Abstract
Currently, microbiological techniques such as culture enrichment and various plating techniques are used for detection of pathogens. These expensive and time consuming methods can take several days. Described below is the design, fabrication, and testing of a rapid and inexpensive sensor, involving the use of microelectrodes in a microchannel, which can be used to detect single bacterial cells electrically (label-free format) in real time. As a proof of principle, we have successfully demonstrated real-time detection of target yeast cells by measuring instantaneous changes in ionic impedance. We have also demonstrated the selectivity of our sensors in responding to target cells while remaining irresponsive to nontarget cells. Using this technique, it can be possible to multiplex an array of these sensors onto a chip and probe a complex mixture for various types of bacterial cells.
INTRODUCTION
Due to increasing threats such as food borne diseases and bioterrorism, there is a growing need for highly sensitive, rapid, yet inexpensive instrumentation for pathogen detection. Current microbiological techniques used for detection of pathogens involve expensive and time consuming methods such as culture enrichment and plating techniques, which can take several days.1 Recently, for example, E. Coli 0157:h7 is a bacterial strain in which several outbreaks have occurred causing hemorrhagic colitis,2, 3 resulting in death.4 Typically for detection, the sample is enriched in a sorbitol Maconkey Agar medium and stained for lactose fermentation. This process requires each colony in the sample to be tested individually, and each test takes between 24–48 h due to the required incubation time.5 Such a time consuming process is insufficient in meeting the demands for food inspection, given that with such a process, a pathogen could not be identified within the time the food under inspection should have already been shipped out.
The main challenge for rapid detection of a single bacterial cell lies in establishing a procedure which is ultrasensitive and can detect in real time, while, at the same time, being inexpensive and easy to use. Recently, many efforts have been made toward the use of impedance based sensors for detection of bacterial cells. Impedance based sensors are advantageous since they eliminate the need for fluorescence labeling. Several groups have reported pathogen detection using electrical impedance sensors.6, 7, 8, 9, 10, 11, 12 Many of the electrical impedance sensors presented (with some exceptions13) to date require numerous washing steps and lack the ability for real-time detection. As for detection time, flow-cytometry based methods such as the use of coulter counters,14 have provided the ability to analyze the dielectric properties of a cell in real time. With the on-chip integration of microchannels and microelectrodes, the ability to count, sort, and trap cells and analyze their dielectric properties has been demonstrated.15, 16, 17, 18, 19, 20 This type of device would operate on the principle of measuring the current change caused by the displacement in the fluid as the particle passes by two measuring electrodes. A device relying solely on this principle has difficulty in differentiating between two different types of cells which may have similar dielectric properties. Thus, this type of device would have difficulty in detecting a target cell in a complex mixture. The use of cell trapping when used in conjunction with impedance spectroscopy21 is a promising method for detection of targeted particles.
Here we describe a chip-based microfluidic device capable of electrical (label-free format) real-time detection of target cells in a solution. The purpose of this study was to investigate use of the described technique for detecting target cells selectively. We have demonstrated the detection selectivity of this technique using yeast cells as target cells and Concanavalin A (Con A), a glycoprotein with affinity for the sugar molecules on yeast surface, in place of antibodies. Future studies will be directed toward improving the detection limit and also the use of this technique for practical applications like pathogen detection.
DEVICE OPERATION AND THEORY
The basic device (Fig. 1) contains three electrodes. The channel current is monitored between electrodes A and C. The volume between electrodes A and C is the active area of the sensor. A third gold electrode, B, is included in the active area of the channel, allowing for immobilization of antibodies with an affinity to bind to target bacterial cells in the active area of the sensor. Gold electrodes are very suitable for surface chemistry modifications, such as deposition of surface assembled monolayers, which will optimize the immobilization of the antibodies. A sample suspected of containing the target bacterial cells is injected into the microchannel. If the sample contains the targeted bacteria, they will attach to the electrodes, partially clogging the channel thus resulting in solution resistance increase. By monitoring the impedance across microelectrodes A and C, it is possible to detect the channel gating caused by bacteria attached inside the channel. By choosing channel and electrode geometries close to the bacteria size, the probability of bacterial cells being captured by the electrodes and also the impedance changes are maximized.
Figure 1.
Cross section schematic of gated microchannel with electrodes labeled A, B, and C. The targeted bacteria bind to the antibodies which are immobilized on the gold electrode. (Bottom plot) Prediction of current between electrodes A and C after injection of bacteria.
For selective detection to be achieved, this technique would require that the channel geometry closely correspond to that of the target cell and that the target cell contain surface markers specific for the monoclonal antibodies immobilized in the active area of the sensor. Thus, we emphasize that the successful detection of target yeast cells demonstrated in this study can be extended for detection of all types of cells including pathogenic bacteria or even cancer cells in blood. However, the channel geometry must be tailored to the type of cell which is being targeted.
MATERIALS AND METHOD
In this section we describe the design and fabrication of the microfluidic biochip used in this study. We also present the immobilization of the antibodies inside the channel, and also describe the electronic instrumentation used for detection of target cells.
Device design
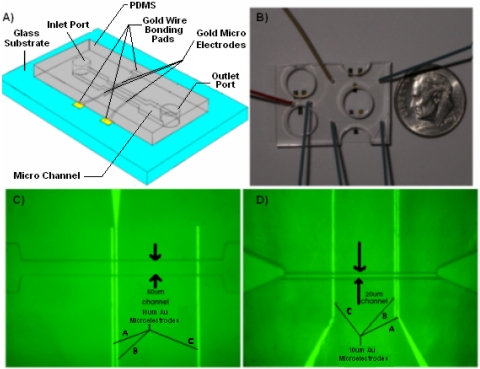
The microfluidic biochip used in this study is shown in Fig. 2A. Multiple channels were fabricated onto a single chip as shown in Fig. 2B. Experiments were conducted on two sets of channel sizes, one 50 μm deep and 50 μm wide [Fig. 2C], and the other 20 μm wide and 10 μm deep [Fig. 2D.
Figure 2.
(A) Schematic of microfluidic chip used in this study. (B) Photograph of a single chip containing three different channels with integrated electrodes. (C) Optical micrograph of top view of 50 μm deep channel device integrated with electrodes labeled A, B, and C. Electrode B was not used in this study. (D) Optical micrograph of top view of 10 μm deep channel.
Electrode fabrication
Au∕Cr electrodes (2000 Å∕150 Å) were fabricated on a glass wafer using traditional photolithography, sputtering, and then lift-off processing. The glass wafer was then diced into individual chips, in order to prepare them for bonding to the PDMS cover.
Channel fabrication in PDMS
The PDMS cover (fabricated by the Stanford Microfluidics Foundry) was made by patterning SU-8 25 photoresist on a silicon wafer. PDMS (10:1 prepolymer: curing agent) was poured into a petri dish with the master mold at the bottom, and then cured. Microbore tubes were inserted to make holes for the inlet and outlet ports of the channels. Individual sets of microchannels were cut and removed for sealing onto the patterned glass chips.
PDMS glass bonding
The glass chips and the PDMS slabs embedded with microchannels were both treated with oxygen plasma. The PDMS slab and the glass chips were then aligned under a microscope so that the electrodes overlap properly with the channel, and subsequently bonded together to make a very tight seal.
Measurement apparatus
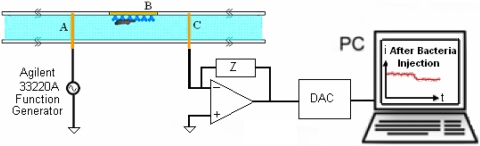
Electrical impedance measurements were collected across electrodes A and C both as a function of frequency and time (Fig. 3). A voltage signal was applied to electrode A using an Agilent 33320A function generator. The current passing across electrodes A and C was converted to a voltage and amplified with an EI-400 Potentiostat (Ensman Instruments, Bloomington, IN), and then sampled with a National Instruments PCI4452 data acquisition card. The data were read and analyzed by a Labview program. In order to confirm the validity of our real-time electrical measurements, we simultaneously monitored our channels using optical microscopy.
Figure 3.
Schematic of experimental setup including amplification circuitry and data acquisition.
EXPERIMENTAL PROCEDURE
In this study, yeast cells were used as target cells, and Concanavalin A (Con A), a glycoprotein with affinity for the sugar molecules on yeast was used in place of antibodies in order to demonstrate the success of the technology in selectively detecting target cells.
Preparation of yeast and Con A
Yeast (S. cervisiae) cells were maintained on YPD (Yeast Extract∕Peptone∕Dextrose) agar plates at 4 °C. An isolated colony was used to inoculate 5 ml of YPD broth, and the culture was grown to saturation for 16 h at 30 °C. Cells were then collected by centrifugation and resuspended in a solution containing 200 mM KCl and 10 mM HEPES in addition to 1 mM MgCl2, 1 mM MnCl2, and 1 mM CaCl2 which are necessary for Con A activity. The cell concentration in the final solution was diluted to 107 cells∕ml.
The Con A, from Calbiochem (San Diego, CA), was diluted to 10 mg∕ml. Immobilization of Con A on the electrodes was carried out by physical adsorption. Con A solution was injected and incubated into the channel for 15 min, then activated by the injection of Mn2+, Mg2+, and Ca2+ ions. A 200 mM KCl solution in 10 mM Hepes buffer with a pH of 6.8 containing yeast was injected into the channel at a flow rate of 100 nl∕min.
RESULTS AND DISCUSSION
Impedance spectrum
The impedance behavior across the channel is dictated by the processes occurring at the electrode-electrolyte interface and also the physical properties of the electrolyte, which can be represented with an electrical equivalent circuit consisting of a network of resistances and capacitances.13 Of particular importance are the bulk solution resistance and also the double layer capacitance, the latter of which results from hydrolyzed ions accumulating at the surface of the electrodes.22 Reactions occurring at the interface such as electron-transfer and ion diffusion from the bulk electrolyte to the electrode surface (Warburg impedance) may also affect the impedance at lower frequencies. However, such effects become negligible at higher frequencies.23 Due to the small separation of the layer of accumulated ions with the electrode surface, the double layer capacitance becomes very large to the extent that it dominates the impedance at low frequencies, which explains the drop in impedance as frequency increases. At higher frequencies, as the impedance resulting from the combined effects of the double layer capacitance, the electron transfer resistance, and also the Warburg impedance diminishes, the solution resistance dominates the impedance, which consequently becomes flat with frequency.
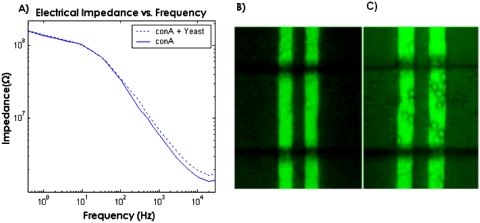
It was necessary to measure the impedance spectrum across the channel in order to gain a proper understanding of the impedance behavior as a function of frequency as shown in Fig. 4A. Figure 4B shows the channels before the binding of yeast, and Fig. 4C shows the channel after the yeast cells have been attached inside the channel. As seen in Fig. 4C, yeast cells bind on both the gold electrodes and the glass base of the channel. However, no yeast cells were observed to bind to the PDMS top layer. This demonstrates that the above mentioned method of Con A immobilization results in the Con A adsorbing onto both the gold electrodes and on the glass base of the channel. This would limit the sensitivity of the device since some targeted cells would bind to the channel wall outside the active area of the sensor. We will address this problem by construction of self-assembled monolayer on gold. Fabrication of a layer of protein G on this surface and attachment of the antibody onto the protein G layer will be carried out, essentially as described by Oh et al.24
Figure 4.
(A) Magnitude of ionic impedance across electrodes. The impedance levels off above 10 kHz indicating the solution resistance is dominant at these frequencies. The binding of yeast to Con A on the electrode results in an increase in ionic impedance at frequencies above 10 kHz indicating that impedance changes can be achieved resulting from ionic solution resistance increase. (B) Optical micrograph of electrodes A and B before and (C) after yeast cells binding to electrodes.
Of particular interest is to find the frequency at which the ionic resistance in the channel begins to dominate the impedance. It may be feasible to detect the binding of a targeted cell based on a change in capacitance, which can be done by measuring impedance at low frequencies where the electrical impedance is dominated by the double layer capacitance of the electrodes. However, we had difficulties achieving this due to a drift in impedance which occurred during our experiments at low frequencies, which made it difficult to get consistent results. It is likely that this impedance drift is due to an accumulation of ions at the surface of the electrode at low frequencies resulting from dc input bias offset currents of the circuitry connected to the electrodes, an effect which seems to have been frequently observed.25 As seen in the impedance spectrum, the binding of yeast cells on the channel walls in the region between electrodes A and C results in an increase in impedance at frequencies above 100 Hz. Based on the impedance curve, it can be seen that the solution resistance begins to dominate the impedance at frequencies above 10 kHz. The binding of yeast to Con A on the electrode results in an increase in ionic impedance at frequencies above 10 kHz indicating that impedance changes can be achieved resulting from ionic solution resistance increase.
Binding specificity
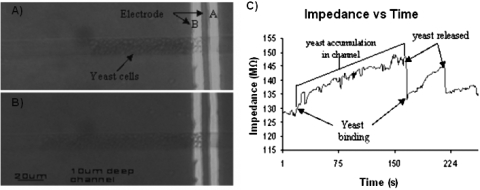
In order to achieve real-time detection, the electrical impedance was measured over time between electrodes and C at a frequency of 29.8 kHz in the 50 μm deep channel. This frequency turned out to be optimum for our system, since the ionic impedance is dominated by solution resistance. We refrained from working at frequencies higher than this, in order to avoid the effects of parasitic inductances. Figure 5C shows a clump of approximately 30 yeast cells binding onto electrode A resulting in an instantaneous increase in impedance at time t=59 s as shown in Fig. 5A. In a separate experiment [Fig. 5B], impedance measurements were taken as a clump of yeast was already bound onto the electrodes. At time t=155 s, the yeast cells were removed by increasing the pressure slightly, which resulted in an instantaneous decrease in impedance. As seen in Fig. 5B, the noise level is 0.02 MΩ, which is 1% of the base value of 2.22 MΩ. A change of 0.8 MΩ resulted from the binding of a clump of approximately 30 cells. This means that with the current device geometry at least eight cells need to bind to the electrodes in order to cause a change greater than the noise level. In order to increase the electrical sensitivity to the single cell level, a potential optimization would consist of decreasing the cross sectional area of the microchannel by a factor of eight, a modification which can be easily carried out.
Figure 5.
(A) Impedance (at 29.8 kHz) vs time. Impedance jump at t=59 s due to yeast binding. (B) Impedance vs time. Impedance drop at t=155 s due to yeast release (C) Optical micrograph of gold electrode A and B. Yeast clump is bound onto electrodes. Electrode C is not shown.
Large channel experiments
Figure 6A shows yeast cells being captured by the receptors on the electrode surface in the 50 μm deep channel. Results in Fig. 6B show an instantaneous increase in electrical impedance as a small number of cells bind to the surface of the electrodes, demonstrating real-time detection of cell capture. A current change of 2.6% resulted from several cells binding onto the electrode. In order to verify that binding of the cells to the channel walls were as a result of specific antigen-antibody interactions, we performed two different control experiments for the 50 μm deep channels. A 200 mM KCl solution in 10 mM Hepes buffer with a pH of 6.8 containing yeast was injected at a flow rate of 100 nl∕min into a channel in which Con A had not been immobilized on the surface. In order to further confirm the specificity, we ran a separate experiment where we treated the surface of yeast with alpha-mannosidase and alpha-glucosidase for removing the sugars, mannose and glucose which have an affinity for Con A. We immobilized the channel with Con A and injected 50 μl of yeast solution with a flow rate of 100 nl∕min. In both experiments, no binding of yeast occurred anywhere in the channel as predicted and consequently no changes in current occurred either. This confirms that results in Fig. 6 are due to specific binding, and that such a device configuration can be used to detect the presence of a target cell in a complex mixture.
Figure 6.
(A) Optical micrograph of gold electrode A after yeast binding has occurred. Electrodes B and C not shown. (B) Impedance (at 29.8 kHz) vs time. Impedance jump at t=55 s due to yeast binding.
The ability to selectively detect target cells in a complex mixture requires that nonspecific binding of nontarget cells onto the electrodes and the glass base between the electrodes be minimized. Given that nonspecific interactions are weaker than specific binding events, we minimized the nonspecific interactions by using a flow rate high enough to unbind the nonspecifically bound cells. In our 50 μm wide by 50 μm deep channels, at very low flow rates (below 100 nl∕min), we experienced many nontarget cells coming to rest on the electrodes and the glass base of the channel. At flow rates higher than 200 nl∕min, we experienced target cells not having the opportunity to adsorb to the electrodes or the glass base of the channel, thus being undetectable using our technique. Thus, we used 100 nl∕min since we found it to be the optimum flow rate for our system, both minimizing nonspecific binding events, while at the same time providing target cells with a sufficient opportunity to bind to the active area of the sensor.
A number of high-affinity monoclonal antibodies raised against bacterial surface antigens can also be used. The use of a mixture of such antibodies in the system would maximize specific interactions and further increase the strength of specific interactions relative to nonspecific binding. These attempts would further lower the possibilities for nonspecific adsorption.
Small channel experiments
In an attempt to further increase the electrical sensitivity of the sensor and also the probability of a cell being captured by the receptors in the active area, the 20 μm wide by 10 μm deep channels were tested. In this particular experiment, no receptors were immobilized onto the electrodes, so all capture was a result of nonspecific binding. Figures 7A, 7B show cells being captured on the electrodes and clogging the channel. As shown in Fig. 7C, at t=20 s as the first cells were captured by the electrode and the subsequent cells began accumulating in the channel, the impedance ramps up at a relatively steady rate. At t=160 s, the fluid pressure was momentarily slightly increased to unbind the cells from the electrodes and unclog the channel, resulting in an instantaneous drop in impedance. Immediately after the drop, cells began re-accumulating, which resulted in a steady increase in impedance until t=220 s when another momentary slight increase in fluid pressure was applied to release the cells. Beyond this time, no more cells were captured in the channel resulting in relatively constant impedance over time.
Figure 7.
(A) Optical micrograph of yeast cells accumulating in channel at t=75 s. (B) at t=130 s. Electrode C is to the left and is not shown in this figure. (C) Plot of impedance (at 29.8 kHz) vs time. Impedance increases steadily as cells accumulate in channel. Release of cells results in impedance drop at t=160 s. The same cycle is repeated until t=220 s. No cells across electrodes after t=220 s.
For channel sizes comparable to the diameter of yeast (5 μm), nonspecific binding and channel clogging have been shown to be problematic. A channel depth of 10 μm has shown to be too shallow for proper operation of the sensor. Larger channels have shown to be more practical, since we have proven them to be sensitive enough to electrically detect the presence of a small number of cells, while at the same time minimizing channel clogging and nonspecific binding. However, in order to obtain an electrical sensitivity approaching the single cell level, an intermediate channel depth should be used, which we shall explore in future studies.
CONCLUSIONS
We have demonstrated the proof of concept for a direct (label-free format) method for real-time detection of target cells. This method utilizes impedance measurements at 29.8 kHz to probe solution resistance changes associated with the blockage of ionic current due to cell binding on the channel walls in the active area of the sensor. While we focused on yeast cells in this study, the method can also be used for detection of pathogenic bacteria, cancer cells, or even testing water quality for possible contaminations, with appropriate optimization of channel geometries. In order to extend this method to practical applications like the detection of bacterial cells, and maintain the high electrical sensitivity of the device, it would be necessary to scale down the size of the channel geometries, thereby making it more compatible to the smaller dimensions of bacterial cells, in comparison to yeast cells. One particular advantage offered by this device is its selectivity in cell capture, which makes it possible to multiplex an array of these sensors onto a single chip and probe a solution to determine which types of bacterial cells it contains.
We demonstrated the feasibility of this technique for selective detection of target cells in a solution containing 107 cells∕ml. Further studies will be dedicated to measuring the maximum detection limit, and exploring various methods of optimization. The sensitivity of this technique is limited by the probability of a target cell being captured in the active area of the sensor. The binding of target cells to the antibodies immobilized on the glass base in regions outside of the active area of the sensor also limits sensitivity. Detection limit can be enhanced by effectively increasing the active area of the device by integrating multiple sets of electrodes across the channel. Further enhancements of sensitivity may be achieved by adopting an immobilization procedure which results in antibodies being immobilized predominantly on the gold electrodes, as opposed to the entire channel length. Multiple recycling of the solution in the channel may also help with capturing cells which may pass through the channel without attaching to the electrodes. Upon making such improvements to the device, it may be possible to achieve detection limits comparable to those reported most recently in the literature, which is near 101 CFU∕ml in solution.26 This method of detection may be used for development of a handheld device for point of care diagnostics.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health Grant P01 HG000205. The authors would like to thank Jessica Melin and the Stanford Microfluidics Foundry for their invaluable help in fabrication of the devices used in this study, and also Cory Nislow of the University of Toronto, and Bob St. Onge of the Stanford Genome Technology center for their help in yeast preparation.
References
- McClain D. and Lee W. H., Laboratory Communication No. 57. (U.S. Department of Agriculture, FSIS, Microbiology Division, Beltsville, Maryland 1989).
- Griffin P. M., “Infections of the gastrointestinal tract,” in Escherichia Coli O157:H7 and Other Enterohemorrhagic Escherichia Coli, edited by Blaser M. J., Smith P. D., Ravdin J. I., Greenberg H. B., and Guerrant R. L. (Raven, New York, 1995), pp. 739–761. [Google Scholar]
- Kaper J. B. and O’Brien A. D., Escherichia Coli O157:H7 and Other Shiga Toxin-Producing E. Coli Strains (ASM, Washington, D.C., 1998). [Google Scholar]
- Mead P. S., Slutsker L., Dietz V., McCaig L. F., Bresee J. S., Shapiro C., Griffin P. M., and Tauxe R. V., Emerg. Infect. Dis. 5, 607 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- March S. B. and Ratnam S., J. Clin. Microbiol. 23, 869 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke S. M. and Alocilja E. C., IEEE Sens. J. 5, 744 (2005). [Google Scholar]
- Gomez R., Bashir R., Sarikaya A., Ladisch M. R., Sturgis J., Robinson J. P., Geng T., Bhunia A., Apple H. L., and Wereley S., Biomed. Microdevices 3, 201 (2001). [Google Scholar]
- Radke S. M. and Alocilja E. C., IEEE Sens. J. 10.1109/JSEN.2004.830300 4, 434 (2004). [DOI] [Google Scholar]
- Yang L., Li Y., Griffis C. L., and Johnson M. G., Biosens. Bioelectron. 19, 1139 (2004). [DOI] [PubMed] [Google Scholar]
- Yang L.Ruan L. Y. C. and Li Y., Biosens. Bioelectron. 10.1016/S0956-5663(03)00229-X 19, 495 (2003). [DOI] [PubMed] [Google Scholar]
- Liu Y., Walter T. M., Chang W., Lim K. S., Yang L., Lee S. W., Aronson A., and Bashir R., Lab Chip 7, 603 (2007). 10.1039/b702408h [DOI] [PubMed] [Google Scholar]
- Gómez-Sjöberg R., Morisette D. T., and Bashir R., J. Microelectromech. Syst. 10.1109/JMEMS.2005.845444 14, 829 (2005). [DOI] [Google Scholar]
- Sengupta S., Battigelli D. A., and Chang H. C., Lab Chip 6, 682 (2006). 10.1039/b516274b [DOI] [PubMed] [Google Scholar]
- Coulter W. H., Pat US., 2656508 (1953).
- Saleh O. A. and Sohn L. L., Rev. Sci. Instrum. 10.1063/1.1419224 72, 4449 (2001). [DOI] [Google Scholar]
- Saleh O. A. and Sohn L. L., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0337563100 100, 820 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonaro A. and Sohn L. L., Lab Chip 10.1039/b504827c 5, 1155 (2005). [DOI] [PubMed] [Google Scholar]
- Gawad S., Schild L., and Renaud Ph., Lab Chip 10.1039/b103933b 1, 76 (2001). [DOI] [PubMed] [Google Scholar]
- Ayliffe H. E., Frazier A. B., and Rabbitt R. D., J. Microelectromech. Syst. 10.1109/84.749402 8, 50 (1999). [DOI] [Google Scholar]
- Cheng I. F., Chang H. C., Hou D., and Chang H. C., Biomicrofluidics 10.1063/1.2723669 1, 021503 (2007). [DOI] [Google Scholar]
- Wu J., Ben Y., and Chang H. C., Microfluid. Nanofluid. 10.1007/s10404-004-0024-5 1, 161 (2005). [DOI] [Google Scholar]
- Wu J., Ben Y., Battigelli D., and Chang H.-C., Ind. Eng. Chem. Res. 10.1021/ie049417u 44, 2815 (2005). [DOI] [Google Scholar]
- Katz E. and Wilner I., Electroanalysis 10.1002/elan.200390114 15, 913 (2003). [DOI] [Google Scholar]
- Oh B. K., Kim Y. K., Park K. W., Lee W. H., and Choi J. W., Biosens. Bioelectron. 19, 1497 (2004). 10.1016/j.bios.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Felice C. J., Madrid R. E., and Valentinuzzi M. E., Biomed. Eng. Online 4, 22 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Alocilja E., and Downes F. P., Biosens. Bioelectron. 22, 2329 (2007). 10.1016/j.bios.2007.01.013 [DOI] [PubMed] [Google Scholar]