Abstract
Objectives
Low socioeconomic status (SES) is one of the most robust social factors associated with disease morbidity, including more severe asthma in childhood. However, our understanding of the biological processes that explain this link is limited. This study tested whether the social environment could get “under the skin” to alter genomic activity in children with asthma.
Design and participants
Two group design of children with physician diagnosed asthma who came from low or high SES families.
Outcomes
Genome-wide transcriptional profiles from T lymphocytes of children with asthma.
Results
Children with asthma from a low SES background showed overexpression of genes regulating inflammatory processes, including those involved in chemokine activity, stress responses and wound responses, compared with children with asthma from a high SES background. Bioinformatic analysis suggested that decreased activity of cyclic AMP response element binding protein and nuclear factor Y and increased nuclear factor κB transcriptional signalling mediated these effects. These pathways are known to regulate catecholamine and inflammatory signalling in immune cells.
Conclusions
This study provides the first evidence in a sample of paediatric patients diagnosed with asthma that the larger social environment can affect processes at the genomic level. Specifically, gene transcription control pathways that regulate inflammation and catecholamine signalling were found to vary by SES in children with asthma. Because these pathways are the primary targets of many asthma medications, these findings suggest that the larger social environment may alter molecular mechanisms that have implications for the efficacy of asthma therapeutics.
The larger social environment has been found to affect physical health in individuals across the lifespan. Across all social factors, low socioeconomic status (SES) exhibits one of the strongest and most consistent associations with morbidity and mortality across a wide range of diseases,1 including childhood asthma. In particular, children with asthma from a low SES background suffer from more frequent hospitalisations, emergency department visits and more functional impairment (eg, activity limitations, days spent in bed) compared with children from a higher SES background.2–8
In seeking to understand how the larger social environment “gets in the body” to impact on the pathophysiology of asthma, researchers have investigated relationships between the social environment and inflammation. In healthy adults, low SES has been linked to heightened levels of circulating inflammatory markers, such as C reactive protein and interleukin (IL)6.9 10 In children with asthma, low SES has been associated with greater eosinophil counts as well as heightened in vitro stimulated production of inflammatory cytokines implicated in asthma, such as IL5 and IL13.11 12 Another social factor associated with low SES—stress—has been linked to heightened inflammatory responses to allergen challenge in patients with asthma,13 and decreased expression of genes coding hormonal receptors that regulate inflammation, including the glucocorticoid and β adrenergic receptors.14
Although relationships between SES and inflammation are increasingly recognised, the molecular mechanisms underlying this relationship remain unclear. The present study utilised an in vivo genomics based approach to identify differences in T cell gene expression associated with SES in a sample of children with asthma, and to identify candidate signal transduction pathways that may shape those differences. Because the focus was on understanding disparities associated with disease progression, we targeted a sample of children with pre-existing asthma.
Based on previous research linking SES and inflammation, we hypothesised that low SES would be associated with alterations in proinflammatory transcription control pathways. Based on previous data implicating the β adrenergic and glucorticoid pathways in the relationship between social factors and inflammation,14 we hypothesised that transcriptional mediators of these types of hormonal signals would differentiate T cell gene expression profiles in children from low versus high SES backgrounds. As a second aim, we tested whether social factors related to SES, such as cognitive interpretations, could explain associations between SES and genome-wide transcriptional profiles. Our hypothesis was that SES colours the way in which children interpret their social world, and that these interpretations in turn alter neuroendocrine and inflammatory signalling processes indicated by genome-wide transcriptional profiles.
METHODS
Additional details are available in the supplement (online only).
Participants
Participants were recruited from an ongoing longitudinal study of children with asthma aged 9–18 years. A subset of 16 children from low SES and 15 from high SES backgrounds provided blood samples for this study. The low and high SES groups were identified based on those who fell into the bottom and top 15% on parent education and family income variables.
Measures
Background characteristics
Family SES was measured by parent report of annual family income and years of parental education. Medical information included use of asthma medications and asthma severity.
Interpretations of stress
The Cognitive Appraisal and Understanding of Social Events (CAUSE) videos depict age appropriate ambiguous life situations, such as being asked to talk with a teacher after class.15 Children’s interpretations are rated by judges, with higher numbers indicating greater perceived threat.
Gene expression arrays
CD2+ cells were isolated from mononuclear cells by immunomagnetic positive selection (Miltenyi Biotec; Auburn, California, USA), and total RNA was extracted (RNAlater/RNeasy; Qiagen, Valencia, California, USA). Genome-wide transcription profiling was conducted, as previously described.16–18 Differentially expressed genes were identified as those showing ≥30% difference in mean expression levels in children from low SES versus high SES backgrounds (false discovery rate of 15%).16 Functional commonalities among differentially expressed genes were identified using the GOstat bioinformatics tool (http://gostat.wehi.edu.au).19
Transcriptional mediation analyses
We used a two sample variant of the Transcription Element Listening System (TELiS) (http://www.telis.ucla.edu)17 to analyse differential gene expression data in terms of the prevalence of transcription factor binding motifs (TFBMs) within the promoters of differentially expressed genes, as previously described.20
Ancillary analyses used a multivariate generalisation of the analysis of covariance (ANCOVA)21 to control for potential confounders which could explain associations between SES and gene expression profiles. Covariates included demographic variables (age, gender), medical variables (asthma severity, use of inhaled corticosteroid medication, use of β agonist medication) and interpretations of stress (CAUSE video responses). To confirm that natural killer cell prevalence did not affect the results, the relative concentration of mRNAs encoding CD16 and CD56 were included as covariates.
RT-PCR confirmation
Ten transcripts identified as differentially expressed in microarray analyses were independently assayed by quantitative real-time RT-PCR using TaqMan Gene Expression Assays (Applied Biosystems, Foster City, California, USA), as described in the online supplement.
Clinical variables
Parents reported on children’s daytime and night-time symptoms over the previous 2 weeks, and whether their child went to the emergency department because of asthma in the past 6 months.
RESULTS
Descriptive information about the sample is shown in table 1. The low and high SES groups did not differ in terms of asthma severity, use of inhaled corticosteroids, use of β agonists, forced expiratory volume in 1 s or atopic status (all p values >0.15).
Table 1.
Demographic characteristics of the sample
| Low SES |
High SES |
|||||
|---|---|---|---|---|---|---|
| % | Mean | SD | % | Mean | SD | |
| Age (year) | 12.8 | 3.5 | 14.9 | 1.9 | ||
| Gender (% boys) | 63 | 85 | ||||
| Medications (% using each type) | ||||||
| Inhaled corticosteroid | 75 | 80 | ||||
| β agonist | 75 | 87 | ||||
| Atopic asthma (%) | 75 | 93 | ||||
| Asthma severity (1–4 scale) | 2.8 | 1.0 | 2.5 | 0.5 | ||
| FEV1% | 94.9 | 12.6 | 99.8 | 17.6 | ||
The groups did not differ significantly in terms of gender, medications, atopic asthma, asthma severity or FEV1% (p values >.0.15). Children in the high SES group were older than children in the low SES group (p = 0.047).
FEV1, forced expiratory volume in 1 s; SES, socioeconomic status.
Differential gene expression
Sixty transcripts were differentially expressed between the low and high SES groups, representing 56 distinct named human genes. Thirty-four reflected overexpression in the low SES group whereas 26 reflected overexpression in the high SES group (table 2).
Table 2.
Genes showing differential expression in children with asthma from low and high SES backgrounds
| Affymetrix probe ID | Gene symbol | Fold-change |
|---|---|---|
| 205048_s_at | PSPH | 2.49 |
| 202917_s_at | S100A8 | 1.80 |
| 221728_x_at | XIST | 1.65 |
| 220784_s_at | UTS2 | 1.61 |
| 209480_at | HLA-DQB1 | 1.59 |
| 214146_s_at | PPBP | 1.50 |
| 214218_s_at | XIST | 1.49 |
| 209116_x_at | HBB | 1.49 |
| 200782_at | ANXA5 | 1.47 |
| 201743_at | CD14 | 1.45 |
| 211696_x_at | HBB | 1.45 |
| 202912_at | ADM | 1.44 |
| 214414_x_at | HBA2 | 1.44 |
| 202651_at | LPGAT1 | 1.41 |
| 217941_s_at | ERBB2IP | 1.41 |
| 206390_x_at | PF4 | 1.41 |
| 202503_s_at | KIAA0101 | 1.41 |
| 202602_s_at | HTATSF1 | 1.40 |
| 215223_s_at | SOD2 | 1.40 |
| 217022_s_at | IGHA1///IGHA2 | 1.40 |
| 203535_at | S100A9 | 1.39 |
| 202589_at | TYMS | 1.38 |
| 221731_x_at | CSPG2 | 1.37 |
| 213831_at | HLA-DQA1 | 1.37 |
| 213376_at | ZBTB1 | 1.37 |
| 205863_at | S100A12 | 1.36 |
| 209773_s_at | RRM2 | 1.32 |
| 204192_at | CD37 | 1.32 |
| 217811_at | SELT | 1.31 |
| 217414_x_at | HBA1///HBA2 | 1.31 |
| 212406_s_at | PCMTD2 | 1.31 |
| 217232_x_at | HBB | 1.31 |
| 220330_s_at | SAMSN1 | 1.30 |
| 211745_x_at | HBA1 | 1.30 |
| 201464_x_at | JUN | 0.78 |
| 201236_s_at | BTG2 | 0.77 |
| 221833_at | LONPL | 0.77 |
| 205436_s_at | H2AFX | 0.77 |
| 214469_at | HIST1H2AE | 0.77 |
| 205376_at | INPP4B | 0.77 |
| 214131_at | CYorf15B | 0.77 |
| 200664_s_at | DNAJB1 | 0.77 |
| 214805_at | EIF4A1 | 0.77 |
| 206385_s_at | ANK3 | 0.76 |
| 201465_s_at | JUN | 0.75 |
| 206700_s_at | SMCY | 0.72 |
| 200799_at | HSPA1A | 0.72 |
| 204141_at | TUBB2A | 0.69 |
| 201693_s_at | EGR1 | 0.69 |
| 201694_s_at | EGR1 | 0.68 |
| 200800_s_at | HSPA1A///HSPA1B | 0.65 |
| 209189_at | FOS | 0.65 |
| 219629_at | FAM118A | 0.64 |
| 213418_at | HSPA6 | 0.63 |
| 202768_at | FOSB | 0.60 |
| 212671_s_at | HLA-DQA1///HLA-DQA2///LOC650946 | 0.60 |
| 205000_at | DDX3Y | 0.55 |
| 202581_at | HSPA1B | 0.50 |
| 201909_at | RPS4Y1 | 0.48 |
| 209728_at | HLA-DRB4 | 0.46 |
SES, socioeconomic status.
Genes showing overexpression in children with asthma from a low SES background contained themes of the following: immune response and inflammation (eg, S100A8, S100A9, S100A12, gene ontology terms: GO:0006955, GO:0006954, GO:0002376), chemokine activity (eg, PPBP, PF4, GO:0008009, GO:0042379), stress response (eg, SOD2, ANXA5, TYMS, GO:0006950), wound response (eg, ADM, ANXA5, GO:0009611) and antigen processing and presentation (eg, HLA-DQA1, HLA-DQB1, GO:0042613, GO:0042611, GO:0002504, GO:0019882) (all p values <0.05). Some overexpressed genes are linked to more than one GO annotation, so the present results should not be considered statistically independent. However, the converging pattern of results indicates relative activation of inflammatory responses in circulating T lymphocytes from children with asthma from a low SES background.
In contrast, genes that were overexpressed in children with asthma from a high SES background had functional themes related to methylation (eg, FOS, BTG2, GO:0043414, GO:0032259), protein folding (eg, HSPA1A, HSPA6, GO:0006986, GO:0051789), cell differentiation (eg, JUN, GO:0035026), stress response (eg, H2AFX, DNAJB1, GO:0006950) and antigen processing and presentation (eg, HLA-DQA1, HLA-DQA2, GO:0042613, GO:0002504, GO:0042611, GO:0019882) (all p values <0.05). These patterns suggest a relative activation of genes regulating both inflammatory processes and counter-regulatory responses to inflammation.
Confirmatory RT-PCR analyses assayed expression of 10 transcripts identified as differentially expressed in microarray analyses. Consistent with the estimated false discovery rate of <15% in the genome-wide differential expression analyses, all tested transcripts showed statistically significant alterations in expression (at p<0.05) (table 3).
Table 3.
RT-PCR confirmation of differential gene expression
| Gene | Microarray ratio* | RT-PCR ratio* | RT-PCR p value† |
|---|---|---|---|
| EGR1 | 0.68 | 0.52 | 0.0001 |
| FOS | 0.65 | 0.59 | 0.0001 |
| FOSB | 0.60 | 0.79 | 0.0487 |
| JUN | 0.75 | 0.51 | 0.0001 |
| ADM | 1.44 | 1.52 | 0.0114 |
| SOD2 | 1.40 | 1.25 | 0.0006 |
| S100A8 | 1.80 | 1.65 | 0.0001 |
| S100A9 | 1.39 | 1.74 | 0.0001 |
| S100A12 | 1.36 | 2.30 | 0.0001 |
| XIST | 1.65 | 5.58 | 0.0001 |
Low SES/high SES.
p Value from linear model analysis.
RT-PCR, reverse transcription-polymerase chain reaction; SES, socioeconomic status.
Transcription control pathways
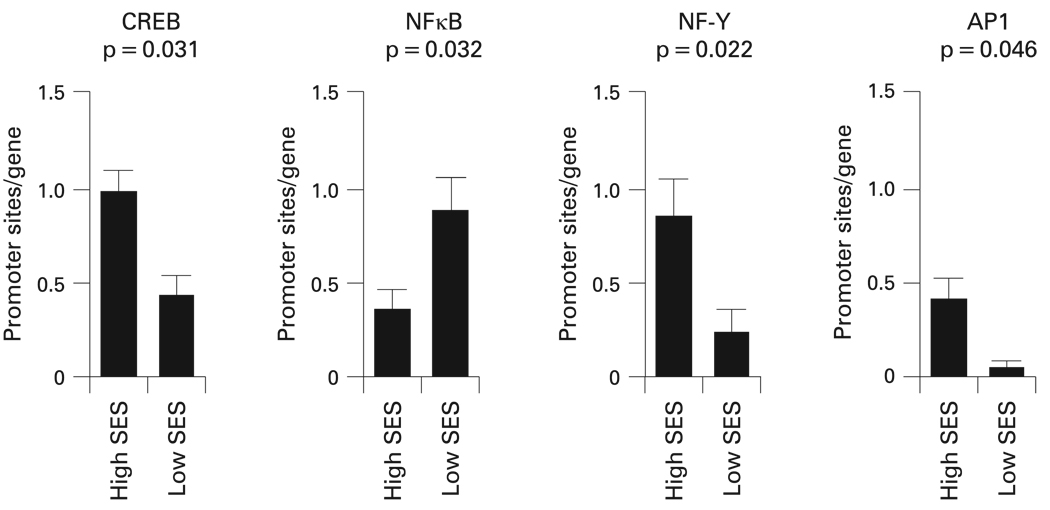
Bioinformatic analyses were used to identify TFBMs within the promoters of differentially expressed genes. Four distinct hormonal or cytokine signalling pathways were consistently linked to low SES (fig 1). Cyclic AMP response element binding protein (CREB) TFBMs were significantly less prevalent in promoters of genes overexpressed in children with asthma in the low SES group (average 55% decrease, p=0.031) (table 4). CREB proteins are key mediators of immune cell transcriptional responses to catecholamines.22
Figure 1.
Transcription factor binding motifs that were differentially represented in the promoters of genes overexpressed in children with asthma from a low socioeconomic status (SES) versus a high SES background. Data represent mean (SEM) prevalence of response elements within promoters from each group. AP1, activator protein 1; CREB, cyclic AMP response element binding protein; NFκB, nuclear factor κB; NF-Y, nuclear factor Y.
Table 4.
Bioinformatic indications of transcription factor activity
| p Value |
|||||
|---|---|---|---|---|---|
| TFBM | Ratio* | Univariate† | Random‡ | Sensitivity§ | |
| V$CP2_01 | 2.33 | 0.0156 | 0.0114 | 0.0943 | |
| V$TAL1BETAITF2_01 | 7.62 | 0.0166 | 0.0084 | 0.0922 | |
| V$NFY_C | 0.28 | 0.0223 | 0.0212 | 0.0485 | |
| V$CREB_Q4 | 0.45 | 0.0306 | 0.0334 | 0.0056 | |
| V$NFKB_Q6 | 2.54 | 0.0316 | 0.0232 | 0.0200 | |
| V$SREBP1_01 | 4.50 | 0.0391 | 0.0316 | 0.0759 | |
| V$COUP_01 | 4.85 | 0.0434 | 0.0360 | 0.0926 | |
| V$HSF2_01 | 0.71 | 0.0435 | 0.0460 | 0.0513 | |
| V$AP1FJ_Q2 | 0.65 | 0.0464 | 0.0484 | 0.0005 | |
| V$VJUN_01 | 0.20 | 0.0477 | 0.0512 | 0.0342 | |
TFBM prevalence in promoters of genes overexpressed in T lymphocytes from the low versus the high SES groups.
p Value from unequal variance Welch t test of TFBM prevalence.
p Value based on 10 000 random samples of 60 human genes.
p Value for average over nine combinations of three promoter lengths (−300 bp, −600 bp, −1000 to +200 bp) and 3 TFBM detection stringencies (MatSim 0.80, 0.90, and 0.95)
SES, socioeconomic status; TFBM, transcription factor binding motif.
A second finding involved decreased prevalence of NF-Y TFBMs in promoters of genes overexpressed in children from a low SES background (average 72% decrease in promoter TFBM prevalence, p=0.022). NF-Y is activated by the same cAMP/PKA signalling pathway as CREB, providing a convergent indication of pathway activation.
Decreased activity of AP1 (Fos/Jun) transcription factors in T cells from children from a low SES background was also found (AP1 p = 0.046; VJUN p = 0.048). Reduced activity of this pathway is consistent with the observed downregulation of the FOS, FOSB and JUN transcripts, which encode key elements of the AP1 transcription factor family.
In addition, a heightened prevalence of NFκB TFBMs in genes overexpressed in the low SES group was found (average 2.54-fold increase in promoter TFBM prevalence, p = 0.032). NFκB activates numerous genes involved with inflammation, wound healing and cellular stress responses.
All of the above findings remained significant in sensitivity analyses parametrically varying promoter length and TFBM detection stringency (table 4). Other TFBMs were identified in initial analyses as potential contributors to the observed SES related differences in gene expression. However, these TFBMs did not show consistent statistical significance across parametric sensitivity analyses (p values >0.05) (table 4). To assess the likelihood that the observed differences might arise by chance, we drew 10 000 random samples of 60 transcripts from the population of genes assayed by the Affymetrix U133A high density oligonucleotide array, and computed the magnitude of differential prevalence for the 10 TFBMs identified in initial analyses. Results confirmed that the observed differential distributions of CREB, NF-Y, AP1 and NFκB were unlikely to occur in a random sample of genes (p<0.05) (table 4).
Confounders
After controlling for age and gender, patterns of associations with CREB, NF-Y and NFκB remained statistically significant (all p values <0.045), with one exception which was the relationship between SES and CREB controlling for age (p = 0.068). Associations also remained significant after controlling for asthma severity, inhaled corticosteroid use and β agonist use (all p values ≤.05). Unlike the other transcription control pathways, indications of association between SES and AP1 activity were rendered non-significant by control for several confounders, including age (p = 0.52), gender (p = 0.15) and β agonist use (p = 0.11).
Controlling for cognitive interpretations reduced the relationship between SES and CREB activity to non-significant (p = 0.25). The same was true for the relationship between SES and NFκB (p = 0.12) and to a lesser extent SES and NF-Y (p = 0.077). These analyses suggest that children from the low SES group were more likely to perceive threat in ambiguous situations, and this tendency may activate neuroendocrine processes (impacting on CREB and NF-Y through the cAMP/PKA pathway) that ultimately impact on inflammatory signalling pathways (eg, NFκB).
Finally, controlling for the prevalence of mRNAs encoding CD16 and CD56 did not significantly alter the results for CREB, NF-Y or NFκB (all p values <0.05), suggesting that results are not simply a function of different relative proportions of T cell versus NK cell subtypes in each group.
Clinical outcomes
The low and high SES groups also differed in clinical profiles, such that children in the low SES group had greater daytime symptoms (low SES mean 5.68 (SD 5.24); high SES mean 1.50 (SD 2.10); t = 2.77, p = 0.010), greater night-time symptoms (low SES mean 2.78 (SD 4.19); high SES mean 0.36 (SD 1.08); t = 2.10, p = 0.046) and showed a trend towards greater emergency department utilisation in the past 6 months because of their asthma (low SES 20%; high SES 0%; χ2 = 3.33, p = 0.068).
DISCUSSION
The present study provides the first empirical evidence linking the larger social environment (SES) to T cell gene expression in the context of a clinical inflammatory disease. Among children with asthma, those from a low SES background showed overexpression of genes regulating a variety of inflammatory processes, including chemokine activity, stress responses and wound responses. Promoter based bioinformatics analyses identified transcription control pathways that may structure the observed patterns of differential gene expression, including decreased CREB, AP1 and NF-Y, and increased NFκB signalling. Children in the low SES group also reported poorer clinical outcomes, such as greater asthma symptoms. Although the cross sectional nature of this study precludes definitive conclusions about causal direction, the findings are consistent with the hypothesis that the larger social environment can get “under the skin” at the level of genomic transcription control pathways.
Genes overexpressed in children in the low SES group included: chemokine ligands such as CXCL4 and CXCL7 which recruit and activate leucocytes;23 those involved in antigen processing and presentation (MHC class II protein complexes), a key feature in asthma inflammatory biology; those related to oxidative stress (SOD2), which recruits and activates immune cells, prolonging inflammation; and those related to calcium binding proteins (S100A8, S100A9, S100A12), which have chemotactic effects on leucocytes and proinflammatory effects on endothelial cells.24 25 These patterns suggest molecular regulatory mechanisms that may heighten inflammation and worsen clinical outcomes in children with asthma from a low SES background.
Genes overexpressed in children in the high SES group included: those encoding heat shock proteins (HSPA1A, HSPA1B, HSPA6, DNAJB1), which protect cells from inflammatory molecules such as reactive oxygen species, and are protective against pulmonary inflammation26 27; those involved in cell differentiation and proliferation, such as c-fos and c-jun, which are increased in response to exposure to reactive oxygen species28; those related to cell cycle control (BTG2), which helps contain the effects of DNA damage29; and those involved in the methylation, or silencing, of genes. Note that FOS, FOSB and JUN gene products also encode key components of the AP1 family of transcription factors, which in promoter based bioinformatic analyses showed increased activity in the high SES group. Taken together, these patterns suggest that among children with asthma from a high SES background, heightened inflammation may be counterbalanced by cellular regulatory mechanisms aimed at containing the damage due to inflammation.
This study also identified candidate transcription control pathways that may orchestrate differential patterns of gene expression as a function of SES. Bioinformatic analyses indicated downregulated activity of CREB, AP1 and NF-Y, and upregulated NFκB mediated transcription in children with asthma from a low SES background. CREB mediates transcriptional responses to β adrenergic receptor signalling through the adenylyl cyclase/cAMP/protein kinase A pathway.22 30 Adenylyl cyclase activity is impaired after allergen challenge in patients with asthma.31 Diminished regulatory signalling from cAMP pathways could lead to increased activation of T cells, and subsequent expression of Th-2 cytokines.31 Reduced CREB signalling may also decrease the efficacy of bronchodilators used as therapeutic agents in asthma. These patterns are consistent with decreased β adrenergic receptor mRNA found in lymphocytes of children with asthma with chronic and acute life stress.14
Bioinformatic analyses also indicated reduced NF-Y mediated transcription in children with asthma from a low SES background. Like CREB, NF-Y is phosphorylated by PKA, and may thus serve as an indicator of signalling along the β adrenergic signalling pathway.32 The fact that both transcription factors showed downregulation in the low SES group suggests multiple parallel deficiencies across catecholamine signalling pathways.
Bioinformatic analyses also indicated upregulated NFκB signalling in children with asthma in the low SES group. NFκB transactivates a wide variety of inflammatory mediators. Some data suggest that elevated cAMP activity can inhibit NFκB activity33; hence increased NFκB and decreased CREB signalling could represent a common regulatory alteration that shifts gene expression profiles towards a more inflammatory phenotype in children from low SES backgrounds.
Our findings are consistent with previous research that has investigated functional immune measures. For example, children with asthma from a low SES background show greater production of Th-2 cytokines and greater eosinophil counts compared with those from a high SES background.12 Greater stress has been linked to in vivo inflammatory responses to allergen challenge and in vitro cytokine production in patients with asthma13 34 and predisposed to allergic disease.35 Furthermore, stress has been linked to reduced expression of genes coding for hormonal receptors that regulate inflammation.14
Statistical analyses revealed that interpretations of stress accounts for some of the SES related differences in indicators of CREB and NFκB activity. This suggests that in order for the social environment to have biological effects, it may have to first be perceived in a threatening manner. In turn, these cognitive perceptions may come with biological costs in transcriptional regulation and inflammatory biology. We note there could be numerous other pathways linking SES to genomic patterns, which should be tested in future studies.
It is unclear whether similar effects would be found in healthy children. However, even if similar effects were evident, these biological mechanisms could still have different implications for those with a pre-existing inflammatory disease. Nonetheless, it would be important for future research to test the effects of SES on gene expression profiles in healthy individuals as well.
Limitations include the observational design, precluding conclusions about causality and directionality. This is a necessary limitation to human work involving social factors that are difficult to randomly assign (eg, socioeconomic status). Secondly, the sample size is small by epidemiological standards, although not unusual for genome-wide microarray studies. One concern about small samples is insufficient power; however, the significant differences in several transcription control pathways suggest that the magnitude of effects is large enough to be detected in a sample of this size. A second concern is whether the findings will be robust and reliable. However, note that the bioinformatics approach used in this study capitalises on the data from thousands of genes to form aggregate and more reliable indicators of transcription factor activity. Thirdly, microarray technology is sometimes criticised as being too exploratory. In this study, however, microarray data were used to test an a priori hypothesis of SES increases in inflammation due to specific hormonal signalling pathways documented in previous research.12 14 Furthermore, the use of false discovery rate analysis is a standard approach in the genomic literature for adjusting statistics to account for multiple comparisons. Nevertheless, replicating these results in other samples would be important in future studies. Finally, selection of the CD2 antigen to isolate T lymphocytes may have permitted contamination by natural killer (NK) cells. (CD3 markers were not used because the immunomagnetic separation process for CD3 would have activated T cells.) However, the present results were not significantly altered in analyses controlling for NK cell marker mRNA.
The present data identified a distinct transcriptional finger-print of low SES environments, and candidate transcription control pathways structuring those differences, in T lymphocytes from children with asthma. Children with asthma from a low SES background showed overexpression of genes related to inflammation, chemokine activity, and stress and wound responses, and bioinformatics indications of reduced CREB, AP1 and NF-Y, and increased NFκB signalling. This study provides the first clinical evidence in asthma that broader social environments affect processes at the genomic level, specifically in terms of transcription control pathways that regulate inflammation and catecholamine signalling. Because these pathways are the primary targets of many asthma medications, these findings suggest that the larger social environment may also affect the efficacy of asthma therapeutics. Finally, perceptions of stress play an important role in explaining how SES gets transduced into alterations at the genomic level. Overall, these findings provide new insights into the mechanisms by which social factors affect the course of inflammatory diseases, and highlight the need for future research investigating how the pathophysiology of asthma is shaped by social environments.
Supplementary Material
Acknowledgments
Funding: This study was supported by NIH grant HL073975 and the Canadian Institutes of Health Research.
Footnotes
Competing interests: None.
Ethics approval: Ethics approval was obtained.
REFERENCES
- 1.Starfield B, Robertson J, Riley AW. Social class gradients and health in childhood. Ambul Pediatr. 2002;2:238–246. doi: 10.1367/1539-4409(2002)002<0238:scgahi>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 2.Chen E, Matthews KA, Boyce WT. Socioeconomic differences in children’s health: How and why do these relationships change with age? Psychol Bull. 2002;128:295–329. doi: 10.1037/0033-2909.128.2.295. [DOI] [PubMed] [Google Scholar]
- 3.Maziak W, von Mutius E, Keil U, et al. Predictors of health care utilization of children with asthma in the community. Pediatr Allergy Immunol. 2004;15:166–171. doi: 10.1046/j.1399-3038.2003.00105.x. [DOI] [PubMed] [Google Scholar]
- 4.Amre DK, Infante-Rivard C, Gautrin D, et al. Socioeconomic status and utilization of health care services among asthmatic children. J Asthma. 2002;39:625–631. doi: 10.1081/jas-120014927. [DOI] [PubMed] [Google Scholar]
- 5.Dales RE, Choi B, Chen Y, et al. Influence of family income on hospital visits for asthma among Canadian school children. Thorax. 2002;57:513–517. doi: 10.1136/thorax.57.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon PA, Zeng ZW, Wold CM, et al. Prevalence of childhood asthma and associated morbidity in Los Angeles County: Impacts of race/ethnicity and income. J Asthma. 2003;40:535–543. doi: 10.1081/jas-120018788. [DOI] [PubMed] [Google Scholar]
- 7.Miller JE. The effects of race/ethnicity and income on early childhood asthma prevalence and health care use. Am J Public Health. 2000;90:428–430. doi: 10.2105/ajph.90.3.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halfon N, Newacheck PW. Childhood asthma and poverty: Differential impacts and utilization of health services. Pediatrics. 1993;91:56–61. [PubMed] [Google Scholar]
- 9.Panagiotakos DB, Pitsavos C, Manios Y, et al. Socio-economic status in relation to risk factors associated with cardiovascular disease, in healthy individuals from the ATTICA study. Eur J Cardiovasc Prev Rehabil. 2005;12:68–74. [PubMed] [Google Scholar]
- 10.Hemingway H, Shipley M, Mullen MJ, et al. Social and psychosocial influences on inflammatory markers and vascular function in civil servants (The Whitehall II study) Am J Cardiol. 2003;92:984–987. doi: 10.1016/s0002-9149(03)00985-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen E, Fisher EB, Jr, Bacharier LB, et al. Socioeconomic status, stress, and immune markers in adolescents with asthma. Psychosom Med. 2003;65:984–992. doi: 10.1097/01.psy.0000097340.54195.3c. [DOI] [PubMed] [Google Scholar]
- 12.Chen E, Hanson MD, Paterson LQ, et al. Socioeconomic status and inflammatory processes in childhood asthma: The role of psychological stress. J Allergy Clin Immunol. 2006;117:1014–1020. doi: 10.1016/j.jaci.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 13.Liu LY, Coe CL, Swenson CA, et al. School examinations enhance airway inflammation to antigen challenge. Am J Respir Crit Care Med. 2002;165:1062–1067. doi: 10.1164/ajrccm.165.8.2109065. [DOI] [PubMed] [Google Scholar]
- 14.Miller GE, Chen E. Life stress and diminished expression of genes encoding glucocorticoid receptor and beta(2)-adrenergic receptor in children with asthmaz. Proc Natl Acad Sci. 2006;103:5496–5501. doi: 10.1073/pnas.0506312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen E, Matthews KA. Development of the Cognitive Appraisal and Understanding of Social Events (CAUSE) videos. Health Psychol. 2003;22:106–110. doi: 10.1037//0278-6133.22.1.106. [DOI] [PubMed] [Google Scholar]
- 16.Cole SW, Galic Z, Zack JA. Controlling false negative errors in microarray differential expression analysis: A PRIM approach. Bioinformatics. 2003;19:1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- 17.Cole SW, Yan W, Galic Z, et al. Expression-based monitoring of transcription factor activity: The TELiS database. Bioinformatics. 2005;21:803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Beissbarth T, Speed TP. GOstat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;6:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 20.Wingender E, Dietze P, Karas H, et al. TRANSFAC: A database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller RG. Beyond ANOVA: Basics of applied statistics. New York, NY: Wiley; 1986. [Google Scholar]
- 22.Kammer GM. The adenylate cyclase–cAMP-protein kinase A pathway and regulation of the immune response. Immunol Today. 1988;9:222–229. doi: 10.1016/0167-5699(88)91220-0. [DOI] [PubMed] [Google Scholar]
- 23.Lukacs NW, Miller AL, Hogaboam CM. Chemokine receptors in asthma: Searching for the correct immune targets. J Immunol. 2003;171:11–15. doi: 10.4049/jimmunol.171.1.11. [DOI] [PubMed] [Google Scholar]
- 24.Lackmann M, Rajasekariah P, Lismaa SE, et al. Identification of a chemotactic domain of the pro-inflammatory S100 protein CP-10. J Immunol. 1993;150:2981–2991. [PubMed] [Google Scholar]
- 25.Hofmann MA, Drury S, Fu CF, et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 26.Jacquiersarlin MR, Fuller K, Dinhxuan AT, et al. Protective effects of HSP70 in inflammation. Experientia. 1994;50:1031–1038. doi: 10.1007/BF01923458. [DOI] [PubMed] [Google Scholar]
- 27.Villar J, Edelson JD, Post M, et al. Induction of heat stress proteins is associated with decreased mortality in an animal model of acute lung injury. Am Rev Respir Dis. 1993;147:177–181. doi: 10.1164/ajrccm/147.1.177. [DOI] [PubMed] [Google Scholar]
- 28.Janssen YMW, Matalon S, Mossman BT. Differential induction of c-fos, c-jun, and apoptosis in lung epithelial cells exposed to ROS or RNS. Am J Physio Lung Cell Mol Physiol. 1997;17:L789–L796. doi: 10.1152/ajplung.1997.273.4.L789. [DOI] [PubMed] [Google Scholar]
- 29.Rouault JP, Falette N, Guehenneux F, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nature Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 30.Sanders VM, Straub RH. Norepinephrine, the beta-adrenergic receptor, and immunity. Brain Behav Immunity. 2002;16:290–332. doi: 10.1006/brbi.2001.0639. [DOI] [PubMed] [Google Scholar]
- 31.Borger P, Postma DS, Vellenga E, et al. Regulation of asthma-related T-cell cytokines by the cyclic AMP-dependent signalling pathway. Clin Exp Allergy. 2000;30:920–926. doi: 10.1046/j.1365-2222.2000.00794.x. [DOI] [PubMed] [Google Scholar]
- 32.Kapatos G, Vunnava P, Wu Y. Protein kinase A-dependent recruitment of RNA polymerase II, C/EBPbeta and NF-Y to the rat GTP cyclohydrolase I proximal promoter occurs without alterations in histone acetylation. J Neurochem. 2007;101:1119–1133. doi: 10.1111/j.1471-4159.2007.04486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elenkov IJ, Wilder RL, Chrousos GP, et al. The sympathetic nerve—An integrative interface between two supersystems: The brain and the immune system. Pharmacol Rev. 2000;52:595–638. [PubMed] [Google Scholar]
- 34.Kang D, Coe C, McCarthy DO, et al. Cytokine profiles of stimulated blood lymphocytes in asthmatic and healthy adolescents across the school year. J Interferon Cytokine Res. 1997;17:481–487. doi: 10.1089/jir.1997.17.481. [DOI] [PubMed] [Google Scholar]
- 35.Wright RJ, Finn P, Contreras JP, et al. Chronic caregiver stress and IgE expression, allergen-induced proliferation, and cytokine profiles in a birth cohort predisposed to atopy. J Allergy Clin Immunol. 2004;113:1051–1057. doi: 10.1016/j.jaci.2004.03.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.