Abstract
Bioluminescent reporter genes are sensitive in situ tools for following disease progression in preclinical models, albeit they are subject to scattering and absorption in deep tissues. We have generated a bicistronic Cre/LoxP reporter mouse line that pairs the expression of firefly luciferase with quantifiable expression of a human placental alkaline phosphatase that is secreted into the serum (SeAP). With the use of this dual-modality bioreporter with a novel, inducible Pax7-CreER line for tracking muscle satellite cells, we demonstrate the longitudinal kinetics of muscle stem cell turnover, accounting for a doubling of the signal from satellite cell and progeny every 3.93 wk in the transition from adolescence to early adulthood. We also show that this dual-modality bioreporter can be incorporated in preclinical cancer models, whereby SeAP activity is reflective of tumor burden. Thus, this dual bioreporter permits both spatial localization and accurate quantification of biological processes in vivo even when the tissue of interest is deep within the animal.—Nishijo, K., Hosoyama, T., Bjornson, C. R. R., Schaffer, B. S., Prajapati, S. I., Bahadur, A. N., Hansen, M. S., Blandford, M. C., McCleish, A. T., Rubin, B. P., Epstein, J. A., Rando, T. A., Capecchi, M. R., Keller, C. Biomarker system for studying muscle, stem cells, and cancer in vivo.
Keywords: optical imaging, reporter gene, satellite cell
Genetically engineered mice are emerging as critical tools to study tissue physiology, stem cell dynamics, and tumor progression because of the reporter genes that can be incorporated into these preclinical models (1). Reporter genes exist that enable both visualization and quantification of a labeled set of cells (1,2,3,4,5,6,7), but the challenge remains to create a reporter system that performs both.
In this study, we introduce a mouse reporter system that facilitates dual spatial detection and quantification for cells of interest in conditional genetic models that use Cre/LoxP technology. Our engineered allele strongly expresses firefly luciferase and a human placental secreted alkaline phosphatase as tandem cistrons from a modified Rosa26 promoter. These reporters are amenable to noninvasive optical imaging and minimally invasive serum microtiter assays, respectively. In this study, we characterize the performance of this mouse line and demonstrate applications in studying cancer biology as well as longitudinal muscle stem cell kinetics using a novel, highly stringent tamoxifen-inducible CreER mouse line specific for muscle satellite cells. This dual-modality bioreporter system promises to have a wide range of application in preclinical models of disease where higher precision and a shorter time to study end point are desirable.
MATERIALS AND METHODS
Generation of dual-modality bioreporter mouse line
For the LUSEAP reporter mouse line, the pRosa26-1 plasmid containing genomic DNA for the Rosa26 locus was obtained from Phillipe Soriano (Mount Sinai School of Medicine, New York, NY, USA). A targeting vector was constructed that consisted of (in 5′ to 3′ order): 1.1 kb of 5′ Rosa26 homology up to the XbaI site in intron 1, the CMV immediate-early promoter/enhancer, and the SV40 late viral protein gene 16S/19S splice donor and acceptor signal sites, a stop cassette consisting of six copies of the SV40 viral early and late polyadenylation signal flanked by LoxP sites, the firefly luciferase gene (pGL3; Promega, Madison, WI, USA), a human internal ribosome entry site (IRES) (8) from the NF-kB repressing factor (8), and the human placental secreted alkaline phosphatase gene (pSEAP2-Basic, Clontech, Mountain View, CA, USA), a flippase recognition target (FRT)-flanked neomycin resistance gene (Neo; ref. 9), 4.2 kb of 3′ Rosa26 homology, and the PYF enhancer driving the thymidine kinase gene (9). The linearized targeting vector was electroporated into R1 mouse embryonic stem cells, and the cells were subjected to positive and negative selection. A correctly targeted clone was identified by a downshift from 11-9 kb by Southern hybridization using a 5′ external probe (10) and digestion by EcoRV. The EcoRV site within the construct was contained in the FRT-flanked Neo cassette (9). Cells from this embryonic stem cell clone were microinjected into C57BL/6 blastocysts to generate chimeric mice. Chimeric mice were mated to C57BL/6 dams, and their agouti offspring were confirmed to harbor the targeted allele by Southern hybridization (data not shown).Germline mice were designated to have the genotype Rosa26LUSEAPp/WT. The FRT-flanked Neo cassette was removed by breeding Rosa26LUSEAPp/WT mice to transgenic mice expressing Flp-e (11), thereby generating Rosa26LUSEAPm/WT mice (i.e., Neo−).
Generation of tamoxifen-inducible Pax7 Cre mouse line
For the Pax7-CreER mouse line, a targeting vector was constructed that placed an ires-CreERTM-FRT-Neo-FRT cassette in the ClaI site within the 3′ untranslated region of the Pax7 gene following the stop codon in exon 9. CreERTM gene was graciously provided by Andrew McMahon (Harvard University, Boston, MA, USA) (12). The Pax7-CreER targeting vector was electroporated into R1 embryonic stem cells that were then subjected to positive and negative selection. One of 144 clones was identified as correctly targeted by screening with EcoRI genomic DNA digests with Southern hybridization using a 3′external probe, as described previously for a similar Pax7-Cre mouse line (13). Germline Pax7CreER mice were generated as described above for the LUSEAP reporter mouse line.
Other mouse lines
All mice used were maintained, crossed, genotyped, injected, and sacrificed in accordance with an approved Institutional Animal Care and Use Committee protocol at the University of Texas Health Science Center at San Antonio or the Veterinary Medical Unit guidelines at the Veteran’s Administration Health Care System in Palo Alto. Z/RED Cre/LoxP reporter mice expressing the monomeric DsRED-MST red fluorescent protein (mRFP) (14) were graciously provided by the Andras Nagy laboratory (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, ON, Canada). Myf6ICNm (Myf6-Cre) and Pax7ICNm (Pax7-Cre) mouse lines were generated as described previously (9, 13). The MCre-TgCre mouse line expressing Cre in the hypaxial subdomain of Pax3 has been described previously (15). The HPRTCre mouse line expressing Cre ubiquitously (16) was generously provided by the Jeff Mann laboratory (City of Hope Beckman Research Institute, Durarte, CA, USA). The Rosa26tm1(EYFP)Cos and Rosa26tm1Sor Cre/LoxP reporter mice that express eYFP or β-galactosidase (βGal), respectively, after Cre expression have also been previously described (10, 17).
Genotyping
For genotyping the LUSEAP mouse line, the 5′ primers were ck360, 5′-AAAGTCGCTCTGAGTTGTTATCA-3′; ph49, 5′-CCGCCAGATTCTGACATGGA-3′, and ph51, 5′-GCGCACCCGGGTTACTCTA-3′. The 3′ primers were ph50, 5′-TTCCAGGAACCAGGGCGTAT-3′; ph52, 5′-CAGAAGACTCCCGCCCATCT-3′; and ba97, 5′-GATCTGGACGAAGAGCATCA-3′. The primer ba97 is only necessary for detection of the Rosa26LUSEAPp allele. DNA was extracted from tails, and 2 μl (5-20 ng) was used in the subsequent PCR reaction. Each 25-μl PCR reaction contained 1× buffer, 2 mM MgCl2, 200 μM deoxynucleotides, 0.2 μM primers, and 0.4 U of Taq DNA polymerase (Promega). Cycling conditions were as follows: 95°C for 5 min, 32 cycles of 95°C for 30 s/64°C for 20 s/72°C for 120 s, followed by 72°C for 7 min. The wild-type, LUSEAPp (Neo+), or LUSEAPm (Neo−), and GoLUSEAP (Cre-activated) alleles resulted in 238-, 668-, 354-, or 391-bp bands, respectively.
For genotyping the Z/RED mouse line, the 5′ primers were js001, 5′-CAACGTGCTGGTTATTGTGC-3′, and ck167, 5′-AAAGTCGCTCTGAGTTGTTAT-3′. The 3′ primers were kn181, 5′-TCAGGAAGATCGCACTCCAG-3′, js002, 5′-ACCTTGAAGCGCATGAACTC-3′, and ck166, 5′-GGAGCGGGAGAAATGGATATG-3′. The ck166 and ck167 primers amplify a 584-bp PCR control band. DNA was extracted from tails, and 2 μl (5-20 ng) was used in the subsequent PCR reaction. Each 25-μl PCR reaction contained 1× buffer, 2.5 mM MgCl2, 200 uM deoxynucleotides, 0.2 μM primers, and 0.4 U of TaqDNA polymerase (Promega). Cycling conditions were as follows: 95°C for 5 min; 30 cycles of 95°C for 30 s, 60°C for 20 s, and 72°C for 60 s; followed by 72°C for 7 min. The unactivated BGeo allele or activated GoZRED alleles generated 390- or 225-bp bands, respectively.
For genotyping the Pax7-CreER mouse line, animals were genotyped using the 5′ primers ck118, 5′-GCTCTGGATACACCTGAGTCT-3′, and ck378, 5′-CGCATGGAGCATCTCTACAAC-3′; and the 3′ primers ck256, 5′-TCGGCCTTCTTCTAGGTTCTGCTC-3′, and ck379, 5′-GGATCGGATATCGAAGTTCC-3′. Reaction conditions were as described above except that 0.4 μM of each primer was used. Cycling conditions were as follows: 95°C for 5 min; 32 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 50 s; followed by 72°C for 7 min. The presence of the wild-type allele resulted in a 465-bp band, whereas presence of the CreERp or CreERm alleles resulted in 352- and 266-bp bands, respectively.
Genotyping for the Myf6ICNm, Pax7ICNm, MCre-TgCre, HPRTCre, Rosa26tm1(EYFP)Cos, and Rosa26tm1Sor mice have all been previously described (9, 10, 13, 15,16,17).
Tumor model
The Pax3:Fkhr and p53 alleles for conditional models of alveolar rhabdomyosarcoma have been previously described (9). Mice were visually inspected every 2 d for tumors because of the fulminant onset in this model.
Optical imaging for luminescence
Luminescent imaging of live female mice was performed using Xenogen IVIS-Spectrum system (Caliper; Xenogen, Hopkinton, MA, USA). The animals were maintained under inhaled anesthesia using 2% isoflurane in 100% oxygen at the rate of 2.5 L/min. For firefly luciferase imaging, the image acquisition parameters were 50-s exposure time, 2 × 2 binning, 12.6-cm field of view, and f/stop of 1/4. Data were acquired and analyzed using the manufacturer’s proprietary Living Image 2.5 software.
Tamoxifen induction for CreER animals
To induce Cre in satellite cells in vivo for single myofiber isolation experiments, 200 μl of 10 mg/ml tamoxifen (Sigma-Aldrich, St. Louis, MO, USA) suspended in corn oil was injected intraperitoneally into 6-wk-old Pax7CreERp/WTRosa26tm1Sor/WT mice 1×/d for 5 d at a dose of 2 mg/20 g body weight/d. Rosa26tm1Sor mice (10) express βGal in cells that have expressed Cre, which in this case includes Pax7-expressing satellite cells that have been exposed to tamoxifen. Uninduced littermates of the same genotype were injected with corn oil as a vehicle control. Mice were sacrificed 9 d after the final tamoxifen dose, and extensor digitorum longus muscles were harvested for myofiber preparation and immunocytochemistry as described below. The same tamoxifen injection procedure was used in the induction of eYFP expression in satellite cells of Pax7CreERp/WTRosa26tm1(EYFP)Cos mice (4 wk old) for the lineage tracing experiment. Mice were sacrificed at time 0 (before tamoxifen injection) and at 2 (14 d after the first tamoxifen injection), 6, and 12 wk, and the tibialis anterior muscles were harvested for immunohistochemistry. The number of cells expressing eYFP, Pax7, and MyoD in each section was counted at each time point. Fields used in cell counting were randomly chosen from different muscle sections, and at least 100 of the cells expressing Pax7 or MyoD were counted.
To trigger the dual-modality bioreporter in Pax7CreERp/WT Rosa26LUSEAPm/WT mice for longitudinal studies, 4-wk-old animals were given tamoxifen 2 mg/20 g body weight for 5 d. After tamoxifen, animals were imaged serially at the stated time points, whereby time 0 and time 2 wk are 0 and 14 d relative to the first tamoxifen injection, respectively.
Single myofiber preparations
Extensor digitorum longus muscles were isolated from injected Pax7CreERp/WTRosa26tm1Sor/WT mice as described previously (18, 19). Single myofibers were plated in each well of an ECM (Sigma-Aldrich)-coated 8-well chamber slide (Invitrogen, Carlsbad, CA, USA) in plating media (DMEM with 10% horse serum and 0.5% chick embryo extract) for either 2 or 24 h before being processed.
Primary satellite cell culture
Hind limb muscles were dissected from Pax7CreERp/WTRosa26LUSEAPm/WT mice (2 wk after tamoxifen injection), and satellite cells were isolated from these muscles as described previously (20). Cells were plated on poly-l-lysine (Sigma-Aldrich)- and fibronectin (Invitrogen)-coated 4-well chamber slides in 10% FBS/DMEM for 10 d. Cells were fixed at days 0, 1, and 10 for immunocytochemistry.
Immunocytochemistry
Cells isolated from Pax7CreERp/WTRosa26LUSEAPm/WT mice were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature. After being washed with PBS, cells were incubated in 5% normal goat serum (Invitrogen) and 0.1% Triton-X (Sigma-Aldrich) to inhibit nonspecific binding of antibodies. Primary antibodies were applied, and cells were incubated overnight at 4°C. The primary antibodies used were anti-Pax7, mouse monoclonal [1:200; Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA, USA]; anti-MyoD1, mouse monoclonal (1:100; 5.8A; Novocastra, Newcastle on Tyne, UK); anti-myogenin, mouse monoclonal (1:200; F5D; DSHB); anti-myosin heavy chain (MHC), mouse monoclonal (1:50; MF20; DSHB); and anti-luciferase, rabbit polyclonal (1:500: Abcam, Cambridge, MA, USA). After cells were washed with PBS, Alexafluor 594 conjugated anti-mouse IgG and Alexafluor 488 conjugated anti-rabbit IgG (1:200) were added and the cells were incubated for 60 min at room temperature. Myofibers were fixed for immunocytochemical analysis using 2% paraformaldehyde. Antibodies used were Syndecan4 (1:1500 dilution; a kind gift of Dr. Brad Olwin, University of Colorado, Boulder, CO, USA), Pax7 (1:100 dilution; DSHB), and βGal (1:2500 dilution; Cappel Laboratories, Cochranville, PA, USA; Covance, Berkeley, CA, USA). Fibers were visualized using an Axioskop 2 Plus microscope (Zeiss, Thornwood, NY, USA), and pictures were taken using an AxioCam camera operated with Axiovision software (Zeiss).
Immunohistochemistry
For immunohistochemistry of frozen skeletal muscle, staining was performed using the M.O.M. Immunodection Kit Staining Procedure (Vector Laboratories, Burlingame, CA, USA) following the manufacturer’s instructions.
The anti-Pax7 antibody (DSHB), anti-MyoD antibody (Novocastra), and anti-myogenin antibody (DSHB) were used at a concentration of 1:50. The anti-α2a laminin antibody (rabbit polyclonal, a kind gift from Dr. Peter D. Yurchenco, University of Medicine and Dentistry of New Jersey, Newark, NJ, USA) was used at a concentration of 1:500. The anti-GFP antibody (chicken polyclonal; Chemicon, Temecula, CA, USA) was used at a concentration of 1:200. Biotinylated anti-mouse IgG (1:250 dilution; Vector Laboratories), FITC-labeled anti-chicken IgG (1:200; Jackson ImmunoResearch Laboratories, West Grove, PA, USA), and Cy5-labeled anti-rabbit IgG (1:200; Jackson ImmunoResearch Laboratories) were used as secondary antibodies. Nuclei were counterstained with DAPI and hematoxylin. Slides were coverslipped with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL, USA) or DAPI mounting medium (Vector Laboratories) and visualized on an Olympus IX81 confocal microscope equipped with Fluoroview 1.6A software (Olympus America, Center Valley, PA, USA).
Serological bioassay measurement
For detection of SEAP in the bloodstream, a blood sample was isolated from the animal through saphenous vein puncture or retro-orbital bleed with minimal hemolysis. The blood was allowed to clot at room temperature for 30-60 min (min) and was centrifuged at 2500 g for 15 min at 4°C. The clear/yellow supernatant serum was assayed with the BD Great EscAPe SEAP chemiluminescent assay (Clontech) according to the manufacturer’s instructions, which includes a 30 min 65°C heating step to inactivate endogenous murine serum phosphatases. Assay samples containing 12.5 μl serum each were measured for luminescent signal using a Xenogen IVIS-Spectrum (Caliper; Xenogen). Imaging was performed 15 min after sample preparation with the standard settings of 60-s exposure time, 2 × 2 binning, 12.6-cm field of view, and f/stop of 2/4.
Statistical analysis
Comparisons between groups were done using a 2-tailed Student’s t test assuming equal variances.
RESULTS
Design and generation of a conditional dual-modality bioreporter and Pax7-CreER
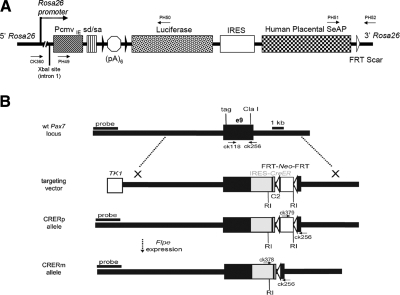
The conditional LUSEAP dual-modality bioreporter allele was designed to express both firefly luciferase and human placental secreted alkaline phosphatase constitutively at high levels after activation by Cre recombinase (Fig. 1A). The two reporter genes were expressed as tandem cistron by means of a human IRES (8). The construct was targeted to the Rosa26 locus in a manner similar to previous strategies (2, 10, 17) except that the native Rosa26 promoter was augmented by the CMV immediate-early promoter/enhancer and the SV40 late viral protein gene 16S/19S splice donor and acceptor signal sites (21) to ensure activity in muscle (22) and to maximize ubiquitous expression. Germline mice were established carrying the Neo-containing allele (Rosa26LUSEAPp/WT). After the removal of the Neo positive selection cassette by Flpe-mediated recombination, Neo-excised Rosa26LUSEAPm/WT mice proved to be viable and fertile as heterozygotes or homozygotes.
Figure 1.
Structure of targeted mouse lines and AAVcre. A) LUSEAP dual-modality bioreporter was targeted to the Rosa26 locus at the XbaI site in intron 1. In addition to the native Rosa26 promoter, the CMV immediate-early promoter/enhancer (PcmvIE) and the SV40 late viral protein gene 16S/19S splice donor and acceptor (sd/sa) signal sites (21) were added to strengthen reporter gene activity. To minimize leakage, a stop cassette consisting of 6 tandem copies of the SV40 viral early and late polyadenylation signal sequences (35) [(pA)6] were inserted downstream of the promoter elements, flanked by LoxP sites (black arrowheads). Stop cassette is followed by firefly luciferase, a human IRES (8), and human placental secreted alkaline phosphatase. White arrowhead indicates FRT scar. B) For the Pax7-CreER allele, a targeting vector was designed for insertion of a complex cassette into the 3′ region of the Pax7 gene. At the ClaI site in exon 10, an IRES-CreER was inserted to allow bicistronic expression of Pax7 and CreER. An FRT-Neo-FRT cassette was inserted 3′ to IRES-CreER. The CRERp allele indicates the presence of the neomycin resistance gene (Neo+), whereas the CRERm allele indicated the absence of the neomycin-resistance gene (Neo−) after breeding to a Flp-e deleter mouse line (11). RI, EcoRI restriction site.
The satellite cell specific tamoxifen inducible CreER allele (Fig. 1B) was designed to allow Cre expression from the Pax7 locus without interfering with the normal Pax7 function. Germline mice were established carrying the Neo-containing allele (Pax7CreERp/WT). Pax7CreERp/WT and Pax7CreER/CreERp mice were phenotypically normal and fertile. Sequencing of the 3′ untranslated region of the Pax7CreERp cDNA from skeletal muscle of these mice confirmed insertion of the ires-CreER cassette into exon 9 of Pax7 at the mRNA level (data not shown).
Ubiquitous activation of the reporter allele leads to the strong expression of luciferase
The optimal timing for imaging after luciferin injection was found to be 15-30 min when using a focally activated AAVcre with LUSEAP mice (Supplemental Figs. S1 and S2; Supplemental Results). To determine the whole body intensity and time course of luciferase expression, reporter mice carrying the Rosa26LUSEAPm/WT allele were bred to HPRPCre/WT mice expressing Cre ubiquitously (16). A 9-mo-old double heterozygote HPRTCre/WT Rosa26GoLUSEAP/WT mouse and wild-type control were injected with a single dose of luciferin (Supplemental Fig. S3). The maximum signal intensity from the luciferase in our bicistronic reporter was 2.55 × 1010 photons/cm2/s/sr, which is comparable to or greater than previously reported, useful monocistronic luciferase reporters (2, 3, 23). The dual-modality reporter was also found to have stepwise increments in luciferase and serum SeAP levels with increasing size of Cre activation domains (Supplemental Fig. S4), while outperforming a red fluorescent protein gene for signal-to-background (Supplemental Figs. S6 and S7).
Pax7-CreER is a temporally inducible tool for the study of satellite cell kinetics
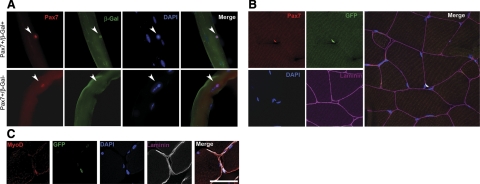
To determine the specificity of our Pax7-CreER mouse line for marking satellite cells, we first generated Pax7CreERp/WTRosa26tm1Sor/WT mice that would express βGal in the presence of tamoxifen-induced Cre activity. Adolescent mice were intraperitoneally injected with tamoxifen for 5 d, and then 9 d later, single myofibers were isolated, cultured, and examined by immunocytochemistry for coexpression of βGal and the satellite cell marker Pax7 (Fig. 2A; Table 1). Under these conditions, 46.9% of cells staining positive for Pax7 also stained positive for βGal 2 h after myofiber isolation. After 24 h in culture, βGal reactivity was seen in a greater proportion (82.7%) of Pax7-positive cells (see Discussion). All βGal+ cells examined coexpressed the satellite cell marker Pax7 (Table 1). Therefore, by this approach, Pax7-CreER mice were suggested to have Cre induction in approximately half of satellite cells in vivo.
Figure 2.
Expression of reporter protein in satellite cells and their progeny using a stringent satellite cell-specific Cre allele. A) Whole-mount immunofluorescence of single myofibers explanted from Pax7CreERp/WTRosa26tm1Sor/WT mice treated with tamoxifen. Satellite cell specificity for the Pax7CreER allele is demonstrated by coexpression of the Cre reporter βGal in cells that also express the satellite cell marker Pax7 (arrowheads, top 4 panels). Some satellite cells (Pax7+) did not express βGal (arrowheads, bottom 4 panels). Scale bar = 100 μm. B) Immunohistochemistry of skeletal muscle from Pax7CreERp/WTRosa26tm1(EYFP)Cos mouse (2 wk after tamoxifen injection). Pax7-expressing cell (red) and eYFP-expressing cell (green) are colocalized beneath basement membrane. C) Immunohistochemistry for MyoD and eYFP of skeletal muscle from Pax7CreERp/WTRosa26tm1(EYFP)Cos mouse (2 wk after first tamoxifen injection). These myoblasts/satellite cell progeny (green) express MyoD (red) and localizes inside of basal lamina. Scale bar = 50 μm.
TABLE 1.
Pax7 and βGal immunoreactivity of satellite cells isolated from single myofibers from Pax7CreERp/WT Rosa26 tm1Sor/WT mice harvested 14 d after initial tamoxifen administration
| Time after isolation | βGal+Pax7+ cells | Pax7+ cells counted | Proportion of βGal+ cells (%) |
|---|---|---|---|
| 2 h (induced) | 333 | 699 | 46.9 ± 2.36 |
| 2 h (uninduced) | 31 | 258 | 12.0 ± 3.72 |
| 24 h (induced) | 87 | 105 | 82.7 ± 0.6 |
| 24 h (uninduced) | 43 | 371 | 10.3 ± 1.7 |
One hundred percent of cells that express βgal stained positively for Pax7.
As a secondary in situ method to determine the stringency, inducibility, and myogenic lineage specificity of the Pax7-CreER mouse line in living muscle under noninjury conditions, we next generated Pax7CreERp/WTRosa26tm1(EYFP)Cos mice that would express eYFP in the presence of tamoxifen-induced Cre activity. Tibialis anterior muscles of Pax7CreERp/WTRosa26tm1(EYFP)Cos mice were dissected at times 0, 2, 6, and 12 wk relative to the first tamoxifen injection and then analyzed by immunohistochemistry for coexpression of eYFP and the myogenic markers Pax7 (for satellite cells) and MyoD (for myoblasts; Table 2). All Pax7+/eYFP+ cells were localized beneath the basement membrane, which is characteristic of satellite cells (24), whereas MyoD+/eYFP+ cells were located either inside or outside muscle fibers (Fig. 2B, C). In quantitative analysis, <9.4% of the Pax7+ or MyoD+ cells were eYFP-positive at time zero (Table 3), suggesting reasonable stringency in the absence of tamoxifen. At 2 wk after tamoxifen, 61% of Pax7+ cells had been induced to express eYFP. Conversely, at the same time point, 60% of eYFP+ cells were Pax7− (Table 4), and this population likely overlaps with the 55% of myoblasts detected to be both MyoD+ and eYFP+ (Table 3). With respect to myoblasts, even as late as 12 wk, 47% of eYFP+ cells were Pax7− (Table 4), with 67% of eYFP+ cells being MyoD+ (Table 3), suggesting that marked MyoD+ myoblasts persist even 3 mo after tamoxifen injection, at least in the context of the tested adolescent/young adulthood stage of life under noninjury conditions.
TABLE 2.
Pax7, MyoD, and eYFP immunoreactivity of satellite cells and myoblasts in situ for Pax7CreERp/WT Rosa26tm1(EYFP)Cos/WT mice after tamoxifen administration
| Week | Pax7+ or eYFP+ cells counted | Pax7+ and eYFP+ | Pax7+ and eYFP− | Pax7− and eYFP+ | MyoD+ or eYFP+ cells counted | MyoD+ and eYFP+ | MyoD+ and eYFP− | MyoD− and eYFP+ |
|---|---|---|---|---|---|---|---|---|
| 0 | 106 | 0 | 106 | 0 | 106 | 9 | 96 | 1 |
| 2 | 192 | 61 | 39 | 92 | 155 | 55 | 45 | 55 |
| 6 | 142 | 50 | 50 | 43 | 172 | 68 | 32 | 72 |
| 12 | 150 | 57 | 43 | 50 | 171 | 68 | 33 | 70 |
At least 100 Pax7+ or MyoD+ cells were counted in several randomly chosen fields in muscle cross section.
TABLE 3.
Proportions and timing of eYFP reactivity for Pax7+ satellite cells and MyoD+ myoblasts
| Week | eYFP+ Pax7+ cells | eYFP+ MyoD+ cells |
|---|---|---|
| 0 | 0 of 106 (0%) | 10 of 106 (9.4%) |
| 2 | 61 of 100 (61%) | 55 of 100 (55%) |
| 6 | 50 of 100 (50%) | 68 of 100 (68%) |
| 12 | 57 of 100 (57%) | 68 of 101 (67%) |
These are the same experimental results as Table 2.
TABLE 4.
Proportions and timing of eYFP reactivity for Pax7− or MyoD− cell populations
| Week | Pax7− eYFP+ cells | MyoD− eYFP+ cells |
|---|---|---|
| 0 | 0 of 0 (0%) | 1 of 10 (10%) |
| 2 | 92 of 153 (60%) | 55 of 110 (50%) |
| 6 | 43 of 93 (46%) | 72 of 140 (51%) |
| 12 | 50 of 107 (47%) | 70 of 138 (51%) |
These are the same experimental results as Table 2.
Application of LUSEAP dual-modality bioreporter in the study for satellite cell kinetics
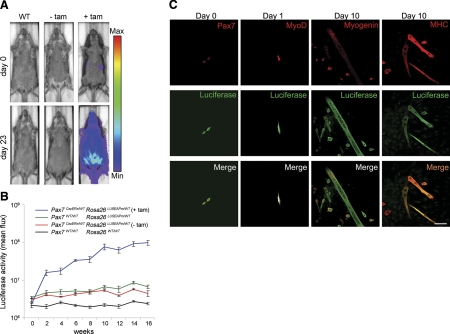
To monitor satellite cell longitudinal kinetics in growing postnatal muscle, tamoxifen was intraperitonealy injected into 4-wk-old Pax7CreERp/WTRosa26LUSEAPm/WT mice and luciferase activity was measured every 2 wk after tamoxifen injection. As a representative time point, these Pax7CreERp/WTRosa26LUSEAPm/WT mice had a very low background level of luciferase expression in the absence of tamoxifen, but their luciferase expression increased within 23 d after tamoxifen administration (Fig. 3A). Luciferase activity of tamoxifen-injected Pax7CreERp/WTRosa26LUSEAPm/WT mice gradually increased in comparison with controls over time (Fig. 3B). From weeks 2 to 12 (relative to the first tamoxifen injection), a 5.84-fold increase in luminescence was measured, suggesting a doubling of signal from satellite cells and progeny every 3.93 wk during the transition from adolescence to young adulthood. Unfortunately, however, incremental changes in serum SeAP did not reach statistical significance for these muscle stem cell tracing experiments (data not shown, see Discussion).
Figure 3.
Application of the dual-modality bioreporter for stem cell biology using a stringent satellite cell-specific Cre allele. A) Representative example of qualitative luciferase signal from satellite cells in Pax7CreERp/WTRosa26LUSEAPm/WT mice treated with a tamoxifen pulse at 6 wk of age vs. controls. Bioluminescence is seen predominantly from the ventral (abdominal) musculature but only in tamoxifen-treated animals. Images are displayed at a minimum-maximum scale of 2 × 105 to 5 × 106 photons/s/cm/sr. B) Time course of quantitative bioluminescence for Pax7CreERp/WTRosa26LUSEAPm/WT mice after tamoxifen injection. Tamoxifen was intraperitoneally injected at 4 wk after birth and then luciferase signal was analyzed every 2 wk. Lines represent Pax7CreERp/WTRosa26LUSEAPm/WT (with tamoxifen; blue; n=7), Rosa26LUSEAPm/WT (green; n=4), Pax7CreERp/WTRosa26LUSEAPm/WT (without tamoxifen; red; n=5), and wild type (black; n=4), respectively. C) Immunocytochemistry of cultured myogenic progenitors from Pax7CreERp/WTRosa26LUSEAPm/WT mice. Luciferase expression was detected in Pax7-expressing satellite cells (column 1), MyoD-expressing myoblasts (column 2), myogenin-expressing immature myotubes (row 3), and MHC-expressing mature myotubes (row 4).
To demonstrate that luciferase expression from Rosa26 promoter is maintained in myogenic lineages after commitment to differentiation, myogenic progenitors were isolated from tibialis anterior muscle of Pax7CreERp/WTRosa26LUSEAPm/WT mice (Fig. 3C) and cultured under conditions that allow differentiation. Luciferase expression was detected in Pax7-expressing satellite cells and MyoD-expressing myoblasts. Luciferase expression was also confirmed in myogenin-expressing immature myotubes and MHC-expressing mature myotubes 10 d after isolation. Furthermore, direct detection of luciferase luminescence from isolated satellite cells, myoblasts, and myofibers after luciferin administration indicated the same level of luminescence of all cell types when normalized for cross-sectional area, but an absolute increase in luminescence as satellite cell derived cells become more differentiated and larger (Supplemental Fig. S5). In these in vivo experiments, myoblasts and myotubes had 2.5- and 7-fold more total cellular luminescence than satellite cells. However, we were careful not to extrapolate such findings immediately to the in vivo setting, wherein the size and behavior of marked myofibers may differ from in vitro myotubes.
LUSEAP dual-modality bioreporter can be applied to preclinical cancer models
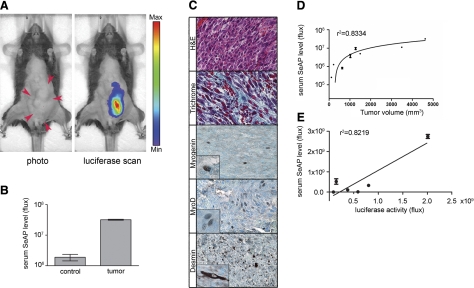
To demonstrate the utility of the dual-modality bioreporter for preclinical cancer models, we incorporated the dual-modality bioreporter into new models of the childhood muscle cancer rhabdomyosarcoma. Previous rhabdomyosarcoma models combined Myf6-Cre mediated activation of the translocation-mediated oncogene Pax3:Fkhr with concurrent inactivation of p53 in maturing skeletal muscle (9, 25). In this experiment, Myf6-Cre was replaced with Pax7-CreER and tamoxifen was given to the mice 4 wk after birth to activate Pax3:Fkhr and inactivate p53 in postnatal satellite cells. In a litter of Pax7CreERp/WTPax3P3Fm/P3FmTrp53F2-10/F2-10Rosa26LUSEAPm/WT mice, an animal developed an invasive abdominal tumor 6 mo after tamoxifen administration (Fig. 4). A whole body luciferase scan of the mouse clearly demonstrated the presence of the tumor with minimal background in other parts of the body (Fig. 4A). In addition, serum SeAP assay of the tumor mouse showed a 26-fold increase relative to an age-matched control mouse with the same genotype (Fig. 4B). Histological diagnosis of rhabdomyosarcoma was confirmed by hematoxylin and eosin staining and positive immunohistochemical staining for myogenin, MyoD, and desmin (Fig. 4C). The collagen-rich stroma of human rhabdomyosarcoma is also reflected in this model, as demonstrated by Masson Trichrome staining of the mouse tumor (Fig. 4C). We have also tested another Cre line, MCre (15), incorporated into a rhabdomyosarcoma mouse model for the purpose of comparing mRFP (Z/Red) and the LUSEAP reporter alleles. This MCre line activates Cre recombination in the Pax3 hypaxial (limb) muscle domain. In a litter of MCre-TgCre/WTPaxP3Fm/P3FmTrp53F2-10/F2-10 Rosa26LUSEAPm/WTZ/RED-TgGoZRED/WT mice, an animal developed an invasive right thigh tumor at age 5 ½ mo. The tumor appeared small at the surface, but at necropsy the tumor was found to nearly replace the thigh, measuring 3.978 cc. A photograph of the tumor-bearing animal is shown alongside mRFP imaging and luciferase imaging in Supplemental Fig. S8A. In the mRFP image of this partially shaved brown mouse, a higher intensity signal is seen from the region of the tumor, but a strong background signal is seen both from midline abdominal muscles as well as unshaved fur. Luminescence imaging of the dual-modality bioreporter showed a high intensity signal from the tumor, a moderate background signal from skeletal muscle in the shaved regions, but no background from the fur coat. Diagnosis of alveolar rhabdomyosarcoma was confirmed by histology and immunohistochemistry for myogenin (Supplemental Fig. S8B, C). With the use of the SeAP serum assay, alkaline phosphatase activity measured by a chemiluminescence assay was nearly 100-fold higher in the tumor-bearing animal than an age-matched littermate control with the same genotype but no tumor (Supplemental Fig. S8D). Normalized to tumor size, the SeAP activity correlated to 2.85 × 108 photons/s/cc of tumor.
Figure 4.
Application of dual-modality bioreporter for cancer biology. A) Abdominal tumor arising in a Pax7CreERp/WTPax3P3Fm/P3Fm Trp53F2-10/F2-10Rosa26LUSEAPm/WT 6-allele mouse allows direct comparison of luciferase and SeAP from dual-modality bioreporter. Tumor is indicated in photograph by red arrows. In luciferase scan panel, bioluminescence from the primary tumor is clearly observed with minimal background luciferase signal in the normal tissue. Image is displayed at a minimum-maximum scale of 3 × 107 to 2 × 108 photons/s/cm2/sr. B) Serum SeAP activity of the tumor-bearing animal (same animal as in A) is 26-fold higher than an age-matched nontumor animal with the same genotype (P<0.001). C) Histopathology confirmed the diagnosis of rhabdomyosarcoma. Hematoxylin and eosin staining shows the primary tumor. MyoD and Desmin are strongly positive, whereas myogenin is focally positive. Gomori trichrome staining indicates a collagen-rich stroma. D, E) Positive correlation was seen between serum SeAP level and tumor volumes (D) and between serum SeAP level and luciferase intensity (E) in Pax7CreERp/WTPax3P3Fm/P3FmTrp53F2-10/F2-10Rosa26LUSEAPm/WT tumor mice.
To further validate the usefulness of this dual-modality reporter in cancer mouse models, we investigated correlations between tumor volumes, tumor luciferase signal, and serum SeAP levels. Seven tumor mice with the Rosa26LUSEAPm/WT were studied. A positive correlation was observed in SeAP level vs. tumor volume (r2=0.8334; Fig. 4D), SeAP vs. luciferase signal (r2=0.8219; Fig. 4E), and luciferase signal vs. tumor volume (r2=0.5805; graph not shown). The relationship between SeAP and tumor size was nonlinear, but the relationship between luciferase and SeAP levels was linear. We have observed that discordance between the reporter gene level of viable tumor cells and overall tumor size in cases where necrosis and intratumoral hemorrhage are present (Supplemental Fig. S9).
DISCUSSION
In this study, we report a dual-modality bioreporter mouse strain that pairs semiquantitative, spatially specific expression of firefly luciferase with quantitative expression of a serum biomarker, human placental secreted alkaline phosphatase. By modifying the Rosa26 native promoter, we achieved strong and sustained expression of firefly luciferase that is comparable to or greater than previously reported Cre/LoxP luciferase reporter alleles (2, 3). The time course of luciferase activity permits optimal detection of luminescence within 15 to 30 min after luciferin injection. Furthermore, both the luciferase and SeAP reporters have a broad range of signal over background: 4.4 × 102 for luciferase, and 1.1 × 102 for SeAP (Supplemental Fig. S4). Comparison with biocompatible red fluorescent protein reporter mouse strains suggests selected advantages of our dual-modality bioreporter in terms of signal over background. In addition, we demonstrate the application of this dual-modality bioreporter to measuring longitudinal stem cell kinetics and tumor burden. For the former, we have examined the satellite cell kinetics in the adolescent and young adult muscle growth phase in vivo using newly developed dual reporter mice. We show that signal from the muscle stem cell lineage doubles every 3.93 wk during this growth phase (to equate this to a number of cell divisions will require future in vivo studies quantifying luciferase expression from satellite cells vs. myoblasts vs. myofibers). Our studies also reveal the surprising result that a significant MyoD+ population remains expanded as late as 12 wk after satellite cell marking, at least in the context of adolescence and young adulthood under noninjury conditions. Thus, the Pax7-CreER mouse line, being stringent and highly inducible, is a unique and powerful tool that will extend the study of satellite cell injury kinetics (26) to new investigations of noninjury kinetics.
For our ex vivo experiments evaluating Pax7-CreER stringency and inducibility, we observed that substantial activation of a βGal reporter gene occurs in the absence of tamoxifen during the exposure to culture conditions; possible explanations include but are not limited to carryover of tamoxifen or its active metabolites 9 d after administration (terminal half-life ∼7-11 d; refs. 27, 28), culture-dependent ongoing ex vivo Cre/Lox recombination and reporter gene transcription and translation, increased Rosa26 promoter activity in the absence of additional Cre/LoxP recombination (as satellite cells go from a quiescent to an activated state), or improved βGal immunoreactivity as a result of extended cell culture. The variability of ex vivo results led us to perform in situ experiments from skeletal muscle, which affirm a leakiness of 10% or less and an inducibility of 61% in Pax7+ satellite cells. In related experiments, we have found that Pax7-CreER recombination can be higher or lower for floxed alleles at other loci; therefore, we recommend that tamoxifen dose be adjusted for each experimental design with the Pax7-CreER mouse line, being careful not to grossly exceed 2 mg/20 g body weight × 5 d, which is near the limit of that which is tolerated in mice in our experience (data not shown).
Reporter systems for conditional genetic mouse models of disease are of increasing importance because conditional models have the potential to more accurately reflect the behavior of human disease. Murine reporter systems for positron emission tomography (1) and magnetic resonance imaging (29) have been reported and shown to have outstanding sensitivity and resolution, respectively. Nevertheless, the availability, lower cost, and simplicity of noninvasive optical imaging instruments make luminescence and fluorescence more practical for many preclinical experiments in academia and industry. Fluorescent proteins have lower quantum yields and depths of penetration than luciferases, even when red shifted (14). Luminescence is generally the most sensitive optical reporter modality, even though scattering in living tissue limits spatial resolution and reduces the signal from deep structures. A disadvantage of luminescence, which makes quantification problematic, is that luciferases require oxygen and an intact blood supply to deliver their exogenous substrate, luciferin (for firefly luciferase) or coelenterazine (for Renilla luciferase or the robust humanized Gaussia luciferase; ref. 30). For preclinical tumor studies or after stem cell niches (31), quantitative measurement of luciferase could be skewed by transient or sustained hypoxia. This theoretical concern may not be a practical problem for the study of muscle stem cells, which we measured by luminescence very readily. Nevertheless, at necropsy only a short window of time exists to image organs for luminescence ex vivo, and at such times when the skin can be removed a red-shifted fluorescent protein is probably preferred (14, 32). Thus, luciferase-RFP fusion proteins hold special promise for a wider range of applications (33). For in situ detection (immunohistochemistry) of cytoplasm-poor satellite cells, nuclear-localized reporter gene products are probably preferred (34).
Quantitative detection and serial monitoring of cells or tissue of interest can be significantly improved using genetically engineered serological biomarkers because these biomarkers do not necessarily depend on oxygen as a cofactor, can be quantified easily using microtiter assays, and may permit shorter term experiments because they obviate the need to have survival as an experimental end point (6). A wide range of clinically measurable, engineered ectopic biomarkers are possible, including α-fetoprotein (7), β-chorionic gonadotropin, prostate-specific antigen, β-2-microglobulin, vanillylmandelic acid, and CA125. In our studies, signal background allowed secreted alkaline phosphatase to be a useful serum biomarker for cancer studies but not stem cell studies. We are currently pursuing antigen-based detection of SeAP in lieu of SeAP enzymatic activity to improve serum biomarker measurement. We envision that this or other serological and urine biomarker reporter systems (“BioReporters”) will become important tools in both xenograft and conditional genetic preclinical models in academia and industry. With the increasing acceptance of conditional genetic models of disease, dual-modality bioreporter systems such as the one reported here are anticipated to expand the scope of what can be achieved in the study of postnatal stem cell physiology as well as in preclinical models of human disease, including muscular dystrophy, sarcopenia, and cancer.
Supplementary Material
Acknowledgments
An application of this reporter system for discovery of spontaneous biomarkers is being patented by University of Texas Health Science Center, San Antonio, and has been licensed for commercial distribution by Numira Biosciences (www.numirabio.com), of which C.K. is a cofounder. Numira Biosciences provided partial funding for these studies. A.T.M. was supported by National Institute of General Medical Sciences grants NIGMS MARC-U*STAR GM-07717 and NIGMS MBRS-RISE GM-60655.
References
- Massoud T F, Gambhir S S. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- Safran M, Kim W Y, Kung A L, Horner J W, DePinho R A, Kaelin W G., Jr Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- Lyons S K, Meuwissen R, Krimpenfort P, Berns A. The generation of a conditional reporter that enables bioluminescence imaging of Cre/loxP-dependent tumorigenesis in mice. Cancer Res. 2003;63:7042–7046. [PubMed] [Google Scholar]
- Nilsson E E, Westfall S D, McDonald C, Lison T, Sadler-Riggleman I, Skinner M K. An in vivo mouse reporter gene (human secreted alkaline phosphatase) model to monitor ovarian tumor growth and response to therapeutics. Cancer Chemother Pharmacol. 2002;49:93–100. doi: 10.1007/s00280-001-0396-0. [DOI] [PubMed] [Google Scholar]
- Chaudhuri T R, Cao Z, Krasnykh V N, Stargel A V, Belousova N, Partridge E E, Zinn K R. Blood-based screening and light based imaging for the early detection and monitoring of ovarian cancer xenografts. Technol Cancer Res Treat. 2003;2:171–180. doi: 10.1177/153303460300200214. [DOI] [PubMed] [Google Scholar]
- Bao R, Selvakumaran M, Hamilton T C. Use of a surrogate marker (human secreted alkaline phosphatase) to monitor in vivo tumor growth and anticancer drug efficacy in ovarian cancer xenografts. Gynecol Oncol. 2000;78:373–379. doi: 10.1006/gyno.2000.5925. [DOI] [PubMed] [Google Scholar]
- O'Neal W K, Rose E, Zhou H, Langston C, Rice K, Carey D, Beaudet A L. Multiple advantages of alpha-fetoprotein as a marker for in vivo gene transfer. Mol Ther. 2000;2:640–648. doi: 10.1006/mthe.2000.0198. [DOI] [PubMed] [Google Scholar]
- Oumard A, Hennecke M, Hauser H, Nourbakhsh M. Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol Cell Biol. 2000;20:2755–2759. doi: 10.1128/mcb.20.8.2755-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel B R, Coffin C M, El-Bardeesy N, DePinho R A, Capecchi M R. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Rodriguez C I, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart A F, Dymecki S M. High-efficiency deleter mice show that FLPe is an alternative to Cre- loxP [letter] Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon A P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Keller C, Hansen M S, Coffin C M, Capecchi M R. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, Nagy A. Mouse in red: red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- Brown C B, Engleka K A, Wenning J, Min Lu M, Epstein J A. Identification of a hypaxial somite enhancer element regulating Pax3 expression in migrating myoblasts and characterization of hypaxial muscle Cre transgenic mice. Genesis. 2005;41:202–209. doi: 10.1002/gene.20116. [DOI] [PubMed] [Google Scholar]
- Tang S H, Silva F J, Tsark W M, Mann J R. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin C S, William C M, Tanabe Y, Jessell T M, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Dev Biol. 1986;115:129–139. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J D, Lunt A I, Parry D J, Partridge T A. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- Allen R E, Rankin L L, Greene E A, Boxhorn L K, Johnson S E, Taylor R G, Pierce P R. Desmin is present in proliferating rat muscle satellite cells but not in bovine muscle satellite cells. J Cell Physiol. 1991;149:525–535. doi: 10.1002/jcp.1041490323. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton-Davis E R, Shoturma D I, Musaro A, Rosenthal N, Sweeney H L. Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc Natl Acad Sci U S A. 1998;95:15603–15607. doi: 10.1073/pnas.95.26.15603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrbom L, Nerio E, Holland E C. Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nat Med. 2004;10:1257–1260. doi: 10.1038/nm1120. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Capecchi M R. New genetic tactics to model alveolar rhabdomyosarcoma in the mouse. Cancer Res. 2005;65:7530–7532. doi: 10.1158/0008-5472.CAN-05-0477. [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau H M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregorio M W, Coronado E, Osborne C K. Tumor and serum tamoxifen concentrations in the athymic nude mouse. Cancer Chemo Pharmacol. 1989;23:68–70. doi: 10.1007/BF00273519. [DOI] [PubMed] [Google Scholar]
- Lien E A, Solheim E, Lea O A, Lundgren S, Kvinnsland S, Ueland P M. Distribution of 4-hydroxy-N-desmethyltamoxifen and other tamoxifen metabolites in human biological fluids during tamoxifen treatment. Cancer Res. 1989;49:2175–2183. [PubMed] [Google Scholar]
- Louie A Y, Huber M M, Ahrens E T, Rothbacher U, Moats R, Jacobs R E, Fraser S E, Meade T J. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- Tannous B A, Kim D E, Fernandez J L, Weissleder R, Breakefield X O. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Ceradini D J, Kulkarni A R, Callaghan M J, Tepper O M, Bastidas N, Kleinman M E, Capla J M, Galiano R D, Levine J P, Gurtner G C. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling H J. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2006;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Ray P, De A, Min J J, Tsien R Y, Gambhir S S. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoller J Z, Degenhardt K R, Huang L, Zhou D D, Lu M M, Epstein J A. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis. 2008;46:200–204. doi: 10.1002/dvg.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell I H, Harrison G S, Wood W M, Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques. 1989;7:276–280. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.