Introduction
Prolonged stimulus of the adult heart by hypertension, arrhythmias, or other pathogenic conditions can result in myocardial hypertrophy and subsequently heart failure. Hypertrophy is characterized by an increase in the size of cardiomyocytes and this is associated with an elevated rate of RNA synthesis and protein synthesis in these cells. Although mechanisms responsible for this increased RNA and protein synthesis remain to be fully elucidated, a publication from Espinoza-Derout et al.1 in this issue of Circulation Research provides additional evidence that dysregulation of a general RNA polymerase II elongation factor known as P-TEFb can be an important mechanism in the development of hypertrophy.
Regulation of RNA Polymerase II Elongation
RNA polymerase II transcribes all protein-coding genes and its function is regulated both at the levels of transcription initiation and elongation. Initiation is positively regulated by transcription factors that bind to specific cis-regulatory sequences in the promoters of individual genes. These transcription factors function to recruit both co-activators and the RNA polymerase II complex and transcription initiation is enhanced at these genes. After initiation, transcriptional elongation can be defective due to the action of two negative factors, DSIF and NELF, which limit elongation2;3. These negative factors may function as a quality control mechanism to ensure capping of nascent mRNA, thereby stabilizing the RNA for subsequent processing events and transcriptional termination4. While almost all protein-coding genes appear to undergo some level of constitutive transcription initiation, only a subset actually produce full-length transcripts, indicating that many genes are defective in transcriptional elongation5. This block to productive elongation is overcome by the action of P-TEFb -- positive transcription elongation factor b. P-TEFb is a protein kinase that can hyperphosphorylate the carboxyl terminal domain (CTD) of the large subunit of RNA polymerase. The CTD of mammalian RNA polymerase II contains 52 repeats of the sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser and phosphorylation of the second serine (Ser2) in this sequence is a hallmark of P-TEFb activity. P-TEFb also phosphorylates protein subunits of both the DSIF and NELF negative factors. Phosphorylation of the CTD, DSIF and NELF by P-TEFb converts RNA polymerase II into a processive enzyme that is capable of transcribing entire genes.
P-TEFb Complexes in Cells
P-TEFb is actually a family of distinct molecular complexes, each containing CDK9 as the catalytic subunit. CDK9 is a serine-threonine kinase and it is expressed as two isoform, a major 42 kDa protein and a minor 55 kDa protein that differ by an amino terminal extension on the larger protein. The catalytic function of Cdk9 requires the association of a cyclin protein, either Cyclin T1, T2, or K. In cells and tissues examined to date, Cyclin T1 and T2 appear to be the major cyclin partners of Cdk9 and the significance of Cyclin K to P-TEFb function remains to be determined.
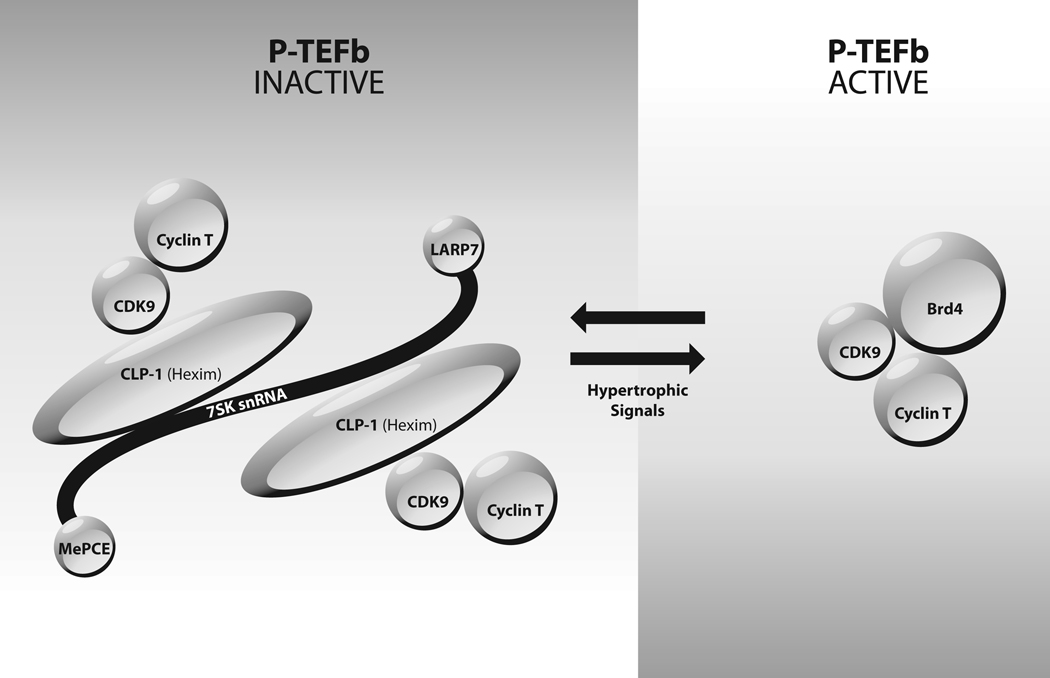
P-TEFb is found in two complexes in cells, a small complex and a ribonucleoprotein complex termed the 7SK snRNP. As illustrated in the Figure, the small complex consists of Cdk9, Cyclin T (either T1 or T2) and Brd4, and this small complex is the form of P-TEFb that is recruited to genes to activate elongation. Brd4 is a bromodomain protein that is capable of binding to acetylated histones in regions of the genome that are marked as transcriptionally active. Brd4 is thought to bind to acetylated histones and direct CDK9/Cyclin T to active genes, where the catalytic activity of CDK9 activates transcriptional elongation.
Figure.
Equilibrium between small P-TEFb complex and 7SK snRNP. Catalytically active P-TEFb exists as a complex of CDK9, Cyclin T (either Cyclin T1 or Cyclin T2) and Brd4. The catalytically inactive 7SK snRNP contains 7SK snRNA, CLP-1/HEXIM1, MePCE, LARP7, CDK9 and Cyclin T (either Cyclin T1 or Cyclin T2). Hypertrophic signals result in disruption of the 7SK snRNP and conversion of P-TEFb to the small active complex.
A significant fraction of P-TEFb is found in the 7SK snRNP complex that contains a small nuclear RNA known as 7SK snRNA, two CDK9/Cyclin T dimers and several other proteins6. The kinase activity of CDK9 in the 7SK snRNP is repressed and this is therefore an catalytically inactive form of P-TEFb. Two proteins are bound to 7SK snRNA in the snRNP -- MePCE (methylphosphate capping enzyme) is bound to the 5’ cap of 7SK and PARP7 (also named PIP7S) is bound to the 3’ end of 7SK. A key protein in the 7SK snRNP is CLP-1 (cardiac lineage protein 1), also known as HEXIM1 (hexamethylene bis-acetamide induced protein). CLP-1/HEXIM1 can exist as a homodimer in the snRNP, or it can exist as a heterodimer with the paralogous HEXIM2 protein. The HEXIM2 protein is generally much less abundant than CLP-1/HEXIM1. Extensive biochemical evidence has shown that CLP-1/HEXIM1 functions as a scaffold to assemble the 7SK snRNP (reviewed in 6).
Disruption of P-TEFb 7SK snRNP and Myocardial Hypertrophy
There is a dynamic equilibrium between the small form of P-TEFb and the 7SK snRNP. A number of stimuli have been identified which disrupt the snRNP and convert the bulk of CDK9/Cyclin T into the small and catalytically active P-TEFb complex. The biological significance of some of these disrupting stimuli, such as Actinomycin D or drugs such as flavopiridol, is questionable. However, an important study from the Schneider laboratory showed that hypertrophic signals such as calcineurin or mechanical stress cause the dissociation of the 7SK snRNP in cultured cardiac myocytes7. This dissociation was associated with an elevated level of CTD Ser2 phosphorylation, indicating that overall P-TEFb function is enhanced by hypertrophic signals. This study suggested that the increase in P-TEFb function caused by disruption of the 7SK snRNP is a mechanism involved in the increase in the global rate of RNA synthesis that is associated with myocardial hypertrophy.
Role of CLP-1/HEXIM1 in Myocardial Hypertrophy
Previous work from the Siddiqui laboratory showed that deletion of the CLP-1/HEXIM1 gene in mice results in lethality at the late fetal stages of development8. An analysis of CLP-1−/− fetal hearts indicated a hypertrophic phenotype, indicating that dysregulation of the 7SK snRNP by the genetic ablation of CLP-1/HEXIM1 can also contribute to hypertrophy. In their publication in this issue of Circulation Research, the Siddiqui laboratory have used genetically modified mice to extend their analysis of the role of CLP-1/HEXIM1 in hypertrophy. Espinoza-Derout et al. analyzed a mouse strain in which an activated form of calcineurin is expressed in the heart1. Calcineurin is a calmodulin-dependent phosphatase that promotes myocardial hypertrophy9. The association of CLP-1/HEXIM1 with P-TEFb was decreased in these mice, suggesting that the calcineurin hypertrophic stimulus results in dissociation of the 7SK snRNP. Additionally, calcineurin expression in the heart resulted in an increase in the overall level of CTD Ser2 phosphorylation, showing that P-TEFb activity is enhanced in these mice and this likely contributes to an increase in total RNA synthesis in cardiomyocytes.
Espinoza-Derout et al. also utilized a previously generated mouse strain (MHC-Cyclin T1) in which the Cyclin T1 subunit of P-TEFb is over-expressed in the heart7. Cyclin T1 levels are apparently limiting for P-TEFb function in wild type mice, as P-TEFb catalytic activity is elevated in the heart of these mice that over-express Cyclin T1. Espinoza-Derout et al. crossed the MHC-Cyclin T1 mouse with the CLP-1/HEXIM1-deleted mouse to generate a MHC-Cyclin T1/CLP+/− mouse. CLP-1+/− mice in a wild type Cyclin T1 background appear to be normal and have no phenotypic abnormalities8. The significance of the MHC-Cyclin T1/CLP-1+/− strain is that Cyclin T1 is over-expressed in the heart in the presence of a reduced level of CLP-1/HEXIM1 – a situation in which the level of the small P-TEFb complex should be considerably increased. There was indeed a striking enhancement of ventricular hypertrophy in the MHC-Cyclin T1/CLP-1+/− mice. Additionally, there was an increase in the overall level of CTD Ser2 phosphorylation in the heart of the mice. Thus, the combination of elevated P-TEFb levels by overexpression of Cyclin T1 and the reduction CLP-1/HEXIM1 levels by the CLP-1+/− genotype strongly enhances the development of hypertrophy. These new observations highlight the consequence of the dysregulation of the equilibrium between the small P-TEFb complex and the 7SK snRNP to myocardial hypertrophy.
Future Studies
The publication by Espinoza-Derout et al. adds further support to the idea that the ability to maintain an appropriate equilibrium between the small P-TEFb complex and the 7SK snRNP in myocardial myocytes may have therapeutic potential for treatment of hypertrophy. A better understanding of signals and mechanisms that control this equilibrium is essential if this potential is to be realized. The formation of the 7SK snRNP requires that Cdk9 be phosphorylated at threonine 186 in a region of the protein termed the T-loop10. This phosphorylation induces a conformational change in CDK9 or related Cyclin-dependent kinases that allows substrates to access the catalytic core of the enzyme11. Phosphorylation of the CDK9 T-loop was recently reported to occur by an auto-activation mechanism12, meaning the CDK9 can activate itself in cis, a mechanism that has been described for a number of protein kinases13. Phosphatases involved in the dephosphorylation of the CDK9 T-loop have been identified, PP2B, PP1alpha and PPM1A, and these enzymes are likely to be key regulators of the equilibrium between P-TEFb complexes14;15. A potentially important area of investigation will be to determine if hypertrophic signals regulate the functions of these phosphatases. Additionally, the majority of CLP-1/HEXIM1 is not associated with P-TEFb and it is therefore possible that this P-TEFb-free CLP-1/HEXIM1 may play a role in hypertrophy. Might the dysregulation of P-TEFb by hypertrophic signals also contribute to the increased rate of protein synthesis that is associated with hypertrophy? It is notable that CDK9 has been found to associate with polysomes16. It is therefore possible that disruption of the 7SK snRNP by hypertrophic stimuli may result in elevated levels of CDK9 on polysomes and this may contribute to increased protein synthesis. Further studies of the role of P-TEFb in myocardial hypertrophy should continue to shed mechanistic insight into this pathogenic process and may suggest strategies for novel therapeutics.
Acknowledgments
Sources of Funding
Supported by the National Institutes of Health
Footnotes
Disclosures
None.
References
- 1.Espinoza-Derout J, Wagner W, Salciccioli L, Lazar JM, Bhaduri S, Mascareno E, Chaqour B, Siddiqui MAQ. Positive transcription elongation factor b (P-TEFb) activity in compensatory myocardial hypertrophy is regulated by Cardiac Lineage Protein-1. Circ Res. 2009 doi: 10.1161/CIRCRESAHA.108.191726. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol Mol Biol Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 5.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michels AA, Bensaude O. RNA-driven cyclin-dependent kinase regulation: when CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotechnol J. 2008;3:1022–1032. doi: 10.1002/biot.200800104. [DOI] [PubMed] [Google Scholar]
- 7.Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 8.Huang F, Wagner M, Siddiqui MA. Ablation of the CLP-1 gene leads to down-regulation of the HAND1 gene and abnormality of the left ventricle of the heart and fetal death. Mech Dev. 2004;121:559–572. doi: 10.1016/j.mod.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem. 2004;279:4153–4160. doi: 10.1074/jbc.M310044200. [DOI] [PubMed] [Google Scholar]
- 11.Morgan DO, De Bondt HL. Protein kinase regulation: insights from crystal structure analysis. Curr Opin Cell Biol. 1994;6:239–246. doi: 10.1016/0955-0674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 12.Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN, Debreczeni JE, Knapp S, Johnson LN. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008;27:1907–1918. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lochhead PA. Protein kinase activation loop autophosphorylation in cis: overcoming a Catch-22 situation. Sci Signal. 2009;2:e4. doi: 10.1126/scisignal.254pe4. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, Zhou Q. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008;22:1356–1368. doi: 10.1101/gad.1636008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, Sung TL, Lin X, Rice AP. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J Biol Chem. 2008 doi: 10.1074/jbc.M807495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rother S, Strasser K. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 2007;21:1409–1421. doi: 10.1101/gad.428407. [DOI] [PMC free article] [PubMed] [Google Scholar]