1. Introduction
We would like to thank all the discussants for their insightful comments on our work. It is clear to all of us that the topic is timely and of great interest, because more and more genetic data sets are collected with a large number of phenotypes that are potentially related to the complex disease under study. At the same time, all of us also agree that the methods of involving multiple traits in genetic analysis are inevitably more complicated than the single-trait based approaches, and hence will raise statistical challenges and opportunities. Because of the methodological complexity, we decided to focus on a very specific, yet we believe important, question. We wanted to understand when and how testing multiple traits jointly may be more powerful than testing a single trait at a time. Hopefully, this understanding can provide some insights into directions for developing promising methods for multiple traits analyses. Although the goal of our current article is not to develop new methods, we are pleased that all discussants provided some new and exciting ideas for developing multiple traits analysis methods.
2. Further Simulation Studies
Wu, Berg, and Li described some biological mechanisms of trait dependence and gave an example about the metabolic rate (M).. Their comments were well taken that we should take into account biological mechanisms of trait dependence in our multiple traits analyses.
We also agree with Wu, Berg, and Li that our model needs to be extended to be useful. Specifically, we considered a linear model
and they suggested an additive model
where f1 (Y1) and f2(Y2) are the linear or nonlinear functions that describe biological dependence of Y3 on traits Y1 and Y2.
The additive model is obviously more general and flexible. Again, we deliberately chose three traits and linear models to focus on the comparison of a univariate test and multivariate test.
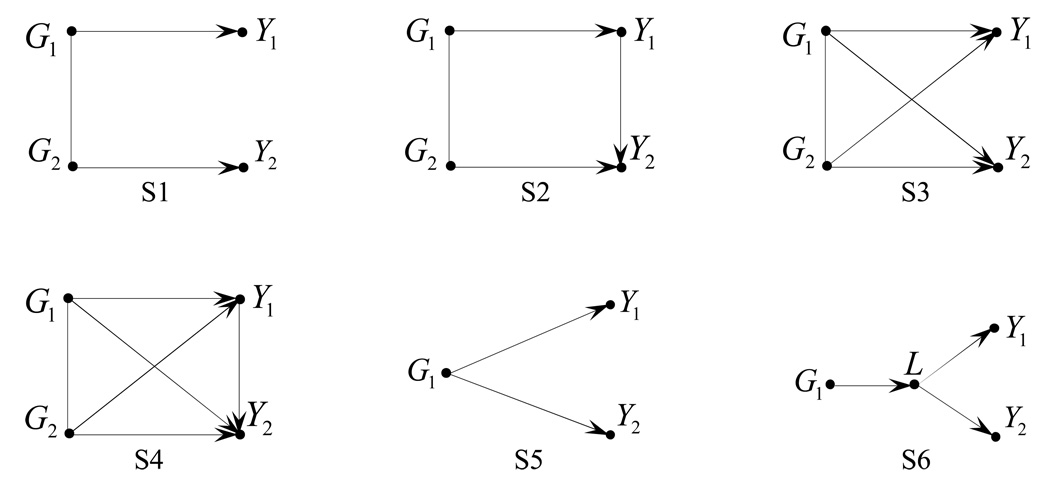
Their suggestion to include pleiotropic and linkage mechanisms for trait dependence in simulations is also worthy pursuing. We considered pleiotropic mechanisms in our previous simulations. Here, we discuss how to compare the multiple-trait test with the single-trait test by considering linkage mechanisms for trait dependence. For clarity, let us consider two traits Y1 and Y2, and two disease loci G1 and G2. Although there exist many possible structures among Y1, Y2, G1 and G2, we select four structures displayed as S1 to S4 in Figure 1. An arrow between any two elements represents a causal relationship. A line between two loci G1 and G2 implies they are in linkage disequilibrium (LD). S1 and S3 illustrate the cases where the trait dependence is due to linkage mechanisms, and S2 and S4 the cases where the trait dependence is due to pleiotropic and linkage mechanisms together.
Figure 1.
Relationship structures between two traits Y1, Y2 and one gene G1 or two genes G1 and G2. An arrow between any two elements represents a causal relationship. A line between two loci G1 and G2 implies that they are in LD. L represents an unobserved latent variable.
We also impose linear structural equation models on each DAG. Without loss of generality, we use S4 as an example, leading to the following models,
where is the number of disease allele D at jth locus, μi denotes the intercept, and represents the additive effect of disease allele D for the ith trait at the jth locus for i, j = 1,2. If extraneous variables are included, (ε1,ε2)' is distributed as N(0,Σ) ; otherwise ε1 and ε2 are mutually independent and respectively distributed as and . Epistasis between G1 and G2 can be added to the above models, as Wu, Berg, and Li suggested. With this setting, simulations can be performed similarly to what we have done in the original article.
3. Analyzing Related Multiple Traits
Feng gave several interesting examples for the relationships between traits and genes in her discussions, and demonstrated that “…these relations are detectable using appropriate models and thus can be considered in the future”. Those examples can be illustrated by graphical models. For example, S1 and S5 in Figure 1 consider the examples with two traits Y1 and Y2 and at most two disease loci G1 and G2. Interestingly, these examples coincide with the pleiotropic and linkage mechanisms for trait dependence mentioned by Wu, Berg, & Li in their discussions.
To perform multiple traits analyses based on S1, S3, and S5, we may consider the models in the framework of generalized linear model (GLM) by applying the generalized estimating equations (GEE) method (Liang & Zeger, 1986). Similar to Wu, Berg, & Li’s discussions, a suitable model based on S3 allows us to test the relative importance of pleiotropy and linkage in trait correlations. Another example of relationship considered by Feng is about latent factors, say S6, where L represents an unobserved latent variable. A simple method for multiple traits analyses based on S6 is to combine all the observed traits into a single trait and then to perform univariate association analysis. An alternative method is to use a latent variable model. Feng also pointed out that “environmental factors may interact with genes to affect some of traits”. To include environmental factor E in multiple traits analyses, we can replace G2 with E in S1 and S3 if we only consider one disease locus, and interactions between G1 and E are added.
4. Analyzing Causally Related Multiple Traits
Fang, Luo, Reveille & Xiong pointed out that there may be a gross lack of the traditional statistical methods and algorithms for multiple traits analyses based on causal structures. The challenge is to identify the existence of the causal effects. They presented structural equations as one of useful tools. Specifically, they provided a two-stage method to perform multiple quantitative traits analysis. They first used structural equations to model genotype and phenotype networks, and then used structural equations again to connect these two networks and derive a larger network, which provided us with the causal relationships among traits and between traits and markers, and pair-wise LD relations among markers. We believe their method may be very useful for multiple traits analyses in the future.
5. Haplotype-Based Multiple Traits Analyses
Wu, Berg & Li and Feng addressed the possibility of involving haplotype into multiple traits analyses. We also think developing haplotype-based method for analyzing multiple traits jointly will become another promising research direction. Schaid (2004) and Clark (2004) discussed the advantages of using haplotypes over using a single marker in terms of biologic function, statistical power improvement, and LD mapping. Haplotypes usually contain more LD information than what pair-wise LD explained, which allow us to use intra-marker relationships nships easily. On the other hand, although there are 2N different haplotypes in theory for N SNP markers, haplotypes usually arise as an intrinsic attribute of population genetic variation, and as a result, only a few haplotypes are exist in practice. The reduction in the number for haplotypes reduces the data dimension in genetic analyses.
6. Conclusion
Multiple traits analyses are becoming more and more important and have brought us with both opportunities and challenges, as clearly pointed out by the discussants. We hope that our paper, along with all discussions and this rejoinder, will encourage the statisticians and geneticists to conduct further research on multiple traits analyses and other related issues in statistical genetics.
Finally, we thank all discussants for their comments. We also would like to thank the Co-Editor Qiwei Yao for inviting our submission and organizing the discussions.
Acknowledgements
This research is supported in part by grants K02DA017713 and R01DA016750 from the National Institutes on Drug Abuse.
References
- Clark AG. The role of haplotypes in candidate gene studies. Genetic Epidemiology. 2004;27:321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;3:13–22. [Google Scholar]
- Schaid DJ. Evaluating associations of haplotypes with traits. Genetic Epidemiology. 2004;27:348–364. doi: 10.1002/gepi.20037. [DOI] [PubMed] [Google Scholar]