus agglutinin I (UEAI) or that of Lotus tetragonolobus, each of which binds to Fucα(1−2)Gal moieties, also enhanced hippocampal neurite outgrowth (8).
Recent reports suggest that Fucα(1−2)Gal glycoproteins are expressed in the adult OB and developing OB (10−13) and may help direct the targeting of OSNs to discrete areas (10,11). The differential expression of specific glycan structures that serve as chemotactic agents would provide a largely uncharacterized mechanism for OSN pathfinding. Previous work has revealed that Fucα(1−2)Gal and N-acetylgalactosamine glycoconjugates show distinct patterns of expression in OSNs of the adult OB, consistent with the notion that a glycocode may contribute to the complex topographical arrangement of OSNs (10). Moreover, mice lacking the α(1−2)fucosyltransferase (FUT1) gene displayed impaired development of the olfactory nerve and glomerular layers of the OB (11).
Despite the involvement of Fucα(1−2)Gal sugars in synaptic plasticity, neurite outgrowth, and development, only one Fucα(1−2)Gal glycoprotein has been well-characterized from the mammalian brain. Synapsin I, a presynaptic phosphoprotein involved in neurotransmitter release and synaptogenesis (14), was identified as a prominent Fucα(1−2)Gal glycoprotein in the adult hippocampus (9). Fucosylation was shown to regulate the stability and turnover of synapsin I, protecting it from proteolytic degradation by the calcium-activated protease calpain. Inhibition of protein fucosylation using 2-dGal reduced synapsin expression and delayed synapse formation in hippocampal cultures (9). The neural cell adhesion molecule (NCAM) has also been suggested to be fucosylated (15−17), although no functional studies have been performed.
Here, we report the first proteomic studies of the plasticity-relevant Fucα(1−2)Gal epitope. Using lectin affinity chromatography (LAC) coupled to mass spectrometry (MS), we identified five classes of putative Fucα(1−2)Gal glycoproteins: cell adhesion molecules, ion channels and solute transporters/carriers, ATP-binding proteins, synaptic vesicle-associated proteins, and mitochondrial proteins. In addition, we provide evidence that fucosylation of NCAM and other glycoproteins by the α(1−2)fucosyltransferase FUT1 contributes to OB development, consistent with the notion of a glycocode for the regulation of OSN targeting. Together, our studies significantly expand the number of Fucα(1−2)Gal glycoproteins from two to more than 30 and suggest new roles for protein fucosylation in mediating neuronal communication and development.
Materials and Methods
Lectin Blotting of Brain Regions
Adult male mice (3−4 months of age) and postnatal day 3 (P3) pups were anesthetized with CO2 and dissected to remove the cerebellum, cortex, hippocampus, hypothalamus, olfactory bulb, striatum, and thalamus. For lectin blotting, dissected tissues were cut into small pieces and placed immediately on ice and then lysed in boiling 1% SDS (5 volumes/weight) with sonication until the mixture was homogeneous. Proteins were resolved via 10% SDS−PAGE and transferred to polyvinylidene fluoride (PVDF, Fisher) membranes. The membranes were blocked with 3% periodated BSA in PBS (9) and then incubated with horseradish peroxidase (HRP)-conjugated UEAI [Sigma; 50 μg/mL in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 (TBST)] for 2 h at room temperature (RT). After being washed (3 × 10 min in TBST), the membranes were developed as described previously (9).
LAC
UEAI lectin conjugated to agarose (Vector Laboratories) or control Protein A conjugated to agarose (Vector Laboratories) was packed into three separate minicolumns [∼333 μL bed volume each (Bio-Rad)] and run in parallel. The resin was equilibrated with 10 column volumes (CV) of lectin binding buffer [100 mM Tris (pH 7.5), 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 0.5% NP-40, and 0.2% sodium deoxycholate supplemented with EDTA-free Complete protease inhibitors (Roche)]. Cell lysate was prepared as follows. The OBs from 30−50 P3 pups were isolated and homogenized in lectin binding buffer by being passed through a 26-gauge needle five times and then sonicated to homogeneity. Samples were clarified by centrifugation at 12000g for 10 min. The total protein concentration of the lysate was determined using the BCA protein assay (Pierce). The lysate (3 mL per column, at 6−10 mg/mL) was bound batchwise with gentle end-over-end mixing at RT for 4 h. The agarose was then allowed to settle, and the flow-through was passed over the column three additional times. The columns were washed with 40 CV of lectin binding buffer, followed by 10 CV of lectin binding buffer lacking detergent. Proteins were eluted in 10 CV of lectin binding buffer lacking detergent and supplemented with 200 mM l-Fuc and protease inhibitors. Protein eluates were concentrated to a volume of 100 μL in 10000 molecular weight cutoff (MWCO) Centricons (Millipore), followed by 10000 MWCO Microcons. Following concentration, samples were boiled with 35 μL of 4× SDS loading dye [200 mM Tris (pH 6.8), 400 mM DTT, 8% SDS, 0.2% bromophenol blue, and 40% glycerol] and loaded onto 10% SDS gels for electrophoresis as described previously (18).
Silver Staining, In-Gel Digestion, and LC−MS Analysis
All silver staining reagents were prepared fresh before they were used. The staining and destaining, in-gel tryptic digests, and peptide extractions were performed as described previously (19). NanoLC−MS of in-gel tryptic digests was performed on a Thermo Fisher LTQ Orbitrap mass spectrometer using a modified vented column setup and data-dependent scanning (20). Samples were loaded onto a 360 μm × 100 μm precolumn (2 cm, 5 μm Monitor C18) and desalted before the precolumn was placed in-line with the analytical column. Peptides were then eluted with a linear gradient from 0 to 40% B over 30 min (A, 0.1 M aqueous HOAc; B, 0.1 M HOAc in CH3CN), with a flow rate of approximately 250 nL/min, and using a 360 μm × 75 μm self-packed column with an integrated electrospray emitter (10 cm, 5 μm Monitor C18). For data-dependent experiments, the mass spectrometer was programmed to record a full-scan ESI mass spectrum (m/z 650−2000, ions detected in the Orbitrap mass analyzer with a resolution set to 100000), followed by five data-dependent MS/MS scans in the ion trap (relative collision energy of 35%, 3.5 Da isolation window). Dynamic exclusion parameters were set as follows: repeat count = 1, repeat duration = 15 s, and exclusion duration = 30 s.
MS/MS spectra were searched against a mouse subset of the European Bioinformatics Institute-International Protein Index (EBI-IPI) database (downloaded August 1, 2007), with an appended reversed database using Sequest 3.0. A fixed modification of Cys (+57), a variable modification of Met (+16), and trypsin cleavage were specified. Search results were compiled and filtered in Scaffold 1.0 (Proteome Software, Inc., Portland, OR). A protein identification was accepted if a minimum of five unique peptides matched to the protein, which corresponded to a ≥99% probability of a correct identification (see Table 1). Although a peptide tolerance of 3 Da was used in the search, the mass accuracy of the precursor ion scan for each identified peptide was manually verified as being within 20 ppm of the theoretical value. Proteins were considered nonspecific if any corresponding peptides were isolated from the control column or from FUT1-deficient mice. Cytosolic proteins identified were removed from the data set because they are not likely to be glycosylated. As the specificity of FUT1 is not known, the list of identified proteins could not be filtered for the presence of a FUT1 consensus sequence. Note that LAC may also result in the isolation of proteins that associate with Fucα(1−2)Gal glycoproteins, which may also be of interest biologically but are themselves not directly glycosylated. Thus, further validation of individual proteins of interest by immunoprecipitation (see Figure 2B) and other methods is recommended to confirm the presence of the Fucα(1−2)Gal epitope.
Table 1. Fucα(1−2)Gal Glycoproteins Identified from the Mammalian Olfactory Bulb.
| protein | function | accession no. | MW (Da) | peptide no. | sequence coverage (%) |
|---|---|---|---|---|---|
| Cell Adhesion Molecules | |||||
| NCAM1 | cell adhesion, axonal outgrowth, and fasciculation | IPI00831465.1 | 93474 | 21 | 33.5 |
| IgSF3 | cell adhesion and neuronal fasciculation | IPI00420589.2 | 134605 | 21 | 18.6 |
| contactin-1 precursor | cell adhesion and neuronal fasciculation | IPI00123058.1 | 113372 | 10 | 10.2 |
| NCAM L1 | cell adhesion and neuronal fasciculation | IPI00406778.3, IPI00785371.1, IPI00831568.1 | 140413 | 9 | 10.7 |
| OCAM (NCAM2) | cell adhesion and neuronal fasciculation | IPI00127556.1, IPI00322617.1 | 93187 | 7 | 9.0 |
| NRCAM | cell adhesion and neuronal fasciculation | IPI00120564.5, IPI00338880.3, IPI00395042.1, IPI00403536.1 | 122736 | 7 | 6.3 |
| kirrel2 | cell adhesion and neuronal fasciculation | IPI00471420.1, IPI00623939.2 | 72754 | 5 | 10.3 |
| Ion Channels and Solute Transporters/ Carrier Proteins | |||||
| slc12a2 | chloride transport | IPI00135324.2 | 130654 | 15 | 14.9 |
| cacna2d1 | calcium channel | IPI00230013.3, IPI00319970.1, IPI00407868.1, IPI00410982.1, IPI00626793.3 | 122505 | 13 | 13.0 |
| aralar1 | Asp, Glu exchanger | IPI00308162.3 | 74553 | 12 | 16.7 |
| slc3a2 | cationic amino acid transporter | IPI00114641.2 | 58805 | 8 | 16.7 |
| slc7a5 | cationic amino acid transporter | IPI00331577.3 | 55856 | 5 | 8.6 |
| VDAC3 | voltage-dependent anion channel | IPI00122548.3, IPI00762642.1 | 30867 | 5 | 16.2 |
| ATP-Binding Proteins/ ATP Synthase | |||||
| Na+/K+-ATPase α1 | maintenance of the electrochemical gradient | IPI00311682.5 | 112967 | 34 | 35.4 |
| ATP synthase α subunit | ATP synthesis | IPI00130280.1 | 59736 | 11 | 23.7 |
| Na+/K+-ATPase β1 | maintenance of the electrochemical gradient | IPI00121550.2 | 35771 | 6 | 25.2 |
| vacuolar ATP synthase catalytic subunit A | ATP synthesis | IPI00407692.3 | 68309 | 6 | 13.1 |
| ATP synthase γ chain | ATP synthesis | IPI00313475.1, IPI00750074.1, IPI00751391.1, IPI00775853.1, IPI00776084.1, IPI00776275.1 | 30239 | 6 | 24.1 |
| ATP synthase B chain | ATP synthesis | IPI00341282.2 | 28931 | 6 | 19.9 |
| Na+/K+-ATPase α3 | maintenance of the electrochemical gradient | IPI00122048.2 | 111676 | 6 | 18.3 |
| ATP-binding cassette, subfamily C, member 9 | efflux of endogenous and xenobiotic molecules | IPI00111686.5 | 148816 | 5 | 3.4 |
| Synaptic Vesicle Proteins | |||||
| munc18 (syntaxin-binding protein 1) | synaptic vesicle cycling | IPI00415402.3 | 67553 | 13 | 23.6 |
| synaptotagmin-1 | synaptic vesicle cycling | IPI00129618.1, IPI00750142.1 | 47400 | 9 | 24.0 |
| NSF vesicle-fusing ATPase | synaptic vesicle cycling | IPI00656325.2 | 82598 | 8 | 11.7 |
| Mitochondrial Proteins | |||||
| prohibitin-2 | protein folding | IPI00321718.4 | 33279 | 7 | 24.4 |
| prohibitin | protein folding | IPI00133440.1 | 29802 | 7 | 29.4 |
| ubiquinol−cytochrome c reductase complex core protein 2 | enzyme | IPI00119138.1 | 48218 | 6 | 19.0 |
| mitochondrial inner membrane protein isoform 1 | unknown | IPI00228150.1, IPI00381412.1, IPI00554845.1 | 83883 | 6 | 11.1 |
| Other | |||||
| plexin B2 | cell signaling receptor | IPI00405742.6, IPI00666301.1, IPI00752396.1 | 206212 | 8 | 4.2 |
| LINGO1 | cell signaling coreceptor | IPI00134200.1, IPI00750473.1 | 69085 | 6 | 10.3 |
| MARCKS | cell signaling | IPI00229534.5 | 29643 | 5 | 17.2 |
| insulin-like growth factor 1 receptor | cell signaling receptor | IPI00120225.1 | 155772 | 5 | 3.1 |
Proteins from C57BL/6 mice were captured by UEAI affinity chromatography in three separate experiments.
The number of sequence unique, full tryptic peptide identifications returned from a reversed database searching strategy. See for procedures and analysis criteria.
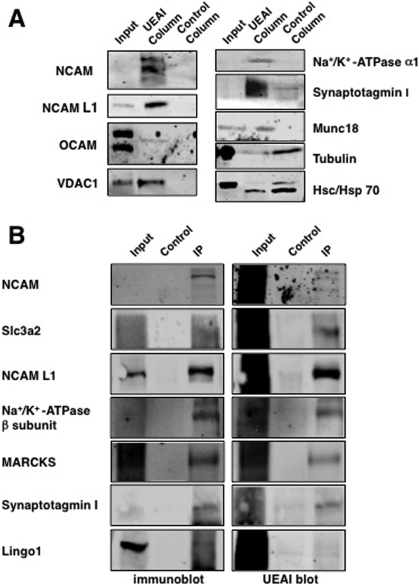
Figure 2.
Confirmation of the major classes of Fucα(1−2)Gal glycoproteins by immunoblotting and immunoprecipitation. (A) Eluates from the UEAI affinity column and the control agarose column were analyzed by Western blotting using antibodies against the indicated proteins. OCAM, NCAM, NCAM L1, synaptotagmin I, munc18, VDAC1, and the Na+/K+-ATPase α1 subunit were specifically captured by UEAI LAC, whereas tubulin and hsc/hsp70 were nonspecifically captured. (B) Confirmation of the Fucα(1−2)Gal moiety on specific proteins by immunoprecipitation and protein detection by Western blotting (left) or sugar detection by UEAI lectin blotting (right).
Immunoblotting To Confirm Proteome Hits
To further validate putative proteome hits, proteins were isolated by lectin affinity chromatography as described above. Lysates from the UEAI column and the control column were resolved on Bis-Tris 4 to 12% NuPage gradient gels (Invitrogen) according to the manufacturer’s protocol in MOPS running buffer. Gels were transferred to PVDF, and the membranes were incubated with infrared blocking buffer (Rockland Immunochemicals) for 1 h at RT. The following primary antibodies were incubated with the membranes overnight at 4 °C in TBST: mouse anti-tubulin (1:50000, Sigma), mouse anti-hsc/hsp70 (1:2000, Stressgen), rabbit anti-munc18 (1:2000, Synaptic Systems), goat anti-VDAC1 (1:100, Santa Cruz), mouse α5 anti-Na+/K+-ATPase α subunits (5 μg/mL, Iowa Hybridoma Bank), mouse anti-OCAM (1:100, R&D Systems), mouse 5e anti-NCAM (10 μg/mL, Iowa Hybridoma Bank), mouse ASCS4 anti-NCAM L1 (2 μg/mL, Iowa Hybridoma Bank), and rabbit anti-synaptotagmin I (1:2000, Synaptic Systems). After the incubation, blots were washed for 3 × 10 min in TBST and then incubated with secondary antibodies for 1 h (1:5000 in TBST with 0.02% SDS). The following secondary antibodies were used: Alexa 680-conjugated donkey anti-mouse (Molecular Probes), Alexa 800-conjugated donkey anti-rabbit (Rockland Immunochemicals), and Alexa 680-conjugated donkey anti-goat (Molecular Probes). Membranes were washed for 3 × 10 min in TBST and imaged using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunoprecipitation To Confirm Proteome Hits
Embryonic rat OB tissue was homogenized in 1% SDS and neutralized by the addition of NETFD [50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM EDTA, 50 mM NaF, and 6% NP-40 supplemented with protease inhibitors] as described previously (9). NCAM was immunoprecipitated using an anti-NCAM antibody (15 μg, Chemicon), and a control experiment was performed under similar conditions in the absence of antibody. Lysates were incubated with the antibody for 3 h at 4 °C with end-over-end mixing; protein A/G Sepharose (Pierce) was added, and the lysates were incubated for an additional 3 h. Immunoprecipitates were washed three times with binding buffer (SDS/NETFD) and eluted with 4× SDS loading dye for SDS−PAGE analysis. The following proteins were immunoprecipitated from OB lysates as described previously: NCAM L1 (21), Slc3a2 (22), Na+/K+-ATPase β1 subunit (23), MARCKS (24), synaptotagmin 1 (25), and LINGO1 (26).
Immunohistochemistry and Quantification of Glomeruli
The brains of P3 mouse pups were removed and immersion-fixed overnight in 4% paraformaldehyde in PBS at 4 °C. The following day, the solution was replaced with an ice-cold solution of 15% sucrose in PBS at 4 °C until the brains sank, followed by 30% sucrose in PBS. The brain tissue was mounted in OCT medium (Tissue Tek) and frozen in a dry ice/MeOH bath. Frozen brains were stored at −80 °C until they were processed for sectioning. Fixed tissues were cryogenically sliced on a Leica CM1800 cryostat in coronal or sagittal sections (20 μm sections for P3 pups and 50 μm sections for adult brain). Sections were dried at 37 °C for 20 min and then blocked in 10% donkey serum and 0.3% Triton X-100 in PBS for 1 h at RT. Sections were incubated with a mouse anti-NCAM antibody (1:100 in 2% donkey serum and 0.1% Triton X-100 in PBS, Sigma), overnight at 4 °C, followed by incubation for 2 h at RT. Sections were washed three times in PBS for 10 min at RT and then incubated with an Alexa 568-conjugated goat anti-mouse secondary antibody (1:500 in 10% donkey serum and 0.1% Triton X-100 in PBS, Molecular Probes) and with UEAI conjugated to fluorescein (50 μg/mL in PBS, Sigma) for 1 h at 37 °C. After being stained, slices were covered with Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories) and a coverslip. Glomeruli were quantified by determining the total number of glomeruli per coronal section labeled by NCAM staining or UEAI staining or surrounded by DAPI nuclear staining for three to six animals. Glomeruli in anterior sections were counted from coronal slices approximately 50−100 μm from the tip of the OB, and posterior sections were counted from sections approximately 50−100 μm rostral to the AOB. The thickness of the ONL was determined by measuring the distance midway between the most ventral point and the medial axis. The thickness was measured in Photoshop CS4, and statistical analysis was performed using the Scheffe test in Kaleidagraph.
Antibody Purification
Ascites (500 μL) containing mouse monoclonal antibodies specific for the α5 Na+/K+-ATPase α subunit, 5e NCAM, and ASCS4 NCAM L1 were purified over Immunopure Protein A agarose columns (300 μL) following the manufacturer’s protocol (Pierce). Eluted antibodies were dialyzed into PBS, and the antibody concentration was adjusted to 1 mg/mL.
Results
Fucα(1−2)Gal Glycoproteins Are Enriched in the Developing OB
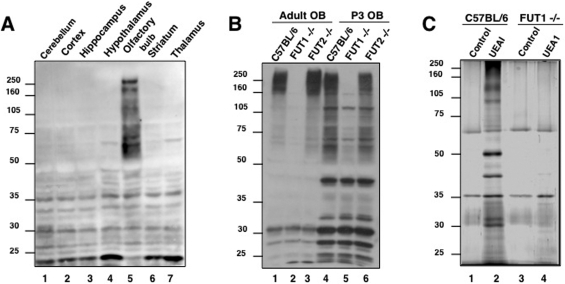
LAC/MS has been extensively used to identify glycosylation-specific subproteomes and provides a rapid enrichment approach for the identification of glycoproteins (27−31). However, the low abundance of fucosylated sugars relative to other glycans in the brain has challenged efforts to identify Fucα(1−2)Gal glycoproteins. To facilitate their capture and identification, we examined the expression levels of Fucα(1−2)Gal in different brain regions. Strong expression of Fucα(1−2)Gal glycans was observed in the OB of adult rats (Figure 1A), as detected using the Fucα(1−2)Gal-specific lectin UEAI (32,33). By comparison, Fucα(1−2)Gal expression levels were significantly lower in the cerebellum, cortex, hippocampus, hypothalamus, striatum, and thalamus. Multiple proteins ranging in size from 50 to 250 kDa were observed specifically in the OB, while several lower-molecular mass proteins between 20 and 50 kDa were detected in all brain regions.
Figure 1.
Fucα(1−2)Gal glycoproteins are enriched in mammalian OB, and their expression is regulated by FUT1. (A) Fucα(1−2)Gal glycoproteins are highly expressed in the adult rat OB relative to other brain regions. (B) Comparison of glycoproteins present in wild-type C57BL/6, FUT1-deficient, and FUT2-deficient mouse OBs from adult and P3 mouse brain tissue. Fucα(1−2)Gal glycoproteins were detected with UEAI conjugated to horseradish peroxidase. An equal amount of protein (200 μg) was loaded in each lane. (C) Silver-stained gel of Fucα(1−2)Gal proteins isolated by UEAI lectin affinity chromatography from the OBs of P3 wild-type C57BL/6 and FUT1-deficient animals.
We next compared the expression of Fucα(1−2)Gal glycoproteins in the developing and mature OBs of mice. Distinct patterns of expression were observed in the OBs of P3 and adult C57BL/6 mice (Figure 1B, lanes 1 and 4). Strong labeling of glycoproteins between 160 and 280 kDa and below 32 kDa was observed in the adult OB, whereas additional labeling of proteins between 50 and 160 kDa and at 45, 38, and 33 kDa was observed in the developing OB. Moreover, the overall expression levels of the Fucα(1−2)Gal epitope were significantly upregulated in the developing P3 OB compared to the adult.
FUT1 Regulates the Fucα(1−2)Gal Proteome of the OB
We next examined whether the expression of Fucα(1−2)Gal sugars in the OB was regulated by FUT1 or FUT2, two known α(1−2)fucosyltransferases (34). FUT1-deficient mice exhibited a significant reduction in the level of Fucα(1−2)Gal expression on glycoproteins in both the developing and adult OB, as detected by UEAI blotting (Figure 1B, lanes 2 and 5). In contrast, deletion of the FUT2 gene had no observable effect on protein fucosylation in the OB (Figure 1B, lanes 3 and 6). Interestingly, several proteins, mostly those of <50 kDa, showed no appreciable change in either FUT1- or FUT2-deficient mice, which suggests the possible existence of an unidentified α(1−2)fucosyltransferase or redundant enzymatic functions. Overall, the results indicate that the Fucα(1−2)Gal modification is primarily regulated by the FUT1 enzyme in the developing and mature OB.
Identification of the Fucα(1−2)Gal Proteome from Mouse Brain
To capture and identify Fucα(1−2)Gal glycoproteins, we exploited UEAI LAC. Studies have reported that UEAI binds specifically to Fucα(1−2)Gal disaccharides, displaying 200-fold greater affinity for the disaccharide than for l-Fuc alone (32,33). On the other hand, recent data from glycan microarrays made available by the Consortium for Functional Glycomics suggest that this difference in binding affinity may be only 3−4-fold. Nonetheless, the glycan microarray analyses have revealed that UEAI binds strongly to Fucα(1−2)Gal-containing glycans and exhibits a preference for glycans containing an additional β(1−4)GlcNAc linked to the reducing end of the Gal residue. Our initial attempts at UEAI LAC led to limited isolation of the desired glycoproteins, which was likely due to the relatively weak carbohydrate binding affinity of the lectin [KA = (1.8 ± 0.8) × 106 M] (33). By performing room-temperature incubations and optimizing the buffer and wash conditions, we developed a method for enhancing the binding of glycoprotein to the lectin column, while minimizing nonspecific interactions (see ). As a control, we used columns containing agarose beads to identify proteins that interacted nonspecifically with the lectin column. Most importantly, we exploited FUT1-deficient mice as a critical validation of the presence of the Fucα(1−2)Gal moiety on the proteins identified because this sugar moiety should be absent in the OB of FUT1-deficient mice.
Using this approach, we observed the selective capture of multiple proteins from the developing OB (Figure 1C, lanes 1 and 2). Very few proteins were isolated from the agarose control columns (lanes 1 and 3) or from FUT1-deficient brain lysates (lane 4). Thirty-three bands from each lane of the wild-type (C57BL/6) control, wild-type UEAI column, and FUT1−/− UEAI column lanes were digested with trypsin and subjected to LC−MS/MS analysis. The data acquisition and subsequent database searching methodologies employed are detailed in . In total, 32 proteins specifically enriched with UEAI (Table 1) were identified on the basis of both a minimum of five unique matching peptides, which corresponds to a ≥99.9% probability of correct protein identification, and their absence from the wild-type control and FUT1−/− UEAI lanes (lanes 1 and 4). Interestingly, the proteins have a broad range of biological roles and can be categorized into five major functional classes: cell adhesion molecules, ion channels and solute carriers/transporters, ATP-binding proteins, synaptic vesicle-associated proteins, and mitochondrial proteins (Table 1).
We further confirmed the identification of proteins from the major classes by immunoblotting eluates from the lectin affinity columns where suitable antibodies were available. Cell adhesion molecules of the immunoglobulin superfamily, including NCAM, NCAM L1, and OCAM (also known as NCAM2), were detected in UEAI column eluates, but not control column eluates, corroborating the mass spectrometry results (Figure 2A). Similarly, voltage-dependent anion channel 1 (VDAC1), the α1 subunit of the Na+/K+-ATPase, and the synaptic vesicle proteins synaptotagmin I and munc18 (syntaxin-binding protein 1) were also confirmed to be captured specifically by UEAI. In contrast, tubulin and hsc/hsp70 were present in both the UEAI and control column eluates, as determined by MS and immunoblotting, confirming that these proteins were nonspecifically captured. As LAC can capture other proteins complexed to the glycoproteins of interest, we also validated the presence of the Fucα(1−2)Gal epitope on specific proteins by immunoprecipitation and subsequent blotting with UEAI. Using this approach, we were able to detect the Fucα(1−2)Gal epitope on most of the proteins where suitable antibodies were available, including NCAM, solute carrier family 3, member 2 (Slc3a2, also known as CD98), NCAM L1, the Na+/K+-ATPase β subunit, myristoylated alanine-rich protein kinase C substrate (MARCKS), and synaptotagmin I (Figure 2B). Interestingly, LINGO1 appears to associate with Fucα(1−2)Gal glycoproteins or is fucosylated only at a low level.
Fucosylation of NCAM in the Developing OB: Evidence for a Glycocode
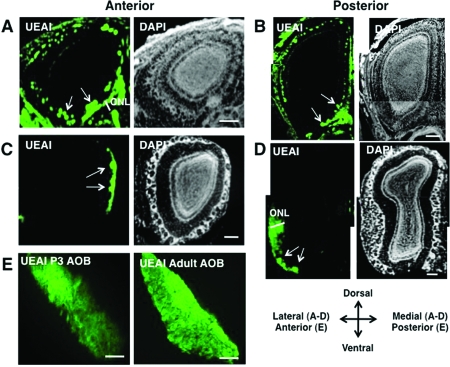
NCAM is a member of the largest functional class of Fucα(1−2)Gal glycoproteins identified (Table 1). To gain insight into the importance of protein fucosylation in regulating NCAM, we compared the expression of both the Fucα(1−2)Gal epitope and NCAM in the mouse OB. Coronal OB sections from developing P3 and adult mice were costained with UEAI, an anti-NCAM antibody, and DAPI to visualize nuclei for glomerular quantification (see for details). Fucα(1−2)Gal sugars were expressed in both the glomerular and olfactory nerve layers (ONL) of the developing and adult OB (Figure 3). We found that Fucα(1−2)Gal exhibited a stronger, more extensive distribution in the developing main OB [MOB (Figure 3A, B)] than in the adult (Figure 3C, D). Only a subset of OSN axons and glomeruli were labeled in the adult MOB, consistent with a previous report (10). In contrast, all faces of the developing MOB exhibited extensive labeling of the ONL and greater numbers of labeled glomeruli. The majority of the glomeruli were UEAI-positive in both the anterior and posterior regions of the developing MOB (94 ± 5% and 80 ± 10% of total glomeruli, respectively), whereas only 11 ± 4% of the total glomeruli were UEAI-positive in the anterior MOB and 7 ± 5% in the posterior MOB of adult animals, suggesting that Fucα(1−2)Gal sugars are significantly enriched in developing glomeruli. Interestingly, the staining was bilaterally symmetrical across multiple animals, indicating a defined spatial organization for Fucα(1−2)Gal.
Figure 3.
Fucα(1−2)Gal sugars are enriched in the developing OB and are present in the ONL and glomerular layers on all faces of the developing OB. (A−D) Coronal sections of the MOB from wild-type P3 mice (A, B) and adult mice (C, D) were labeled with UEAI conjugated to fluorescein and imaged by confocal fluorescence microscopy. Nuclei were stained with DAPI. Extensive staining of the ONL and glomerular layer was observed in the developing MOB, whereas labeling was confined to small clusters of glomeruli in the adult MOB. The arrows indicate the presence of glomeruli in the olfactory bulb. The scale bar is 200 μm. (E) Sagittal sections of the developing AOB were labeled with UEAI conjugated to fluorescein (green). Fucα(1−2)Gal sugars were enriched in the anterior AOB and were present in a gradient from anterior to posterior. The scale bar is 100 μm.
Fucα(1−2)Gal sugars were also highly enriched in both the developing and adult accessory olfactory bulb [AOB (Figure 3E)], a structure involved in the secondary detection of a particular class of chemical signals important for regulating sexual behaviors and detecting pheromones. We detected Fucα(1−2)Gal sugars in both the anterior and posterior regions of the developing AOB, with the strongest staining occurring in the anterior region (Figure 3E). As expected, FUT1-deficient adult and P3 mice exhibited no labeling of the glomerular layer or ONL in either the MOB or AOB, consistent with regulation of the Fucα(1−2)Gal proteome by FUT1 (Figure S1, ).
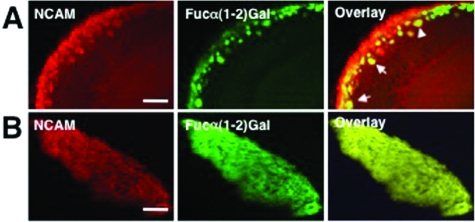
We next compared the colocalization of NCAM with Fucα(1−2)Gal sugars in the developing OB. We observed significant overlap between NCAM and UEAI staining. Approximately 98 ± 5% of NCAM-positive glomeruli in the anterior region and 88 ± 12% of NCAM-positive glomeruli in the posterior region overlapped with UEAI-positive glomeruli. However, NCAM staining only partially colocalized with Fucα(1−2)Gal sugars in the glomerular layer and ONL of the MOB (Figure 4A). The overlap between NCAM and UEAI staining was most prevalent in the dorsal-medial aspect of the developing OB (Figure S2, ). NCAM was also strongly colocalized with Fucα(1−2)Gal sugars in the AOB (Figure 4B), suggesting that OSNs from both the olfactory epithelium and vomeronasal organ contain fucosylated NCAM.
Figure 4.
NCAM partially colocalizes with Fucα(1−2)Gal sugars in the MOB and AOB. (A) NCAM staining partially colocalizes with UEAI staining in the ONL and glomerular layers of the MOB. (B) NCAM staining and UEAI staining show extensive colocalization in the AOB, suggesting that NCAM is strongly fucosylated in this region. Shown are confocal fluorescence microscopy images of coronal OB slices from wild-type P3 mice. Slices were stained with an anti-NCAM antibody (red) and UEAI conjugated to fluorescein (green), and the overlay of the images is colored yellow. The scale bar is 100 μm.
Developmental Defects in FUT1 Knockout (KO) Mice
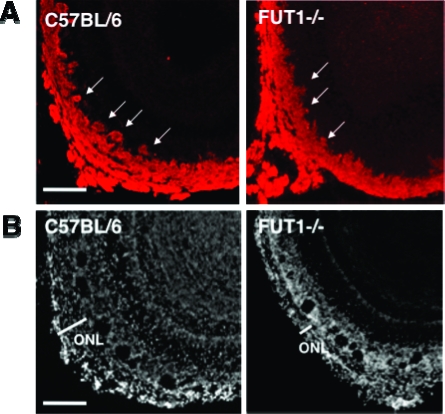
As Fucα(1−2)Gal sugars are abundantly expressed in the developing OB, we investigated whether loss of fucosylation in the FUT1-deficient mice leads to defects in OB development. OB sections from P3 wild-type and FUT1 knockout mice were stained with DAPI. We also costained the sections with an anti-NCAM antibody to probe whether fucosylation might contribute to NCAM function in the OB. Defects in the number and demarcation of glomeruli and in the thickness of the ONL were observed in NCAM-expressing regions of the developing OB (Figure 5A). These deficits were most pronounced in the medial-ventral surface of the posterior MOB. The total number of glomeruli was decreased by 16% (P < 0.05) in the posterior OB of FUT1-deficient versus wild-type C57BL/6 mice (Figure 5B), although no significant decrease in the numbers of glomeruli was found in the anterior region. NCAM expression was localized to the majority of glomeruli (99 ± 3% in the anterior and 96 ± 5% in the posterior regions), and thus, the defects in OB development were strongly associated with NCAM-expressing neurons. In addition, we observed a 24% (P < 0.05) decrease in the thickness of the ONL on the medial-ventral face of the developing FUT1−/− OB (Figure 5B). Interestingly, no obvious defects in the development of OSNs in the AOB were found (data not shown), suggesting that fucosylation may contribute to the development of MOB, but not AOB, topography.
Figure 5.
ONL and glomerular layers of FUT1-deficient mice are defective in areas expressing the Fucα(1−2)Gal glycoprotein NCAM. Coronal OB slices from wild-type C57BL/6 and FUT1-deficient mice were stained with an anti-NCAM antibody (A) or DAPI (B). The arrows point to large glomeruli that are present in wild-type mice but are absent in FUT1 knockout mice. The scale bar is 75 μm.
Discussion
Herein we report the first proteomic studies of the plasticity-relevant Fucα(1−2)Gal epitope. The proteins were identified using an optimized LAC method coupled to MS. Our approach exploited lysate from the developing OB to facilitate enrichment of Fucα(1−2)Gal glycoproteins as well as FUT1-deficient mice to validate identification of the desired glycoproteins. We used UEAI lectin because studies have established that UEAI binds specifically to Fucα(1−2)Gal disaccharides (33) and shows no binding to other sugars, such as d-Fuc, d-Gal, and d-mannose, or l-Fuc present in Fucα(1−3)Gal linkages (32,35). Furthermore, we employed FUT1 knockout mice to confirm the specific identification of Fucα(1−2)Gal glycoproteins because we found that these mice do not synthesize Fucα(1−2)Gal glycans in the OB and therefore the target glycoproteins should be missing from LAC eluates.
In contrast to a previous report that NCAM is the only Fucα(1−2)Gal glycoprotein in the OB (17), our studies suggest the existence of many Fucα(1−2)Gal glycoproteins in the mammalian brain. We identified five major classes of putative Fucα(1−2)Gal glycoproteins, including cell adhesion molecules, ion channels and solute carriers/transporters, ATP-binding proteins, synaptic vesicle-associated proteins, and mitochondrial proteins. We found multiple ATP-binding and mitochondrial proteins, including several subunits of the mitochondrial ATP synthase and Na+/K+-ATPase. Interestingly, the mitochondrial ATP synthase has been reported previously to be N-glycosylated in INS-1 insulinoma cells (36). In addition, the β1 subunit of Na+/K+-ATPase, a protein critical for maintenance of the electrochemical gradient of sodium and potassium ions across the plasma membrane (37), has been shown to contain N-linked fucosylated glycans (38). Although the precise fucose linkage was previously uncharacterized, our studies suggest that the glycans on Na+/K+-ATPase contain the plasticity-relevant Fucα(1−2)Gal epitope.
Cell adhesion molecules represent one of the largest classes of putative Fucα(1−2)Gal glycoproteins identified. These proteins have well-established roles in cell signaling, neuronal development, and synaptic plasticity (39,40). Of the seven proteins identified, all are members of the immunoglobulin superfamily of cell adhesion molecules, suggesting an important functional role for fucosylation in regulating this family. NCAM plays critical roles in axonal fasciculation and neurite outgrowth (40−44) and may contribute to OSN axon guidance (45). Two other cell adhesion molecules identified in this study, kirrel2 and OCAM, have also been reported to play important roles in OB development. Kirrel2 is proposed to participate in the activity-dependent sorting of axons and glomerular formation (46), while knockout studies have implicated OCAM in maintaining the compartmental organization of axodendritic and dendrodendritic synapses within glomeruli (47).
Our discovery of several synaptic proteins, including munc18 and synaptotagmin I, suggests a functional role for the Fucα(1−2)Gal epitope in synaptic signaling. N-Glycosylation has been shown to be important for the targeting of synaptotagmin I to synaptic vesicles (48), suggesting that fucosylation may be involved in this process. Previous studies by our laboratory have demonstrated that fucosylation regulates the expression and turnover of synapsin I, a protein involved in regulating neurotransmitter release and synaptogenesis (9). Our current results suggest that Fucα(1−2)Gal sugars may modify additional synaptic proteins, which may reveal an expanded role for these sugars in modulating synaptic signaling. However, given the potential for lectins to recognize l-Fuc monosaccharides and trisaccharide epitopes containing Fucα(1−2)Gal, further studies will be required on individual proteins identified in this study to confirm the presence of the Fucα(1−2)Gal moiety and elucidate its diverse functional roles.
Here, we demonstrate that Fucα(1−2)Gal glycans are enriched in the developing OB and FUT1 is primarily responsible for controlling their expression. Mice deficient in the FUT1 gene showed a significant reduction in the level of Fucα(1−2)Gal expression in the OB, and fucosylation of the proteins identified herein appeared to be lacking in FUT1 knockout mice. We found that fucosylated glycans were highly localized to OSNs and glomeruli within the OB. As the Fucα(1−2)Gal epitope has been implicated in the regulation of neurite outgrowth and neuronal morphology (8), our results raise the exciting possibility that enhanced fucosylation during development may participate in the outgrowth and pathfinding of OSNs.
Each OSN expresses one of more than 1000 different odorant receptors, and its axon connects onto a topographically fixed position in the OB (49−51). Although the cell bodies of the OSNs are arranged in a mosaic distribution within one of four zones in the mammalian olfactory epithelium, the axons of neurons expressing a single type of odorant receptor converge onto only one to three glomeruli (51−55). The sorting of these axons appears to be controlled by the coordinated spatiotemporal expression of proteins, such as axon guidance cues and their cognate guidance receptors (56). However, these molecules alone are not sufficient to account for the highly specific topographical arrangement of OSNs and glomeruli in the OB. Interestingly, specific glycan structures are also expressed in a distinct spatiotemporal arrangement, and the selective expression of certain glycans has been proposed to direct axonal pathfinding of OSNs (10,11,57,58). Our findings lend support to the notion that a glycocode contributes to neuronal pathfinding in the OB. First, we showed that Fucα(1−2)Gal sugars have a distinct, bilaterally symmetrical expression in the developing OB, indicative of a defined spatial organization for Fucα(1−2)Gal sugars. Second, we found that the overall expression of Fucα(1−2)Gal sugars is significantly upregulated during OB development. A stronger, more extensive distribution of Fucα(1−2)Gal sugars was observed in the developing OB, suggestive of specific temporal regulation of the sugars. The majority of glomeruli in the developing OB expressed Fucα(1−2)Gal sugars, whereas Fucα(1−2)Gal expression was confined to small numbers of glomeruli in the adult OB. Third, we observed fucosylated NCAM in only a subset of NCAM-positive OSNs, indicating that NCAM is likely differentially fucosylated. Differential fucosylation of NCAM and other Fucα(1−2)Gal glycoproteins could provide a novel mechanism for distinguishing specific OSNs and help target them to appropriate glomeruli or regions of the developing OB. Consistent with this notion, NCAM glycoforms containing novel carbohydrates NOC-3 and NOC-4 have been shown to define four subpopulations of OSNs whose axons terminate on specific regions of the rat OB (59). Lastly, we found that deletion of the FUT1 gene had critical consequences for OB development, suggesting key roles for Fucα(1−2)Gal sugars and their associated proteins in OB development. Although additional molecular determinants are likely important, the observed spatiotemporal pattern of Fucα(1−2)Gal expression and the developmental defects of FUT1 knockout mice point to the participation of Fucα(1−2)Gal sugars in the targeting of OSN axons to discrete areas within the OB.
Previous studies have shown that NCAM is glycosylated by several different epitopes, including NOC-3 and -4, HNK-1, and polysialic acid residues. Our results corroborate the existence of multiple glycoforms of NCAM and suggest that another significant glycoform of NCAM carries the Fucα(1−2)Gal epitope. Notably, FUT1-deficient mice exhibited defects in OB development similar to those reported for NCAM-180 knockout mice, including fewer and smaller glomeruli (60). Moreover, we found that the developmental defects in FUT1 knockout mice were strongly associated with NCAM-expressing neurons. Thus, fucosylation of NCAM may play a critical role in the regulation of NCAM function during OB development. Given the ability of Fucα(1−2)Gal glycans to modulate neurite outgrowth and neuronal morphology, one possibility is that the Fucα(1−2)Gal epitope on NCAM may assist in OSN outgrowth and guidance. In the future, fucosylation-defective mutants of NCAM will be analyzed to elucidate further the roles of NCAM fucosylation in neuronal development. Moreover, an important implication of this study is that a large number of Fucα(1−2)Gal glycoproteins exist in the developing OB. As such, additional Fucα(1−2)Gal glycoproteins may also contribute to OB development. The analysis of kirrel, OCAM, and other proteins identified herein will shed light on the relative contributions of various Fucα(1−2)Gal glycoproteins in the process. Finally, although this study focused on the identification of glycoproteins, Fucα(1−2)Gal glycans are also found on glycolipids, revealing another interesting facet of Fucα(1−2)Gal biology that remains to be explored.
In summary, our findings significantly expand the number of known candidate Fucα(1−2)Gal glycoproteins and reveal new insights into the functions of the Fucα(1−2)Gal epitope. We identified glycoproteins involved in cell adhesion, synaptic vesicle cycling, membrane excitability, and synaptic plasticity, suggesting broad functional roles for Fucα(1−2)Gal glycans. In addition, we demonstrate that Fucα(1−2)Gal sugars are highly enriched in the developing OB and display a defined spatiotemporal organization that is regulated by FUT1. Deletion of the FUT1 gene results in defects in glomerular formation that are strongly associated with regions of extensive fucosylation and NCAM expression. Cumulatively, our studies suggest that the regulated fucosylation of NCAM and possibly additional Fucα(1−2)Gal glycoproteins within specific neuronal substructures contribute to axon pathfinding during development.
Acknowledgments
The ASCS4 hybridoma developed by P. H. Patterson, the 5e hybridoma developed by U. Rutishauser, and the α5 hybridoma developed by P. M. Fambrough were obtained from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child and Human Development and maintained by the Department of Biology, The University of Iowa, Iowa City, IA 52242. We thank John Lowe for contributing the FUT1- and FUT2-deficient mice used in this study.
Supporting Information Available
UEAI staining of FUT1−/− MOB and AOB and colocalization of NCAM and UEAI staining in the MOB. This material is available free of charge via the Internet at http://pubs.acs.org.
This research was supported by National Institutes of Health Grants RO1 GM084724-05 (L.C.H.-W.), T32 GM08501 (H.E.M.), and 5T32 GM07616-30S1 (C.K.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Pohle W.; Acosta L.; Ruthrich H.; Krug M.; Matthies H. (1987) Incorporation of [H-3] fucose in rat hippocampal structures after conditioning by perforant path stimulation and after LTP-producing tetanization. Brain Res. 410, 245–256. [DOI] [PubMed] [Google Scholar]
- Matthies H.; Staak S.; Krug M. (1996) Fucose and fucosyllactose enhance in-vitro hippocampal long-term potentiation. Brain Res. 725, 276–280. [DOI] [PubMed] [Google Scholar]
- Krug M.; Wagner M.; Staak S.; Smalla K. H. (1994) Fucose and fucose-containing sugar epitopes enhance hippocampal long-term potentiation in the freely moving rat. Brain Res. 643, 130–135. [DOI] [PubMed] [Google Scholar]
- Tiunova A. A.; Anokhin K. V.; Rose S. P. R. (1998) Two critical periods of protein and glycoprotein synthesis in memory consolidation for visual categorization learning in chicks. Learn. Mem. 4, 401–410. [DOI] [PubMed] [Google Scholar]
- Rose S. P. R.; Jork R. (1987) Long-term-memory formation in chicks is blocked by 2-deoxygalactose, a fucose analog. Behav. Neural Biol. 48, 246–258. [DOI] [PubMed] [Google Scholar]
- Lorenzini C. G. A.; Baldi E.; Bucherelli C.; Sacchetti B.; Tassoni G. (1997) 2-Deoxy-d-galactose effects on passive avoidance memorization in the rat. Neurobiol. Learn. Mem. 68, 317–324. [DOI] [PubMed] [Google Scholar]
- Krug M.; Jork R.; Reymann K.; Wagner M.; Matthies H. (1991) The amnesic substance 2-deoxy-d-galactose suppresses the maintenance of hippocampal LTP. Brain Res. 540, 237–242. [DOI] [PubMed] [Google Scholar]
- Kalovidouris S. A.; Gama C. I.; Lee L. W.; Hsieh-Wilson L. C. (2005) A role for fucose α(1−2) galactose carbohydrates in neuronal growth. J. Am. Chem. Soc. 127, 1340–1341. [DOI] [PubMed] [Google Scholar]
- Murrey H. E.; Gama C. I.; Kalovidouris S. A.; Luo W. I.; Driggers E. M.; Porton B.; Hsieh-Wilson L. C. (2006) Protein fucosylation regulates synapsin la/lb expression and neuronal morphology in primary hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 103, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb B. W.; Treloar H. B.; Klenoff J.; Greer C. A. (2003) Cell surface carbohydrates and glomerular targeting of olfactory sensory neuron axons in the mouse. J. Comp. Neurol. 467, 22–31. [DOI] [PubMed] [Google Scholar]
- St John J. A.; Claxton C.; Robinson M. W.; Yamamoto F.; Domino S. E.; Key B. (2006) Genetic manipulation of blood group carbohydrates alters development and pathfinding of primary sensory axons of the olfactory systems. Dev. Biol. 298, 470–484. [DOI] [PubMed] [Google Scholar]
- Salazar I.; Quinteiro P. S. (2003) Differential development of binding sites for four lectins in the vomeronasal system of juvenile mouse: From the sensory transduction site to the first relay stage. Brain Res. 979, 15–26. [DOI] [PubMed] [Google Scholar]
- Salazar I.; Quinteiro P. S.; Lombardero M.; Cifuentes J. M. (2001) Histochemical identification of carbohydrate moieties in the accessory olfactory bulb of the mouse using a panel of lectins. Chem. Sens. 26, 645–652. [DOI] [PubMed] [Google Scholar]
- Ferreira A.; Rapoport M. (2002) The synapsins: Beyond the regulation of neurotransmitter release. Cell. Mol. Life Sci. 59, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M.; Geyer H.; von der Ohe M.; Gerardy-Schahn R.; Schachner M.; Geyer R. (2003) Localization of defined carbohydrate epitopes in bovine polysialylated NCAM. Biochimie 85, 207–218. [DOI] [PubMed] [Google Scholar]
- Liedtke S.; Geyer H.; Wuhrer M.; Geyer R.; Frank G.; Gerardy-Schahn R.; Zahringer U.; Schachner M. (2001) Characterization of N-glycans from mouse brain neural cell adhesion molecule. Glycobiology 11, 373–384. [DOI] [PubMed] [Google Scholar]
- Pestean A.; Krizbai I.; Bottcher H.; Parducz A.; Joo F.; Wolff J. R. (1995) Identification of the Ulex europaeus agglutinin-I-binding protein as a unique glycoform of the neural cell-adhesion molecule in the olfactory sensory axons of adult-rats. Neurosci. Lett. 195, 117–120. [DOI] [PubMed] [Google Scholar]
- Tai H.-C.; Khidekel N.; Ficarro S. B.; Peters E. C.; Hsieh-Wilson L. C. (2004) Parallel identification of O-GlcNAc-modified proteins from cell lysates. J. Am. Chem. Soc. 126, 10500–10501. [DOI] [PubMed] [Google Scholar]
- Clark P. M.; Dweck J. F.; Mason D. E.; Hart C. R.; Buck S. B.; Peters E. C.; Agnew B. J.; Hsieh-Wilson L. C. (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 130, 11576–11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidekel N.; Ficarro S. B.; Clark P. M.; Bryan M. C.; Swaney D. L.; Rexach J. E.; Sun Y. E.; Coon J. J.; Peters E. C.; Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348. [DOI] [PubMed] [Google Scholar]
- Buhusi M.; Schlatter M. C.; Demyanenko G. P.; Thresher R.; Maness P. F. (2008) L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J. Neurosci. 28, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior A.; Denys A.; Deligny A.; Mazurier J.; Allain F. (2008) Cyclophilin B induces integrin-mediated cell adhesion by a mechanism involving CD98-dependent activation of protein kinase C-δ and p44/42 mitogen-activated protein kinases. Exp. Cell Res. 314, 616–628. [DOI] [PubMed] [Google Scholar]
- Zatyka M.; Ricketts C.; Xavier G. D.; Minton J.; Fenton S.; Hofmann-Thiel S.; Rutter G. A.; Barrett T. G. (2008) Sodium-potassium ATPase β1 subunit is a molecular partner of Wolframin, an endoplasmic reticulum protein involved in ER stress. Hum. Mol. Genet. 17, 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. D.; Lejen T.; Zhang L.; Trifaro J. M. (2001) Chromaffin cell F-actin disassembly and potentiation of catecholamine release in response to protein kinase C activation by phorbol esters is mediated through myristoylated alanine-rich C kinase substrate phosphorylation. J. Biol. Chem. 276, 36757–36763. [DOI] [PubMed] [Google Scholar]
- Musch M. W.; Arvans D. L.; Walsh-Reitz M. M.; Uchiyama K.; Fukuda M.; Chang E. B. (2007) Synaptotagmin I binds intestinal epithelial NHE3 and mediates cAMP-and Ca2+-induced endocytosis by recruitment of AP2 and clathrin. Am. J. Physiol. 292, G1549–G1558. [DOI] [PubMed] [Google Scholar]
- Bai Y.; Markham K.; Chen F. S.; Weerasekera R.; Watts J.; Horne P.; Wakutani Y.; Bagshaw R.; Mathews P. M.; Fraser P. E.; Westaway D.; George-Hyslop P. S.; Schmitt-Ulms G. (2008) The in vivo brain interactome of the amyloid precursor protein. Mol. Cell. Proteomics 7, 15–34. [DOI] [PubMed] [Google Scholar]
- Qiu R. Q.; Regnier F. E. (2005) Use of multidimensional lectin affinity chromatography in differential glycoproteomics. Anal. Chem. 77, 2802–2809. [DOI] [PubMed] [Google Scholar]
- Ghosh D.; Krokhin O.; Antonovici M.; Ens W.; Standing K. G.; Beavis R. C.; Wilkins J. A. (2004) Lectin affinity as an approach to the proteomic analysis of membrane glycoproteins. J. Proteome Res. 3, 841–850. [DOI] [PubMed] [Google Scholar]
- Hagglund P.; Bunkenborg J.; Elortza F.; Jensen O. N.; Roepstorff P. (2004) A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566. [DOI] [PubMed] [Google Scholar]
- Plavina T.; Wakshull E.; Hancock W. S.; Hincapie M. (2007) Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J. Proteome Res. 6, 662–671. [DOI] [PubMed] [Google Scholar]
- Vosseller K.; Trinidad J. C.; Chalkley R. J.; Specht C. G.; Thalhammer A.; Lynn A. J.; Snedecor J. O.; Guan S.; Medzihradszky K. F.; Maltby D. A.; Schoepfer R.; Burlingame A. L. (2006) O-Linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934. [DOI] [PubMed] [Google Scholar]
- Pereira M. E. A.; Kisailus E. C.; Gruezo F.; Kabat E. A. (1978) Immunochemical studies on combining site of blood-group H-specific lectin-1 from Ulex europeus seeds. Arch. Biochem. Biophys. 185, 108–115. [DOI] [PubMed] [Google Scholar]
- Hindsgaul O.; Khare D. P.; Bach M.; Lemieux R. U. (1985) Molecular recognition. 3. The binding of the H-type-2 human-blood group determinant by the lectin-I of ulex-europaeus. Can. J. Chem. 63, 2653–2658. [Google Scholar]
- Domino S. E.; Zhang L.; Lowe J. B. (2001) Molecular cloning, genomic mapping, and expression of two secretor blood group α(1,2)fucosyltransferase genes differentially regulated in mouse uterine epithelium and gastrointestinal tract. J. Biol. Chem. 276, 23748–23756. [DOI] [PubMed] [Google Scholar]
- Allen H. J.; Johnson E. A. Z.; Matta K. L. (1977) Comparison of binding specificities of lectins from Ulex europaeus and Lotus tetragonolobus. Immunol. Commun. 6, 585–602. [DOI] [PubMed] [Google Scholar]
- Anello M.; Spampinato D.; Piro S.; Purrello F.; Rabuazzo A. M. (2004) Glucosamine-induced alterations of mitochondrial function in pancreatic β-cells: Possible role of protein glycosylation. Am. J. Physiol. 287, E602–E608. [DOI] [PubMed] [Google Scholar]
- Scheiner-Bobis G. (2002) The sodium pump: Its molecular properties and mechanics of ion transport. Eur. J. Biochem. 269, 2424–2433. [DOI] [PubMed] [Google Scholar]
- Treuheit M. J.; Costello C. E.; Kirley T. L. (1993) Structures of the complex glycans found on the β-subunit of (Na,K)-ATPase. J. Biol. Chem. 268, 13914–13919. [PubMed] [Google Scholar]
- Rougon G.; Hobert O. (2003) New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu. Rev. Neurosci. 26, 207–238. [DOI] [PubMed] [Google Scholar]
- Ronn L. C. B.; Hartz B. P.; Bock E. (1998) The neural cell adhesion molecule (NCAM) in development and plasticity of the nervous system. Exp. Gerontol. 33, 853–864. [DOI] [PubMed] [Google Scholar]
- Baldwin T. J.; Fazeli M. S.; Doherty P.; Walsh F. S. (1996) Elucidation of the molecular actions of NCAM and structurally related cell adhesion molecules. J. Cell Biol. 61, 502–513. [DOI] [PubMed] [Google Scholar]
- Cremer H.; Chazal G.; Goridis C.; Represa A. (1997) NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol. Cell. Neurosci. 8, 323–335. [DOI] [PubMed] [Google Scholar]
- Honig M. G.; Petersen G. G.; Rutishauser U. S.; Camilli S. J. (1998) In vitro studies of growth cone behavior support a role for fasciculation mediated by cell adhesion molecules in sensory axon guidance during development. Dev. Biol. 204, 317–326. [DOI] [PubMed] [Google Scholar]
- Pollerberg G. E.; Becksickinger A. (1993) A functional-role for the middle extracellular region of the neural cell-adhesion molecule (NCAM) in axonal fasciculation and orientation. Dev. Biol. 156, 324–340. [DOI] [PubMed] [Google Scholar]
- Gong Q. Z.; Shipley M. T. (1996) Expression of extracellular matrix molecules and cell surface molecules in the olfactory nerve pathway during early development. J. Comp. Neurol. 366, 1–14. [DOI] [PubMed] [Google Scholar]
- Serizawa S.; Miyamichi K.; Takeuchi H.; Yamagishi Y.; Suzuki M.; Sakano H. (2006) A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell 127, 1057–1069. [DOI] [PubMed] [Google Scholar]
- Walz A.; Mombaerts P.; Greer C. A.; Treloar H. B. (2006) Disrupted compartmental organization of axons and dendrites within olfactory glomeruli of mice deficient in the olfactory cell adhesion molecule, OCAM. Mol. Cell. Neurosci. 32, 1–14. [DOI] [PubMed] [Google Scholar]
- Han W. P.; Rhee J. S.; Maximov A.; Lao Y.; Mashimo T.; Rosenmund C.; Sudhof T. C. (2004) N-Glycosylation is essential for vesicular targeting of synaptotagmin 1. Neuron 41, 85–99. [DOI] [PubMed] [Google Scholar]
- Nagao H.; Yoshihara Y.; Mitsui S.; Fujisawa H.; Mori K. (2000) Two mirror-image sensory maps with domain organization in the mouse main olfactory bulb. NeuroReport 11, 3023–3027. [DOI] [PubMed] [Google Scholar]
- Ressler K. J.; Sullivan S. L.; Buck L. B. (1994) Information coding in the olfactory system: Evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell 79, 1245–1255. [DOI] [PubMed] [Google Scholar]
- Vassar R.; Chao S. K.; Sitcheran R.; Nunez J. M.; Vosshall L. B.; Axel R. (1994) Topographic organization of sensory projections to the olfactory bulb. Cell 79, 981–991. [DOI] [PubMed] [Google Scholar]
- Treloar H. B.; Feinstein P.; Mombaerts P.; Greer C. A. (2002) Specificity of glomerular targeting by olfactory sensory axons. J. Neurosci. 22, 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P.; Wang F.; Dulac C.; Chao S. K.; Nemes A.; Mendelsohn M.; Edmondson J.; Axel R. (1996) Visualizing an olfactory sensory map. Cell 87, 675–686. [DOI] [PubMed] [Google Scholar]
- Vassar R.; Ngai J.; Axel R. (1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74, 309–318. [DOI] [PubMed] [Google Scholar]
- Ressler K. J.; Sullivan S. L.; Buck L. B. (1993) A zonal organization of odorant receptor gene-expression in the olfactory epithelium. Cell 73, 597–609. [DOI] [PubMed] [Google Scholar]
- Cho J. H.; Lepine M.; Andrews W.; Parnavelas J.; Cloutier J. F. (2007) Requirement for slit-1 and robo-2 in zonal segregation of olfactory sensory neuron axons in the main olfactory bulb. J. Neurosci. 27, 9094–9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehrehasa F.; Key B.; St Johcn J. A. (2008) The cell surface carbohydrate blood group A regulates the selective fasciculation of regenerating accessory olfactory axons. Brain Res. 1203, 32–38. [DOI] [PubMed] [Google Scholar]
- St John J. A.; Key B. (1999) Expression of galectin-1 in the olfactory nerve pathway of rat. Dev. Brain Res. 117, 171–178. [DOI] [PubMed] [Google Scholar]
- Dowsing B.; Puche A.; Hearn C.; Key B. (1997) Presence of novel N-CAM glycoforms in the rat olfactory system. J. Neurobiol. 32, 659–670. [PubMed] [Google Scholar]
- Treloar H.; Tomasiewicz H.; Magnuson T.; Key B. (1997) The central pathway of primary olfactory axons is abnormal in mice lacking the N-CAM-180 isoform. J. Neurobiol. 32, 643–658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.