Abstract
Efficient and correct responses to double stranded breaks (DSB) in chromosomal DNA are critical for maintaining genomic stability and preventing chromosomal alterations leading to cancer1. The generation of DSB is associated with structural changes in chromatin and the activation of the protein kinase ataxia-telangiectasia mutated (ATM), a key regulator of the signaling network of the cellular response to DSB 2,3. The interrelationship between DSB-induced changes in chromatin architecture and the activation of ATM is unclear 3. Here we show that the nucleosome-binding protein HMGN1 modulates the interaction of ATM with chromatin both prior to and after DSB formation thereby optimizing its activation. Loss of HMGN1, or ablation of its ability to bind to chromatin, reduces the levels of IR-induced ATM autophosphorylation and the activation of several ATM targets. IR treatments lead to a global increase in the acetylation of Lys14 of histone H3 (H3K14) in an HMGN1 dependent manner and treatment of cells with a histone deacetylase inhibitor bypasses the HMGN1 requirement for efficient ATM activation. Thus, by regulating the levels of histone modifications, HMGN1 affects ATM activation. Our studies identify a new mediator of ATM activation and demonstrate a direct link between the steady-state intranuclear organization of ATM and the kinetics of its activation following DNA damage.
Keywords: DNA damage response, ataxia, telangiectasia mutated (ATM), chromatin, histone modifications, HMG proteins
It is now clear that the action of chromatin modifiers that are recruited to DSB sites plays an important role in the DNA damage response and the repair of these lesions 4,5; however, the chain of events linking the generation of DSB to architectural changes in chromatin and activation of ATM are still not fully understood. Recent electron microscopy studies revealed that the generation of DSB leads to a rapid, ATP dependent, local decondensation of chromatin which occurs in the absence of ATM activation6, supporting the suggestion that architectural changes in chromatin may be involved in the initiation of the DNA damage response 7. In addition, ATM mediates transient, global chromatin decondensation via phosphorylation of the KAP-1 protein 8. The local and global changes in chromatin organization facilitate the recruitment of damage response proteins and remodeling factors that further modify the chromatin in the vicinity of the DSB and propagate the DNA damage response. Given the extensive changes in chromatin structure associated with the DSB response it could be expected that architectural chromatin-binding proteins such as the high mobility group (HMG) proteins would be involved in this process 9,10.
HMG is a superfamily of nuclear proteins that bind to chromatin without any obvious specificity for a particular DNA sequence and affect the structure and activity of the chromatin fiber 9,11. We have previously reported that HMGN1, an HMG protein that binds specifically to nucleosome core particles, affects the repair of DNA damaged by either UV12 or ionizing13 radiation. Hmgn1−/− mice and embryonic fibroblasts (MEFs) are hypersensitive to IR and have an increased tumorigenic potential13.
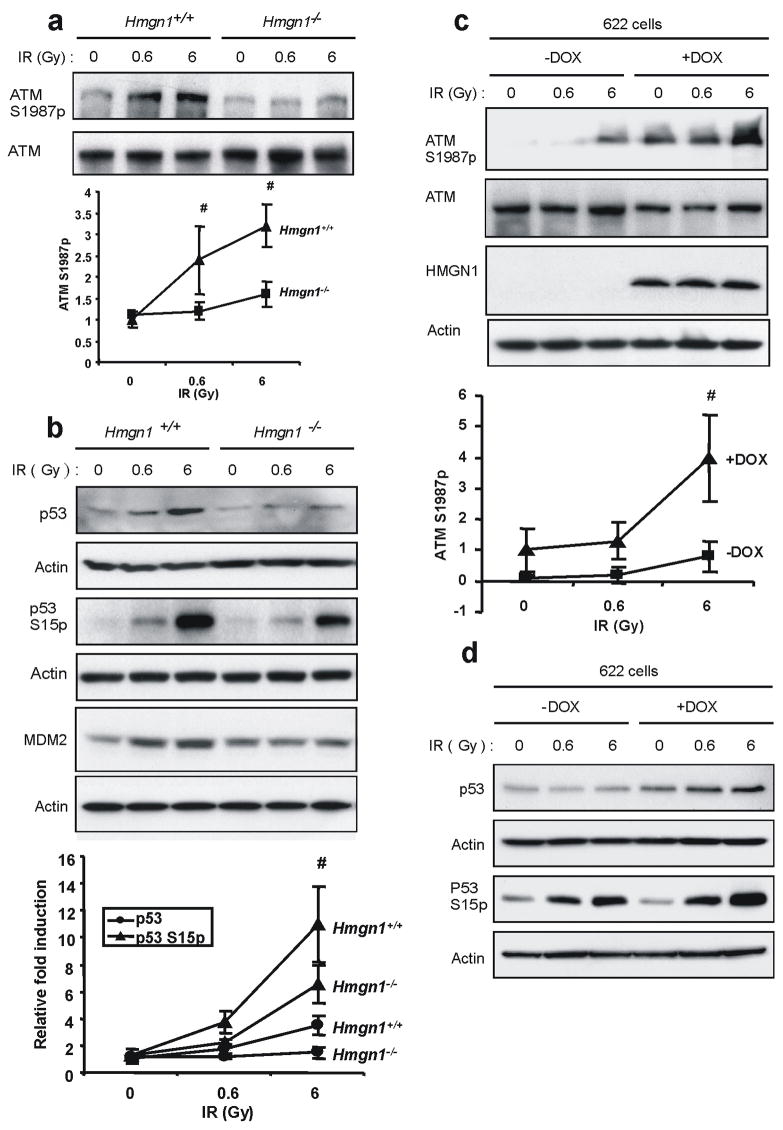
Since DSB are critical DNA lesions induced by IR and since ATM is the chief transducer of the DSB response, we tested whether HMGN1 is involved in IR-induced ATM activation. A convenient marker of mouse ATM activation is its autophosphorylation on Ser1987 14. Analyses of IR-treated primary MEFs isolated from wild type and Hmgn1−/− littermates revealed that loss of HMGN1 does not affect the ATM level, but significantly reduces the IR-induced autophosphorylation of S1987 (Fig. 1a). Loss of HMGN1 also reduced the IR- induced phosphorylation of p53, and reduced the levels of both p53 and MDM2 (Fig 1b). Likewise, loss of HMGN1 reduced the IR-induced phosphorylation of SMC1, CHK1 and CHK2 (Fig. S1), all of which are ATM targets 2.
Figure 1.
Loss of HMGN1 impairs IR-induced ATM autophosphorylation and activation. (a) Western blotting analyses with the indicated antibodies in IR-treated Hmgn1+/+ and Hmgn1−/− littermate MEFs. The graph shows calculated averages with s.d. of 5 independent experiments with MEFs prepared from different sets of littermates. (b) Impaired damage-induced p53 stabilization and MDM2 activation in Hmgn1−/− cells. Western blotting analysis was carried out on extracts from IR-treated MEFs with the indicated genotypes. The graph shows calculated averages with s.d. of 3 independent experiments with MEFs prepared from different sets of littermates. (c) Rescue of ATM activation by re-expression of HMGN1. Hmgn1−/− MEFs stably transformed with a doxycyline (Dox)-inducible vector expressing ectopic HMGN1 (cell line #622) were irradiated and extracts analyzed by western blotting with the indicated antibodies. Dox treatment induces HMGN1 expression and rescues ATM autophosphorylation. The graph shows calculated averages with s.d. of 3 independent experiments. (d) Rescue of p53 activation by re-expressing HMGN1. Experimental details are as in (c).
To verify that the IR-induced ATM activation is indeed HMGN1 dependent, we tested stable transformants of Hmgn1−/− cells ectopically expressing HMGN1 under the control of the tetracycline promoter (cell line #622). Prior to HMGN1 induction, only minor ATM autophosphorylation on S1987 was detected following IR, but doxycline-induced expression of physiological levels of HMGN1 elevated both the IR-dependent autophosphorylation of ATM (Fig 1c) and the p53 phosphorylation and stabilization (Fig 1d).
A key event in the cellular response to DSB is the accumulation of sensors such as Mre11, Rad50 and Nbs1 (the MRN complex), MDC1 and 53BP1, at the damaged sites15. Formation of these multiprotein complexes at DSB sites facilitates the activation of ATM and the amplification of the damage signal. Since in living cells HMGN1 molecules are in constant motion and continuously exchange among chromatin binding sites 16,17 we tested whether HMGN1 accumulates at the damaged sites and affects the recruitment of the damage sensory proteins. Immunofluorescence analyses and examination of live cells expressing HMGN1-GFP indicated that IR treatment did not affect the intracellular distribution of HMGN1 and the protein did not accumulate at the break sites (Fig. S2a, b). Likewise, the IR-induced formation of Nbs1, MDC1, 53BP1 and γH2AX foci in Hmgn1−/− cells was similar to that of wild type MEFs (Fig. S3). Thus, the reduced ATM activation in Hmgn1−/− cells is not due to faulty recruitment of DSB sensors to the damaged sites.
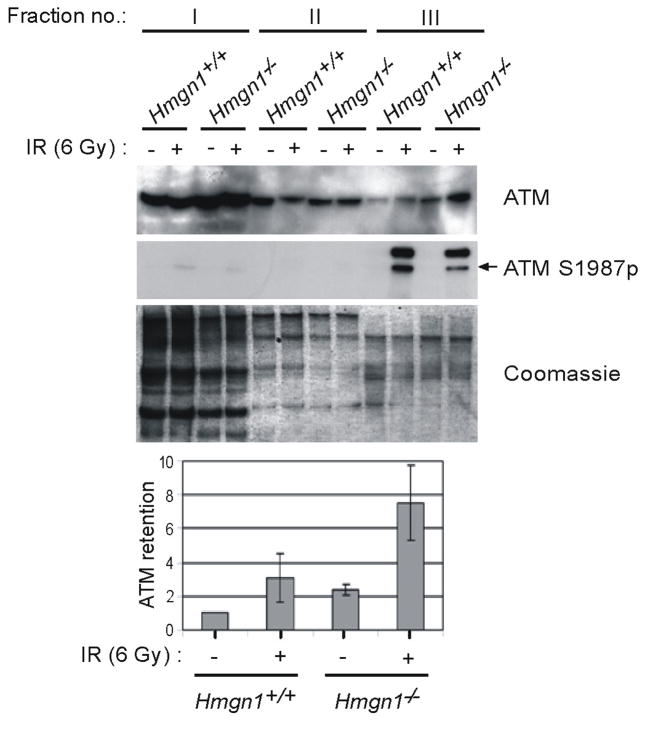
These results raised the possibility that the impaired ATM activation in Hmgn1−/− cells is related to the interaction of ATM with chromatin, which is consistent with the suggestion that changes in chromatin structure activate ATM even in the absence of DSB7. To examine this possibility, we used an established “chromatin retention assay” 18 to follow IR-induced recruitment of ATM to chromatin. As expected, in Hmgn1+/+ cells, induction of DSB led to a three fold increase in the level of chromatin-bound ATM (Fig. 2, fraction III and graph). Surprisingly, prior to irradiation the levels of chromatin-bound ATM were already two fold higher in Hmgn1−/− than in Hmgn1+/+ cells and DSB induction further increased by 3-fold the amount of chromatin-bound ATM in Hmgn1−/− cells (Fig. 2). In agreement with previous data 19, ATM S1987p was detected mainly in the chromatin-bound fraction of the irradiated cells and loss of HMGN1 decreased the levels of this modification. Hence, loss of HMGN1 increases the amount of chromatin-bound ATM but reduces the amount of ATM S1987p thus resulting in a relative phosphorylation level (ATM S1987p/ATM) of ATM that was at least 4 times lower than that in Hmgn1−/− cells (Fig. 2).
Figure 2.
Enhanced ATM chromatin retention in Hmgn1−/− cells. Littermate Hmgn1 +/+ and Hmgn1−/− MEFs were fractionated 1 h following mock treatment or irradiation with 6 Gy. Equal no. of cells from the soluble fraction (I), the washing fraction (II) and from the chromatin enriched fraction (III) were analyzed by Western blotting for ATM and ATM S1987p levels. Coomassie blue staining of a parallel gel shows equal loading of protein. The bar graphs represent the relative levels of ATM retained on chromatin as an average, with s.d. from 3 independent experiments. The retained ATM level in non-irradiated Hmgn1+/+ cells was set as 1.
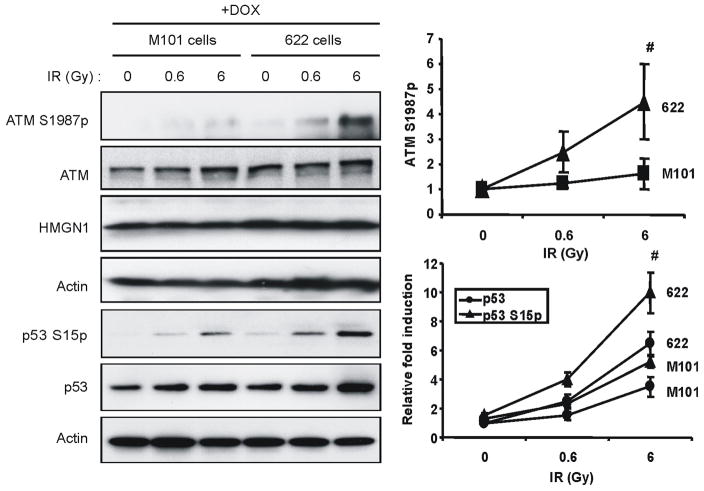
HMGN1 is a structural protein that binds specifically to nucleosome core particles and affects DNA–dependent activities such as transcription and repair, but only in the context of chromatin11,16. HMGN1 mutants that do not bind to chromatin affect neither UV- nor IR-mediated DNA repair 12,13. To test whether the effects of HMGN1 on ATM activation are contingent on chromatin binding, we compared the IR-induced levels of ATM S1987p in Hmgn1−/− cells expressing wild type HMGN1 protein (cell line #622) to cells expressing a double mutant HMGN1S20,24E (cell line #M101) which localizes to the nucleus but does not bind to chromatin 16. Ectopic expression of wild type HMGN1, but not of the HMGN1S20, 24E mutant elevated the DSB-dependent ATM autophosphorylation at S1987 and the activation of p53 (Fig. 3). We therefore conclude that the interaction of HMGN1 with chromatin enhances ATM activation. However, immunofluorescence analyses revealed that HMGN1 does not colocalize with ATM, an indication that ATM does not bind to chromatin regions containing HMGN1 (Fig S4). Thus, the impaired activation of ATM in Hmgn1−/− cells is due to global changes in chromatin organization rather than to loss of specific interaction between HMGN1 and ATM.
Figure 3.
HMGN1 enhances ATM activation by binding to chromatin. Hmgn1−/− MEFs stably transformed with a doxycyline (Dox)-inducible vector expressing either wild type HMGN1 (cell line #622) or the mutant HMGN1 S20, 24E (M101cells) which does not bind to chromatin, were irradiated and extracts analyzed by westerns with the indicated antibodies. The graphs represent calculated averages, with s.d. of 3 independent experiments.
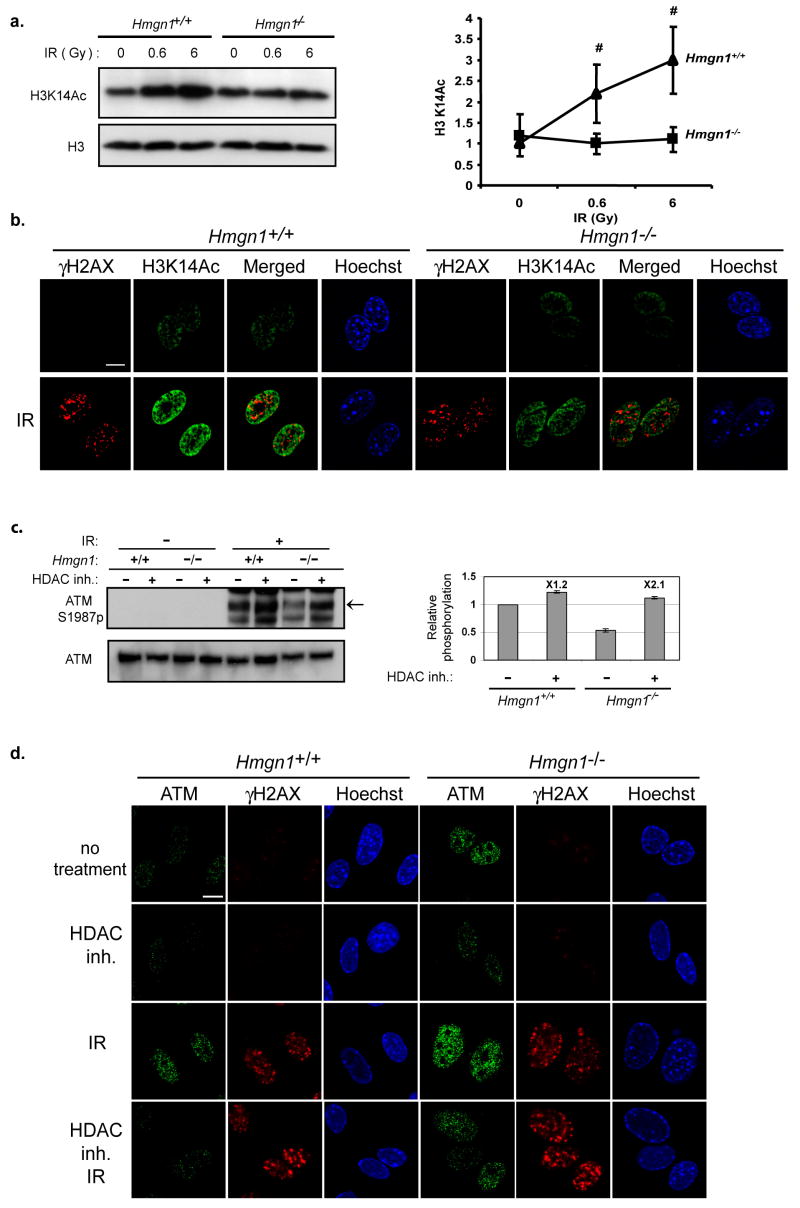
The interaction of HMGN1 with nucleosomes alters not only the “higher order” chromatin structure but also the local accessibility of histone tails to modifying enzymes and therefore the levels of several histone posttranslational modifications in Hmgn1−/− cells are different from those of wild type MEFs 20. Alterations in the levels of histone modifications are known to play an important role in DSB repair 4,21,22. We therefore hypothesized that HMGN1 affects ATM activation via modulation of histone modifications. To test this possibility, we investigated whether IR alters the levels of modifications in the tail of histone H3, at positions known to be affected by HMGN1. Western blotting and immunofluorescence analyses (Fig. 4a, b) revealed that IR treatments enhance the global acetylation of H3K14 in Hmgn1+/+, but less so in Hmgn1−/− thereby providing support for our hypothesis. As a further test of the link between HMGN1-mediated histone modification and ATM activation we examined whether elevation of histone acetylation levels by an HDAC inhibitor, prior to IR treatment, would simulate the effect of HMGN1 on ATM activation. MEFs derived from Hmgn1−/− and Hmgn1+/+ littermate mice were incubated with HDAC inhibitors, exposed to IR and ATM autophosphorylation levels were determined (Fig. 4c). In Hmgn1+/+ cells, pre-incubation with HDAC inhibitors increased the IR-induced ATM phosphorylation levels by approximately 20% (Fig 4c, graph). Significantly, in cells lacking HMGN1, pre-incubation with HDAC inhibitors elevated the levels of IR-induced ATM autophosphorylation by 2-fold. As a result the relative phosphorylation levels of HDAC-treated Hmgn1−/− cells were indistinguishable from those of the wild type cells (Fig. 4c, graph).
Figure 4.
HMGN1 modulates the interaction of ATM with chromatin. (a) Loss of HMGN1 reduces IR-induced acetylation of K14 in histone H3. Shown is Western blotting analysis of H3K14ac in IR-treated Hmgn1−/− and Hmgn1+/+ MEFs. The graphs represent calculated averages with s.d. of 3 independent experiments (b) IR-induced acetylation of H3K14 is HMGN1 dependent and not limited to the DNA breakage points. Shown are confocal images of immunostaining with antibodies to H3K14ac and γH2AX in IR-treated Hmgn1−/− and Hmgn1+/+ MEFs. Note that the H3K14ac in Hmgn1+/+ cells is more intense than in Hmgn1−/− cells. Scale bar is 10 μm. (c) Enhanced histone acetylation prior to IR treatment abrogates the effect of Hmgn1−/− on ATM activation. Shown are Western analyses of the levels of ATM and ATM S1987p in Hmgn1−/− and Hmgn1+/+ MEFs that were incubated with or without 0.5 μg/ml of Trichostatin A and 2mM sodium butyrate (HDAC inhibitors) for 4 h prior to exposure to 6 Gy of IR. The bar graph represents ATM activation as the ratio between autophosphorylated and total ATM(ATM S1987p/ATM), obtained by quantifying the Western signals from 3 independent experiments. This ratio for irradiated Hmgn1+/+ cells that were not exposed to the HDAC inhibitors was set as 1. (d) Enhanced histone acetylation prior to IR treatment abrogates the effect of HMGN1 on ATM chromatin binding. Hmgn1−/− and Hmgn1+/+ MEFs were either treated or not with histone deacetylase (HDAC) inhibitor, exposed to 6 Gy of IR and pre-lysed on ice prior to PFA fixation. The fixed cells were immunostained with antibodies against ATM and γH2AX. DNA was stained with Hoechst. Scale bar is 10 μm
Our finding that incubation of cells with an HDAC inhibitor prior to IR exposure simulates the effects of HMGN1, taken together with the observation that the absence of HMGN1 protein increases the chromatin retention of ATM even without IR treatment (Fig 2), raises the possibility that HMGN1 affects the intranuclear organization of ATM. Indeed, in situ ATM retention assays revealed increased ATM retention in Hmgn1−/− cells both prior and after IR treatment (Fig 4d, top and 3rd row). Significantly, HDAC inhibitors reduced the levels of chromatin-bound ATM both prior to, and following, IR exposure (Fig. 4d). ATM activation has been linked to its acetylation23; however, we find that following HDAC treatment, the acetylation of ATM in IR treated Hmgn1−/− cells was not higher than in similarly treated Hmgn1+/+ cells (Fig S5). Thus, HMGN1 enhances the IR-induced activation of ATM by elevating the acetylation levels of nucleosomal histones. While HMGN1 affects the IR dependent ATM phosphorylation it does not significantly alter the UV-irradiation dependent ATM phosphorylation (Figure S6).
In summary, our results suggest that the activation dynamics of ATM is affected by chromatin interactions that occur prior to the IR-induced generation of DSB. ATM retention assays with purified chromatin, immunofluorescence analyses of permeabilized cells, and analyses of HDAC treated cells (Fig 2, Fig 4c, d) all demonstrate that a fraction of ATM is associated with chromatin prior to IR treatment and that loss of HMGN1 increases this fraction. These results, taken together with the findings that HMGN mutants that do not bind chromatin do not affect ATM activation (Fig 3), and that HMGN1 does not accumulate at DSB (Fig S2), suggest that by modifying chromatin, HMGN1 affects the global organization of ATM throughout the nucleus and not only at the DSB site (Fig 4d). Thus, the properties of the chromatin fiber predetermine the cellular response to IR induced DNA damage.
Methods
Preparation of primary MEFs and MEF cell lines
Primary Hmgn1+/+ and Hmgn1−/− MEFs, and transformed MEF cells stably expressing either wild-type (WT) or mutant HMGN1 protein under the control of the Tet promoter were described previously 12. Cells expressing the ATM-S1987A mutant were described and characterized previously 22.
Irradiation and antibodies for Western blotting
Cells were incubated in 10 mm dishes until confluent within 70–80% for irradiation. The cells were exposed to ionizing radiation from a 137Cs Shepherd Mark II irradiator at a cumulative dose of 0.6 and 6Gy. After irradiation, the cells were incubated for 1 h. For western blotting, the cells were scraped and washed with PBS buffer. Then cell lysates were prepared for sonication with 1× SDS sample buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT), heated to 95°C for 5 minutes and centrifuged for 5 minutes. The proteins were separated by SDS-PAGE and transferred to PVDF membranes. The following antibodies were used for Western blotting: CHK1 (G-4) (Santa Cruz Biotechnology, Santa Cruz, CA), pATM (Ser-1987), pSMC1 (Ser-957) (Rockland, Gilbertsville, PA), p53, SMC1, pp53 (Ser-15), pCHK1 (Ser-345) (Cell Signaling, Beverly, MA), H3K14ac, CHK2 (Upstate, Lake Placid, NY), HMGN1, histone H3 12, MAT3 anti ATM 18. The same membranes were re-blotted for anti-non-phospho-antibodies and β-actin antibodies (Sigma, Saint Louis, MO) to control the amounts of protein loaded.
Immunostaining analysis
For foci formation, control or X-ray irradiated MEFs plated on glass cover slides were further incubated for 30 min or 16 h at 37°C. Following fixation in methanol at −20°C for 20 minutes, the cells were washed three times with PBS and blocked in PBS-BSA (1% BSA in PBS) at R.T. for 1 h. The cells were labeled with antibodies against MDC1, Nbs1 24, 53BP1 (Novus Biologicals, Littleton, CO) and γH2AX (Upstate, Lake Placid, NY) for 1 h at R.T., washed three times with PBS and labeled with secondary antibodies; Cy3 fused donkey anti-rabbit or FITC fused donkey anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at R.T. Following three washings with PBS the DNA was stained with Hoechst 33258 (Molecular Probes, Eugene, OR) and the cover slips were mounted using Vectashield mounting medium (H-1000, Vector Laboratories, Burlingame, CA). For H3K4ac cells were fixed with 3% PFA in PBS for 20 min at RT, treated with 0.1% triton in PBS for 10 min and immunostained as described above.
ATM retention assay
The ATM retention assay was modified from a previous report 18. Briefly, for biochemical fractionation 15 cm plates with 60–70% confluence MEFs were collected, washed twice with PBS and the cell pellets were re-suspended in 150 μl of buffer I (50 mM HEPES at pH 7.6, 150 mM NaCl, 1 mM EDTA, 0.25% Triton X-100, 0.5 μg/ml TSA, Protease inhibitor cocktail, Phosphatase inhibitor cocktail 1 and 2 (Sigma, Saint Louis, MO). Following 5 min incubation on ice and 5 min centrifugation at 1000 × g at 4°C the supernatants were collected (fraction I), the pellets were washed once with 150 μl buffer I and the supernatants were collected (fraction II). Following addition of 150 μl of SDS Sample buffer the nuclei pellets were sonicated and boiled for 5 minutes (fraction III). Equal volumes from the different fractions were separated on 5% SDS-PAGE and immunoblotted for ATM and pATM (Ser-1987). For in situ retention control or X-ray irradiated MEFs grown on glass cover slides were further incubated for 1 h at 37°C and placed on ice. Following one wash with ice cold PBS the cover slides were incubated twice in buffer I supplemented with 1% Triton X-100 for 10 min each. Next, the cells were fixed in 3% paraformaldehyde in PBS at R.T. for 20 min and immunostained with antibodies against ATM (H248, Santa Cruz Biotechnology, Santa Cruz, CA) and γH2AX. The DNA was stained with Hoechst 33258.
Supplementary Material
Acknowledgments
We thank Dr. A. Celeste, NCI for advice and help and to Ms S. Garfield, Confocal Core Facility of the LEC, NCI, for help with imaging. The research was supported by the intramural program of the NCI. Work in the laboratory of Y.S. is supported by the A-T Medical Research Foundation, the A-T Children’s Project, The Israel Science Foundation, the A-T Medical Research Trust, the Israel-Germany Joint Program on Cancer Research, and the A-T Ease Foundation.
References
- 1.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194(77) doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]; Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411 (6835):366. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31 (7):402. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Lavin MF, Kozlov S. ATM activation and DNA damage response. Cell Cycle. 2007;6 (8):931. doi: 10.4161/cc.6.8.4180. [DOI] [PubMed] [Google Scholar]
- 4.Wurtele H, Verreault A. Histone post-translational modifications and the response to DNA double-strand breaks. Curr Opin Cell Biol. 2006;18 (2):137. doi: 10.1016/j.ceb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Koundrioukoff S, Polo S, Almouzni G. Interplay between chromatin and cell cycle checkpoints in the context of ATR/ATM-dependent checkpoints. DNA Repair (Amst) 2004;3 (8–9):969. doi: 10.1016/j.dnarep.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Kruhlak MJ, et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172 (6):823. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421 (6922):499. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 8.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8 (8):870. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 9.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17(2):72. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15 (5):496. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Reeves R, Adair JE. Role of high mobility group (HMG) chromatin proteins in DNA repair. DNA Repair (Amst) 2005;4 (8):926. doi: 10.1016/j.dnarep.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19 (8):5237. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birger Y, et al. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. Embo J. 2003;22 (7):1665. doi: 10.1093/emboj/cdg142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birger Y, et al. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005;65 (15):6711. doi: 10.1158/0008-5472.CAN-05-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellegrini M, et al. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443 (7108):222. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308(5721):551. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]; Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22(20):5612. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298(5597):1435. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]; DiTullio RA, Jr, et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4(12):998. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]; Stewart GS, et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]; Goldberg M, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421 (6926):952. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 16.Catez F, et al. HMGN dynamics and chromatin function. Biochem Cell Biol. 2003;81 (3):113. doi: 10.1139/o03-040. [DOI] [PubMed] [Google Scholar]
- 17.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404 (6778):604. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 18.Andegeko Y, et al. Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem. 2001;276 (41):38224. doi: 10.1074/jbc.M102986200. [DOI] [PubMed] [Google Scholar]
- 19.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21 (2):187. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Lim JH, et al. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol Cell. 2004;15(4):573. doi: 10.1016/j.molcel.2004.08.006. [DOI] [PubMed] [Google Scholar]; Lim JH, et al. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. Embo J. 2005;24 (17):3038. doi: 10.1038/sj.emboj.7600768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murr R, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8 (1):91. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 22.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5 (7):675. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y, et al. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102 (37):13182. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Difilippantonio S, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat Cell Biol. 2005;7 (7):675. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.