Abstract
When a high-fidelity DNA polymerase encounters certain DNA-damage sites, its progress can be stalled and one or more lesion-bypass polymerases are recruited to transit the lesion. Here, we consider two representative types of lesions: (i) 7,8-dihydro-8-oxogua-nine (8-oxoG), a small, highly prevalent lesion caused by oxidative damage; and (ii) bulky lesions derived from the environmental pre-carcinogen benzo[a]pyrene, in the high-fidelity DNA polymerase Bacillus fragment (BF) from Bacillus stearothermophilus and in the lesion-bypass DNA polymerase IV (Dpo4) from Sulfolobus solfataricus. The tight fit of the BF polymerase around the nascent base pair contrasts with the more spacious, solvent-exposed active site of Dpo4, and these differences in architecture result in distinctions in their respective functions: one-step versus stepwise polymerase translocation, mutagenic versus accurate bypass of 8-oxoG, and polymerase stalling versus mutagenic bypass at bulky benzo[a]pyrene-derived lesions.
Lesions and DNA polymerases
Until 1999, it was widely understood that bacteria have three DNA polymerases and mammals have five [1]. In 1999, however, an entirely new category of polymerases was discovered in prokaryotes and eukaryotes [2 – 4] – the lesion-bypass DNA polymerases. The function of lesion-bypass polymerases is to replicate past lesion-containing DNA templates in a process called translesion synthesis [5–7]. Now at least 16 DNA polymerases are known in eukaryotes [8] and five in bacteria [9]. Furthermore, since 1999, crystal structures of DNA polymerases, including those containing lesions, have provided new structural insights into the mechanistic aspects of translesion synthesis and polymerase stalling associated with DNA lesions. For a review of the structures and mechanisms of DNA polymerases up to the year 2005, see Ref. [10].
Here, we discuss the structural and functional features of representative high-fidelity and lesion-bypass DNA polymerases (Box 1) and how these features affect the processing of certain forms of DNA damage. The lesions considered are derived from reactions of intermediates produced by endogenous sources or from the metabolic activation of environmental genotoxic chemicals. An A-family (bacterial polymerase I group) high-fidelity DNA polymerase from the archaeon Bacillus stearothermophilus (large fragment, BF) is compared with the Y-family lesion-bypass DNA polymerase IV (Dpo4) from Sulfolobus solfataricus.
Box 1. High-fidelity and lesion-bypass DNA polymerases
DNA polymerases are grouped into A, B, C, D, X, Y and RT families, based on amino acid sequence homologies [10]. High-fidelity polymerases are members of the A and B families, whereas the lesion-bypass DNA polymerases belong primarily, but not exclusively, to the Y family. The lesion-bypass DNA polymerases have important roles in the temporary replacement of high-fidelity DNA polymerases that are stalled upon encountering damaged DNA sites, an event termed ‘polymerase switch’ [5–7].
Both high-fidelity and lesion-bypass DNA polymerases have right-handed architectures with core domains of palm, fingers and thumb, and active sites formed by the palm and fingers domains (Figure I). Both types of polymerase use a two-metal ion catalytic mechanism [58] for the nucleotidyl-transfer reaction, and have three universally conserved carboxylate-containing residues (aspartate or glutamate) at their active sites. Most Y-family DNA polymerases exhibit low processivity and a high error rate upon replicating normal, unmodified DNA [3,4]. The higher fidelity A- and B-family DNA polymerases have intrinsic error rates in the range of 10−2–10−6, whereas the error rates of lower fidelity Y-family polymerases are in the 1–10−3 range [59]. Specifically, the Y-family polymerase Dpo4 has a mismatch error rate of ~6.5 × 10−3 and a single-base-deletion error rate of ~2.4 × 10−3 [60], whereas the A-family polymerase BF has an error rate of ~1.5 × 10−5 (http://www.freepatentsonline.com/5834253.html). Polymerase processivity, namely the number of nucleotides added per binding event, is only one or two nucleotides for Dpo4 without additional cofactors in vitro [61], whereas, for BF, it is 111 nucleotides [62]. The presence of accessory proteins in the holoenzyme in vivo increases DNA-synthesis processivity thousands of folds for high-fidelity polymerases, but only slightly for lesion-bypass DNA polymerases [63,64].
How polymerases process lesions and whether a mutation ensues are dictated by the type of lesion, the sequence context and the particular lesion-bypass DNA polymerase. The polymerase might be specifically tailored to replicate past the lesion accurately, or translesion synthesis can occur in a low-fidelity, mutagenic manner [3]. Current understanding of in vivo functioning indicates that one lesion-bypass DNA polymerase might be involved in the nucleotide-insertion step, whereas one or more extend beyond the lesion until the lesion-induced distortions are transited and the high-fidelity DNA polymerase resumes synthesis [2–4,65]. Aside from translesion synthesis, it has recently been shown that at least some Y-family DNA polymerases are involved in other functions, including recombination and somatic hypermutation [8,9,66].
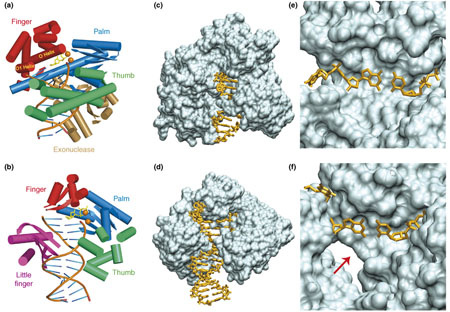
Figure I. Cartoon and molecular surface representations of BF and Dpo4 ternary structures. BF features a sterically tight active site and an exonuclease domain {non-functional in the strain used in crystallographic studies [11,31,62] but functional in Bst 320 ATCC55719 (http://www.patentstorm.us/patents/5747298-claims.html)}, but lacks a little finger domain, whereas Dpo4 lacks an exonuclease domain but has an additional little finger domain, and a sterically more open active site. Cartoon representations of BF (a) and Dpo4 (b). For clarity, only α helices and β sheets of proteins, and DNA backbone and base registers are shown. Incoming dCTP is shown as yellow sticks, and divalent metal ions are shown as orange spheres. The thumb domains are shown in green, palm domains in blue, fingers domains in red, the BF exonuclease domain in gold and the Dpo4 little finger domain in purple. The views are into the major-groove side of the nascent base pair, with the minor-groove side behind. Molecular surface representations of BF (c) and Dpo4 (d). The minor groove adjacent to the replicating base pair is exposed to solvent in Dpo4, but completely buried in BF. The DNA molecules are drawn as sticks. Close-up views of the replicating base pair in BF (e) and Dpo4 (f). The minor-groove side of this base pair indicated by the red arrow is solvent-accessible in Dpo4. (BF PDB code: 1LV5, chains A/C/D [11]; Dpo4 PDB code: 2ATL, chains A/D/E [14].)
Crystallographic structures of BF have been extensively studied and there is considerable understanding of the replication mechanism for unmodified DNA [11,12]. Likewise, X-ray crystallographic structures of Dpo4 in complex with DNA, both with and without dNTP have been determined (for example, see Refs [4,13–17]). Crystal structures of lesion-containing complexes for these two polymerases are also available (for example, see Refs [14,18–27]), which shed light on their contrasting mechanisms for processing damaged DNA templates. We consider two important DNA lesion prototypes as examples of small and bulky lesions, respectively: 7,8-dihydro-8-oxoguanine (8-oxoG; Box 2), a ubiquitous product of reactive oxygen species (ROS), and DNA adducts derived from a tumorigenic metabolite of the environmental pre-carcinogen benzo[a]pyrene (BP; Box 3). We highlight that the tight fit of the BF polymerase around the nascent base pair contrasts with the more spacious, solvent-exposed active site of Dpo4, and these distinct architectures produce distinctions in polymerase translocation mechanisms, and in fidelity and efficiency of lesion processing. Understanding the processing of endogenous and environmental DNA damage by DNA polymerases is key to elucidating how mutations that initiate diseases, including cancer, originate.
Box 2. 7,8-dihydro-8-oxoguanine
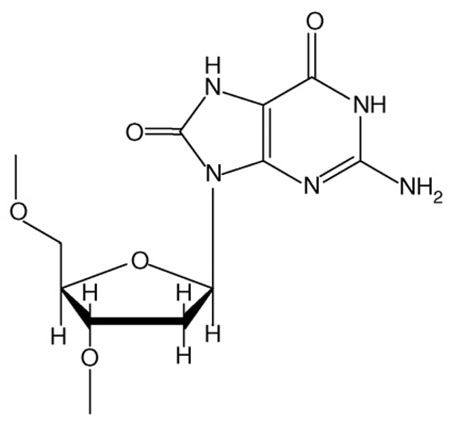
7,8-dihydro-8-oxoguanine (8-oxoG; Figure I) is the most prevalent mutagenic lesion derived from the interaction of ROS with DNA [67]. Approximately 104 such lesions arise per cell per day [68]. ROS are produced endogenously as by-products of normal aerobic respiration and inflammation [69]. In addition, they are produced by ionizing radiation [67], including diagnostic X-rays, and by the metabolism of polycyclic aromatic chemicals present in tobacco smoke, automobile exhaust and broiled proteinaceous foods [70]. Adding to its importance, the 8-oxoG lesion is, itself, a source of numerous types of DNA damage through further oxidative reactions [71]. DNA lesions derived from ROS are implicated in aging and many human diseases, including cancer and several neurogenerative and cardiovascular diseases [67]. Bypass of this lesion is mutagenic in BF and accurate in Dpo4 (Figure II).
Figure I. Structure of 8-oxoG.
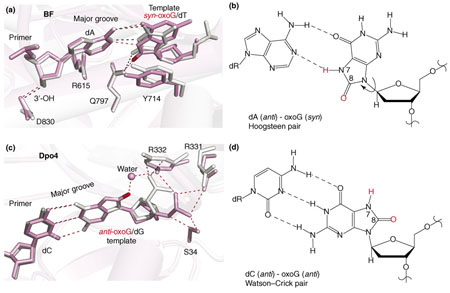
Figure II. 8-oxo-G lesions within the active site regions of BF and Dpo4. The designated syn and anti conformations differ by ~180° rotation about the bond connecting the base with the sugar. BF favors syn-8-oxoG–anti-A mispairing, which mimics a normal anti-A–anti-T Watson–Crick pair, whereas Dpo4 bypasses 8-oxoG with high fidelity through formation of a stabilizing hydrogen-bond network. The 8-oxoG-containing complexes are shown in pink, and the unmodified complexes in silver. (a) BF in binary complex with syn 8-oxo-guanine mismatched with dATP (PDB code: 1U49; pink) mimicking a normal A–T Watson–Crick pair in the unmodified complex (PDB code: 1U48; silver, superimposed). The O8 of the 8-oxoG is shown in red. Note that crucial hydrogen-bonding interactions between BF residues (Gln797, Arg615 and Asp830) and the 8-oxoG–A mispair, and those within the mispair (denoted by pink broken lines) are comparable to those in the unmodified complex. (b) The base-pairing alignment of syn oxoG with dATP. The O8 and H7 of 8-oxoG are shown in red. (c) Dpo4 in ternary complex with anti 8-oxo-guanine, Watson–Crick paired with dCTP (PDB code: 2ASD, chains A/D/E; pink), superimposed with unmodified complex (PDB code: 2ATL, chains A/D/E; silver). Note the water-mediated hydrogen bond between Arg332 and the unique O8 of 8-oxoG, which stabilizes the anti conformation, and the DNA-backbone rotation required to avoid steric close contact in accommodating the O8. (d) The base-pairing alignment of anti oxoG with dCTP. The O8 and H7 of 8-oxoG are shown in red.
Box 3. Benzo[a]pyrene-derived DNA lesions
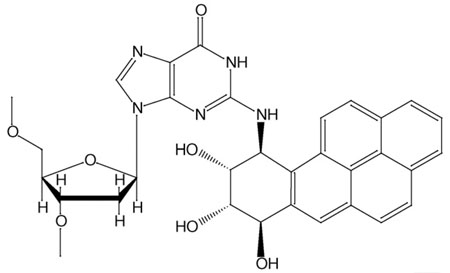
The environmental chemical pollutant benzo[a]pyrene is metabolically activated to form highly reactive intermediates that can subsequently react with DNA, yielding carcinogen–DNA adducts [72,73]. These lesions can produce mutations on replication in vitro and in vivo [74–76]. Such mutations, when present in DNA sequences that regulate the cell cycle, can trigger cancer initiation [72]. The most tumorigenic benzo[a]pyrene (BP) metabolite is the diol epoxide (+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydroben-zo[a]pyrene [(+)-anti-BPDE] [77]; this metabolite reacts largely, but not exclusively, with the exocyclic amino group of guanine to produce the major 10S (+) trans-anti-BP-N2-dG adduct (Figure I). In vitro primer extension studies using purified high-fidelity DNA polymerases have revealed that this lesion stalls the progress of high-fidelity DNA polymerases, although small amounts of bypass have been observed [24,36,74,78–80]. By contrast, although the rates of DNA replication catalyzed by Dpo4 are markedly retarded, translesion bypass by Dpo4 is much more facile than by BF, where polymerase blockage predominates [36] (L. Oum, PhD thesis, New York University, 2007). Notably, in both cases, bypass is frequently mutagenic. BP causes mainly blockage in BF, but mutagenic bypass in Dpo4 (Figure II).
Figure I. Structure of 10S (+) trans-anti-BP-N2-dG.
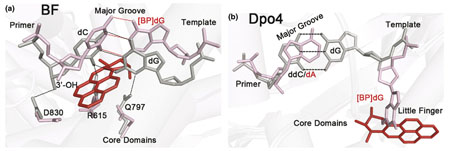
Figure II. The bulky 10S (+) trans-anti-BP-N2-dG adduct within the BF and Dpo4 active-site regions. The adduct stalls BF by disrupting polymerase–DNA interactions on the minor-groove side of the [BP]dG–dC pair, whereas it fits snugly on the minor-groove side in the gap between the Dpo4 core domains and the little finger domain. The 10S (+) trans-anti-BP-N2-dG-containing complexes are shown in pink with the BP moiety in red, and the unmodified complexes in gray. (a) BF binary complex with BP-N2-dG-modified template (PDB code: 1XC9) superimposed with unmodified complex (PDB code: 1L5U). The damaged dG pairs with dC in the primer strand. The BP moiety is positioned on the minor-groove side of the evolving duplex, disrupting the interactions between the minor-groove side of the newly formed base pair and proofreading residues Gln797 and Arg615; the 3′-OH of the primer dC base in the BP-modified complex is too far away to form the normal hydrogen bond with the conserved Asp830 residue. (b) Dpo4 ternary complex with BP-N2-dG-modified template (PDB code: 2IA6, chains B/E/F) superimposed with unmodified complex (PDB code: 2AGQ; silver). The BP moiety and the base of the damaged G are positioned outside the double helix on the minor-groove side of the DNA duplex, with the BP aromatic ring system in a gap between the Dpo4 little finger and core domains. The dA base in the primer strand lacks a base-pairing partner. In the unmodified complex, the dG base forms a base pair with the ddC (2′3′-dideoxycytidine) base of the primer strand.
The high-fidelity DNA polymerase BF
Comprehensive studies of BF crystal structures have uncovered properties that reflect the high fidelity and processivity of the A family, Pol I group DNA polymerases. Moreover, structures have been identified that reveal the different stages in the replication cycle (Figure 1a) and highlight the conformational transitions of the polymerase during this process [11].
Figure 1.
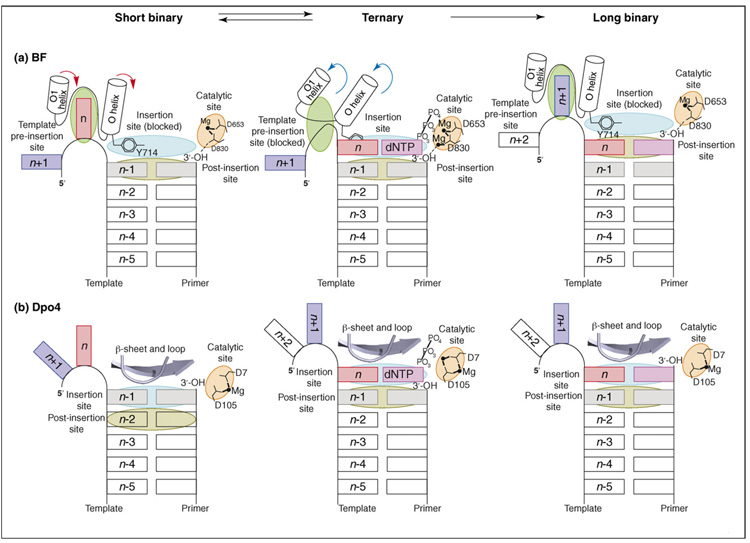
The replication cycle of BF and Dpo4. Schematic overviews of the polymerase active sites derived from the short binary, ternary and long binary structures, which represent a complete round of DNA synthesis. (a) The BF replication cycle starts with the acceptor template base (n; red rectangle) bound at the template pre-insertion site (between the O and O1 helices; green oval); Tyr714 blocks access to the insertion site (light blue oval) and stacks with the n−1 base pair at the post-insertion site (olive green oval). Formation of the closed conformation involves rearrangement (red arrows) of the O and O1 helices, which simultaneously blocks the template pre-insertion site and unblocks the insertion site. These rearrangements move the acceptor template base (n) to the insertion site, where it pairs with an incoming dNTP (pink rectangle) and Tyr714 to a position behind the nascent base pair. Nucleotide incorporation occurs on formation of a cognate base pair and proper assembly of the catalytic site (orange oval). The cycle is completed with translocation of the polymerase with respect to DNA by one base pair. The polymerase resets to the open conformation (blue arrows) in preparation for the next round of DNA synthesis. (b) The reaction cycle of Dpo4 also starts with the acceptor template base (n; red rectangle) positioned immediately 5′to the n–1 base pair. However, the pre-insertion site is absent in Dpo4. Formation of the ternary complex involves polymerase movement, which transits the n–1 base pair from the insertion site (light blue oval) to the post-insertion site (olive green oval), so that the acceptor template base n pairs with an incoming dNTP (pink rectangle) at the insertion site. Nucleotide incorporation occurs on formation of a cognate base pair and proper assembly of the catalytic site (orange oval). The cycle is completed with further translocation of the polymerase relative to DNA. Part (a) modified, with permission, from Ref. [11] (Copyright 2004, National Academy of Sciences, U.S.A.).
Initially, the templating base(n)resides in a pre-insertion site in the short binary complex (Figure 1a), in which the enzyme and the DNA primer-template are bound to one another and the dNTP is absent. The O helix of the fingers domain is in an ‘open’ conformation. Tyr714, at the base of the O helix, occupies the insertion site and stacks with the (n−1) base pair, thus preventing the next upstream 5′-templating (n) base from slipping into the active site. In the ternary complex – containing dNTP and DNA (Figure 1a) – the Tyr714 residue is displaced from the insertion site to a position behind the nascent base pair, thus enabling pairing of the incoming dNTP and the templating (n) base. BF is in a ‘closed’ conformation, resulting from a ~40° rotation of the O helix from its ‘open’ position [11]. Specifically, the O helix of the fingers domain closes down towards the flat surface of the nascent base pair, resulting in a tight-fitting, reaction-ready ternary complex. At this point, the close steric fit of the template base–dNTP pair is tailored for the correct Watson–Crick base pair, favoring accurate replication. This is termed an ‘induced-fit’ mechanism [28]. Following or accompanying the nucleotidyl-transfer chemical reaction, the polymerase translocates so that the nascent base pair proceeds to the post-insertion site and the polymerase complex returns to the open conformation with the Tyr714 residue once again occupying the insertion site (Figure 1a). At the end of this process, the DNA duplex has grown by one base pair, producing the post-insertion long binary complex. The next 5′-template (n+1) base occupies the pre-insertion site, ready for another cycle of replication.
The lesion-bypass DNA polymerase Dpo4
The first crystal structures of Dpo4-containing DNA primer–template complexes with incoming dNTP (ternary complexes), determined in 2001 [13], revealed many features common to lesion-bypass DNA polymerases [3]. For a recent, comprehensive review of the current understanding of Dpo4 and other Y-family DNA polymerase structure–function relationships, see Ref. [4]. Dpo4 has a spacious, highly solvent-accessible active-site region, whereas ternary complexes of high-fidelity DNA polymerases contain active sites with the nascent Watson–Crick pair in tight van der Waals contact with polymerase residues [10,11,29,30]. Although both high-fidelity and lesion-bypass polymerases contain a large solvent-exposed pocket on the evolving major-groove side of the duplex, lesion-bypass polymerases, including Dpo4, also contain a smaller open pocket on the minor-groove side (Box 1, Figure Id). Dpo4 and other lesion-bypass DNA polymerases have an additional flexible domain that is not present in high-fidelity polymerases – the little finger or polymerase associated domain (PAD) – but they lack an exonuclease domain. The Dpo4 active-site region is sufficiently spacious that two nucleotides of the template strand can be simultaneously accommodated [13], which is in contrast to high-fidelity DNA polymerases that can fit only one templating nucleotide in the active site. Furthermore, in the active site region, fewer Dpo4 amino acid residues are in contact with the DNA:Dpo4 forms hydrogen bonds only with the phosphate groups of the DNA duplex region [13]. By contrast, BF forms additional extensive sequence-independent interactions with the minor-groove edges of the first four base pairs at the growing end of the DNA duplex, with hydrogen bonds between the bases and highly conserved protein side chains (Box 4) or oriented water molecules anchored to protein side chains [31]. The sterically open Dpo4 active-site region is structured to enable the accommodation of DNA lesions, while facilitating low-fidelity DNA replication. Stages in the replication cycle of Dpo4 are illustrated in Figure 1b.
Box 4. Key structural differences between BF and Dpo4
Characteristic features of high-fidelity DNA polymerase structures are the close interactions between polymerase amino acid residues and the developing minor-groove side of the growing duplex. These interactions constitute a minor-groove scanning track or ‘reading head’ that enables the polymerase to sense an incorporated mismatch [31]; this proofreading promotes subsequent routing of the error-containing duplex for excision by an intrinsic exonuclease domain or an external exonuclease activity [10,59]. (Proofreading 3′→5′ exonuclease activity catalyzes the removal of mismatched nucleotides from the growing end of the primer strand.) As part of the minor-groove scanning track, two strictly conserved ‘reading’ residues, Arg615 and Gln797 in the case of BF, form hydrogen bonds with the newly formed base pair, ensuring its proper geometry [31]. Gln797 interacts with the template base and Arg615 interacts with the dNTP. By contrast, in lesion-bypass DNA polymerases, there is no exonuclease activity, the minor-groove scanning track is absent and the minor-groove side of the nascent duplex has far fewer interactions with the polymerase [4]. However, the major-groove side is open in both cases. Furthermore, Dpo4 lacks the O and O1 helices and the insertion-site-blocking tyrosine residue positioned at the base of the O helix; instead the nascent base pair is contacted by a β strand and a loop, whose conformation do not change greatly upon ternary complex formation (see Figure 1 of main text). The high-fidelity DNA polymerases are processive, and check for fidelity both before and after the nucleotidyl-transfer reaction. By contrast, the DNA and lesion-bypass DNA polymerases are distributive (non-processive), synthesizing only a few nucleo-tides before dissociating, and the accuracy of replication depends primarily on accurate Watson–Crick base pairing rather than on nucleotide selectivity based on steric fit [4]. This property has been demonstrated by kinetic studies, in which the use of variably sized non-polar thymidine analogs revealed low steric selectivity by Dpo4 [81].
Induced fit and translocation: differences between BF and Dpo4
Translocation is the processive movement of a polymerase relative to the DNA duplex between two rounds of the phosphoryl-transfer reaction during polynucleotide replication [32]; it is essential for polymerase function and necessarily occurs with every polymerase upon completion of one replication cycle (Figure 1). Although the actual dynamic mechanism of translocation is not known for any polymerase, static crystal structures can indicate plausible intermediate states in this process. Recent crystallo-graphic studies with BF [11] and Dpo4 [14] reveal structural rearrangements that accompany dNTP entry into the active site in addition to nucleotide incorporation, thus providing insight into how translocation might occur.
BF: one-step translocation
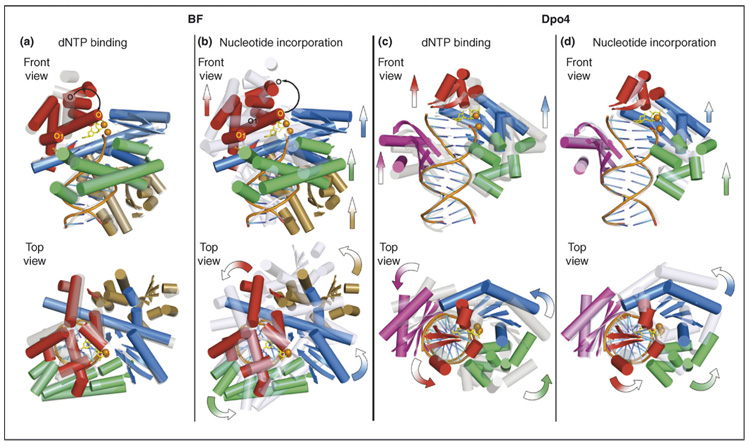
Structures of short binary, ternary, and long binary complexes are available for the high-fidelity DNA polymerase BF [11]; here, we analyze these structures to gain insight into the translocation process. For this purpose, we have selected three previously described structures [11] (PDB codes: 1L3T, 1LV5 and 1L3U), and superimposed them along their DNA phosphodiester backbones at residues in register (Figure 2a,b). The induced-fit conformational transition involving rotation of the O helix occurs upon nucleotide entry into the active site. However, no other translocational motion, that is, rotation or translation of the polymerase with respect to DNA, is apparent (Figure 2a). In proceeding from the ternary to the long binary complex, the polymerase translates by one nucleotide (~3.3 Å), and rotates around 1/10 of a turn of B-DNA (~36°); the fingers domain, including the O helix, returns to its open position after completion of the nucleotidyl-transfer reaction (Figure 2b).
Figure 2.
Different translocation mechanisms for BF and Dpo4 suggested from their crystal structures. BF: a one-step translocation mechanism that takes place following the nucleotide-incorporation phase; Dpo4: a step-wise translocation mechanism that spans both dNTP-binding and nucleotide-incorporation phases. All structures are superimposed according to the backbone atoms in the DNA-duplex region. Ternary complexes are shown in a solid multicolor cartoon representation, and the short and long binary complexes are shown in transparent gray and light blue cartoon representations, respectively. For clarity, only α helices and β sheets of proteins and DNA backbone and base registers are shown. For ternary complexes, the incoming dCTP is shown in yellow sticks, and divalent metal ions are shown in orange spheres. The thumb domains are shown in green, palm domains in blue, fingers domains in red, the BF exonuclease domain in gold and the Dpo4 little finger domain in purple. The front views are into the major-groove side of the nascent base pair; the top views are in the 5′→3′ direction of the template strand. For the dNTP-binding phase, movements are directed from the transparent gray representation towards the solid multi-colored representation; for the dNTP incorporation phase, the movements are directed from the solid multi-colored representation towards the transparent light blue representation. Arrows in the front views denote translational movements, and curved arrows in the top views denote rotational movements. (a) Superimposition of the BF short binary (PDB code: 1L3T) and ternary (PDB code: 1LV5, chains A/C/D) complexes. A predominant protein conformational change is observed in the closing of the O helix (black curved arrow) in the fingers domain during the dNTP binding step, whereas no other marked translational or rotational movement of polymerase domains with respect to DNA is observed. (b) Superimposition of the BF ternary (PDB code: 1LV5, chains A/C/D) and long binary (PDB code: 1L3U) complexes showing the translocational movement of the thumb, palm and fingers domains upon dNTP incorporation. The O helix resumes its position in the binary complex (black curved arrow). (c) Superimposition of the Dpo4 short binary (PDB code: 2ASJ, chains A/D/E) and ternary (PDB code: 2ASD, chains A/D/ E) complexes showing the relocation of the polymerase domains during the dNTP-binding step, using the thumb–DNA contact points as the hinge. Note that no marked translational movement is observed for the thumb domain. (d) Superimposition of the Dpo4 ternary (PDB code: 2ASD, chains A/D/E) and long binary (PDB code: 2ASL, chains A/D/E) complexes showing the relocation of the polymerase domains during the dNTP incorporation step, using the little-finger–DNA contact points as the hinge. Note that no marked translational or rotational movement is observed for the little finger domain. In (c) and (d), the entire polymerase is relocated in two steps as a near rigid body, in which the overall protein conformation undergoes little change.
Dpo4: stepwise translocation
Dpo4 crystal structures containing a short binary complex, a ternary complex, and a long binary complex following the nucleotidyl-transfer reaction have been investigated recently [14]. Superimposition along their DNA phosphodiester backbones at residues in register revealed no conformational changes akin to the induced-fit motif in high-fidelity DNA polymerases. In this case, a stepwise, two-phase translocation process was suggested: first, a predominant movement of the palm, fingers, and little finger domains with respect to DNA occurs during nucleotide binding, generating the space needed for nascent base pair formation (Figure 2c); this is followed by primarily a thumb movement accompanying or following the chemical nucleotidyl-transfer reaction (Figure 2d). In each segment, the entire polymerase is relocated, with respect to DNA, as a nearly rigid body. The rigid-body motion entails movement of the entire unit, for example, movement of the whole hand about the wrist, without marked movement of any fingers. Thus, the overall protein conformation itself is changed little in proceeding from the binary to the ternary complex, and the protein conformations of short and long binary complexes are identical. The crystal structures indicate a screwlike counter-clockwise rotation and translation of the polymerase along the DNA helix axis, when viewed in the 5′→3′ direction of the template strand, during a replication cycle. The right-handed screw of the DNA is employed as a scaffold for threading through the polymerase, advancing the helix by a rise (~3.3 Å) and a twist angle (~36 °) corresponding to approximately one DNA base pair step during each replication cycle.
Thus, the crystal structures indicate differences in the translocation mechanisms used by Dpo4 and BF. Dpo4 translocation displays a two-phase mechanism, whereas a single step is indicated for BF. Specifically, for Dpo4, the first stage of the translocation is suggested to occur as the incoming dNTP enters the active site, and the second stage is associated with the chemical reaction step. In agreement with this mechanism, a stepwise translocation mechanism for a close homolog of Dpo4, Dbh, has recently been inferred through kinetics studies [33]. By contrast, the BF crystal structures indicate that the entire translocation cycle occurs following dNTP entry into the BF active site and the induced-fit polymerase closing. The driving forces that could power translocation have been discussed based on crystal structures of a B-family DNA polymerase, phi29 from Bacillus subtilis [32]: dissociation of pyrophosphate breaks the electrostatic link between the fingers domain and the catalytic Mg2+ ions, enabling the fingers to pivot to the open position. Then, two conserved tyrosine residues enter the insertion site and sterically exclude the nascent base pair. Evolving charge dynamics during the nucleotidyl-transfer reaction is an important element in the replication process and has been computationally elucidated for Dpo4 [34].
Small lesion processing: 8-oxoG
BF favors 8-oxoG-A mispairing
The small 8-oxoG lesion is highly prevalent (Box 2); hence, it is inevitable that high-fidelity processive DNA polymerases will encounter this lesion, despite the activity of specific repair enzymes, which scan DNA to locate and repair it [35]. Biochemical processing and X-ray crystal-lographic studies of DNA containing 8-oxoG in complexes with BF [25] and Dpo4 [14,26] have highlighted striking differences in how these polymerases cope with this oxidative guanine lesion. The high-fidelity DNA polymerase BF incorporates the mismatched dATP opposite the damaged base approximately ninefold more efficiently than it does the normal partner dCTP [25], whereas Dpo4 favors dCTP over dATP by > 100-fold [14]. Following incorporation of 2′-deoxycytidine (dC) or 2′-deoxyadenosine (dA), extension occurs much more readily with adenine than with cytosine in BF [25]. Examination of the BF crystal structures has revealed the basis for the preferential selection of dATP by a templating 8-oxoG base in this enzyme [25]. The syn-8-oxoG-anti-A pair in the BF ternary complex mimics the geometry of a normal Watson–Crick anti-T–anti-A pair. Syn and anti conformations (see Box 2, Figure II) differ by a ~180° rotation about the glycosidic bond connecting base to sugar. The 8-oxoG template adopts the syn glycosidic bond conformation, and forms a Hoogsteen pair with the incoming dATP (see Box 2, Figure IIa,b). In the syn conformation, the steric crowding between the O8 and adjacent sugar–backbone moieties is alleviated, and the syn 8-oxoG mimics a pyrimidine in the active site, leaving the active site essentially undistorted. Crucial hydrogen bonds between the polymerase residues (Arg615, Gln797 and Asp830) and the syn-8-oxoG–anti-A pair remain comparable to those between the polymerase and the anti-T-anti-A pair (see Box 2, Figure IIa,b). However, with dCTP opposite 8-oxoG, the 8-oxoG in the anti glycosidic bond conformation is Watson–Crick-paired with dC. The steric crowding produced by the O8 atom is relieved by torsional rotations in the backbone that lift the 8-oxoG away from the surface of the polymerase. This action disrupts the interaction between 8-oxoG and the Gln797 residue in the minor-groove scanning track (Box 4). The alignments of the O and O1 helices are distorted, thus inhibiting entry of the next template into the pre-insertion site.
Dpo4 bypasses 8-oxoG accurately
The Y-family DNA polymerase Dpo4 exhibits a ~100-fold preference for inserting dCTP over dATP opposite 8-oxoG, with dCTP incorporation nearly as efficient as that opposite an undamaged guanine base [14]. The extension frequency beyond an 8-oxoG–C base pair is approximately eightfold that beyond a normal G–C base pair at the same site [14]. By contrast, the extension frequency beyond an 8-oxoG–A mismatched base pair is ~30 times less than beyond an 8-oxoG–C pair. Thus, the 8-oxoG lesion is bypassed by Dpo4 with relatively high fidelity. The reasons for this bypass fidelity reside in the unique structural features of the 8-oxoG–C base pair in Dpo4. The O8 atom causes relatively little overall structural perturbation of the DNA compared with the structures of the undamaged complex (see Box 2, Figure IIc,d). Moreover, the crystal structure of the ternary complex – containing the templating 8-oxoG lesion Watson–Crick paired with dCTP – shows a unique network of hydrogen bonds, which stabilize the 8-oxoG with the glycosidic torsion angle of the modified guanine in the normal anti conformation (see Box 2, Figure IIc,d): there are two non-specific hydrogen bonds between the phosphodiester backbone of the damaged base with the Arg331 guanidine group, one with the Arg332 guanidine group, two with the hydroxyl group of Ser34 and one key water-mediated hydrogen bond with the Arg332 side chain to the unique O8 atom of the 8-oxoG residue. By contrast, in the case of the unmodified guanine at the same position (see Box 2, Figure IIc), there are only two hydrogen bonds from the phosphate backbone of the unmodified glycine to the Arg332 guanidine group. The hydrogen-bonding network in the 8-oxoG structure is an indirect readout of the unique O8 atom of the 8-oxoG: the steric clash engendered by the O8 forces the backbone to rotate by ~180° around its O3′–P torsion, relocating it in a way that produces the observed hydrogen-bonding network. Analogous crystal structure studies using Dpo4 harboring Arg332 substitutions also support the view that a hydrogen bond between Arg332 and 8-oxoG helps in determining the high fidelity and efficiency of Dpo4-catalyzed bypass of this lesion [26]. The general principle emerges that a templating 8-oxoG residue within high-fidelity DNA polymerases, with their confined active sites, has a greater preference than that within Dpo4 for adopting the abnormal syn glycosidic bond conformation to alleviate the steric crowding. The spacious active site of Dpo4 enables a different structural rearrangement in the DNA backbone that relieves steric crowding without distorting the enzyme structure. This rearrangement produces the unique hydrogen-bonding network that stabilizes the anti Watson–Crick paired conformation.
Processing bulky lesions: BP-derived adducts
BF is stalled by bulky BP-derived adducts
We now focus on lesions produced by the covalent reaction between a tumorigenic metabolite of the pre-carcinogen BP and guanine in DNA (Box 3). A crystal structure of this lesion in the BF post-insertion site [24] has been determined (PDB code: 1XC9), and it demonstrates why BF is predominantly blocked by this lesion [24,36]. This structure is a binary complex and represents BF after nucleotide incorporation opposite the BP–2′-deoxyguanosine (dG) adduct; the BP aromatic rings are 5′-directed along the template strand on the evolving minor-groove side, with Watson–Crick pairing intact at the lesion site (see Box 3, Figure IIa). This conformation is structurally similar to NMR structures observed for this adduct in solution in the absence of protein, indicating that conformational preferences in solution can be maintained with biological relevance in the polymerase [27]. The DNA duplex is extensively distorted over three base pairs on the 3′ side of the lesion and the polymerase is severely perturbed locally. Because the BP rings are positioned in the crowded BF minor-groove scanning track (Box 3), key interactions between protein residues and the [BP]dG–dC base pair are impaired. Notably, the crucial hydrogen-bonding interactions between Arg615 and Gln797 in the minor-groove reading head, and the [BP]dG–dC base pair are disrupted (see Box 3, Figure IIa). Owing to the wedging of the BP rings between the DNA and the polymerase, primer and template strands are separated from the polymerase minor-groove scanning track. Consequently, the primer 3′-OH group is no longer poised for in-line attack during the nucleotidyl-transfer reaction (see Box 3, Figure IIa). Furthermore, the BP ring system is positioned to sterically exclude entry of an incoming dNTP into the active site for the subsequent extension step. It is plausible that the polymerase distortions and attendant stalling are signals for recruiting the lesion-bypass DNA polymerases. The infrequent extension observed in BF probably necessitates a rearrangement of the [BP]dG moiety, likely through a rotation of the glycosidic bond from the normal anti to the syn domain, which places the BP rings on the spacious, open major-groove side of the evolving duplex in the polymerase [36,37].
Dpo4 produces mismatch and frameshift mutations at BP-derived lesions
Dpo4 can accommodate a bulky BP aromatic ring system on both the major- and minor-groove sides of the evolving duplex. The available crystal structures of BP-derived adducts reveal some of these possibilities. In a recent series of crystal structures of the 10S (+)-trans-anti-BP-N2-dG adduct (Box 3) in the post-insertion position, with a dA constructed to be opposite the lesion, one structure has the BP aromatic ring system extruded into a gap between the little fingers and the core domains, on the minor-groove side of the evolving duplex [22] (see Box 3, Figure IIb). The damaged anti-guanine is also extruded into this pocket, leaving adenine of the primer strand without a base-pairing partner. Another structure with BP similarly extruded shows the adducted guanine displaced and looped out from the active site; this presents a structural model of a slipped-frameshift intermediate that, if bypassed, would produce a –1 deletion. In vitro primer extension studies in this work revealed incorporation of adenine opposite the lesion in addition to −1 deletions [22]. The well-stabilized accommodation of the 10S (+)-trans-anti-BP-N2-dG adduct on the minor-groove side in the Dpo4 complex contrasts sharply with its damaging impact on protein–DNA interactions when on the evolving minor-groove side in the high-fidelity DNA polymerase BF. The minor-groove lesion positioning is suggested to be relevant to other Y family DNA polymerases of the DinB branch, including the human homolog Pol κ [22]. The DinB polymerases seem to have a special role in bypassing N2-dG lesions [38], with higher efficiency and accuracy than other DNA polymerases in cellular [39] and in vitro environments [40,41].
When the tumorigenic BP metabolite (+)-anti BPDE (Box 3) reacts chemically with DNA, other lesions are formed in addition to the major reaction product with the amino group of guanine [42]. Among these is the adenine 10R (+)-cis-anti-BP-N6-dA adduct. Two different conformers were obtained in a crystal structure of this adduct at the Dpo4 post-insertion site [23]. In vitro primer extension studies revealed that dA is predominantly misincorporated opposite this adenine lesion, and frameshift mutations were also observed. The first conformation shed light on the observed misincorporation of dA: the BP aromatic rings, located in the open major-groove-side pocket of the polymerase, caused displacement of the damaged adenine towards the major groove so that it could not Watson–Crick hydrogen bond to its partner 2′-deoxythymidine (dT). However, the structure indicated that the dislocated, adducted adenine was well placed for mispairing with another adenine. The second conformer, with the BP ring system intercalated (nearly identical to its NMR solution structure [27]), might have an intermediate role in producing the observed frameshift mutations [23]. Primer extension studies also showed that replication beyond this lesion is rather poor [23]. Modeling studies indicate that extension would be inhibited when bulky adducts are positioned on the evolving major-groove side because movement of the little finger, which is necessary for successful translocation, is hindered [43].
Overall, the spacious Dpo4 active site, with limited contacts to DNA and space to house a bulky lesion on the evolving major- and minor-groove side, facilitates bulky lesion bypass, which is frequently mutagenic and can be undiscriminating in nucleotide selectivity [22,37] (L. Oum, PhD thesis, New York University, 2007). By contrast, the active-site region of BF, with its close fit to the evolving minor groove, is highly disturbed when a bulky lesion is housed in this position, leading primarily to polymerase blockage, which could be a signal for the switch to a lesion-bypass DNA polymerase [24,36].
Concluding remarks and future perspectives
The available lesion-containing crystal structures provide a springboard for molecular modeling and dynamics studies for cases in which crystals are not yet available [36,37,44,45]. Eagerly awaited are future structural and biochemical studies of lesion-containing DNA polymerases, to provide further insights into mechanisms of high- and low-fidelity translesion bypass. Of particular interest are the human lesion-bypass DNA polymerases η, ι, κ and Rev 1. Crystal structures of these enzymes from humans [46,47] and yeast [48,49] have been obtained, as has a lesion-containing yeast Pol η structure [50]. Complemented by biochemical studies [3,50], these structures reveal unique features for each enzyme that reflect striking specificities in their function. One question that arises is whether these human lesion-bypass DNA polymerases, like the archaeal Dpo4, also employ a two-step translocation mechanism [14]. Another challenge involves delineating connections between lesion impact on DNA polymerase function and lesion repair. Here, RNA polymerases provide a potential link: bulky lesions such as those derived from BP [51] cause RNA polymerase stalling, which invokes the transcription-coupled nucleotide excision repair (NER) machinery [52,53], but much remains to be learned about relationships between lesion structure and repair susceptibility by this machinery [54]. Understanding lesion-processing in cells will require the elucidation of concerted interactions between Y-family DNA polymerases with other polymerases and other components of the holoenzyme replication complex; the participation of other proteins such as the eukaryotic ubiquitylation machinery also must be further explicated [6,7,55–57]. To fully decipher how high-fidelity and lesion-bypass DNA polymerases correct the plethora of possible DNA lesions, structural and biochemical information will be needed on a case-by-case basis. However, the structure–function relationships indicated by the model systems described here are plausibly relevant to lesion processing by other high-fidelity and lesion-bypass DNA polymerases, in view of structural similarities within high-fidelity and lesion-bypass polymerase family members [4,10].
Acknowledgements
Our work is supported by NIH Grants CA28038 and CA75449 to S.B., CA99194 to N.E.G. and CA46533 to D.J.P. Molecular images were made with PyMOL (DeLano Scientific LLC, www.delanoscientific.com). We thank Lucy Malinina for her key role in the crystallographic studies from the Patel laboratory. We thank Wei Yang and Hong Ling for critical reading of the manuscript and very helpful discussions. In addition, we thank the reviewers for many helpful suggestions.
References
- 1.Lewin B. Genes V. Oxford University Press; 1994. Primosomes and replisomes: the apparatus for DNA replication; pp. 571–603. [Google Scholar]
- 2.Friedberg EC, et al. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 3.Prakash S, et al. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 4.Yang W, Woodgate R. What a difference a decade makes: Insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pagès V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 6.Friedberg EC, et al. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst.) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Pavlov YI, et al. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- 9.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 10.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SJ, et al. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, et al. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 14.Rechkoblit O, et al. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trincao J, et al. Dpo4 is hindered in extending a G.T mismatch by a reverse wobble. Nat. Struct. Mol. Biol. 2004;11:457–462. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- 16.Vaisman A, et al. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 18.Warren JJ, et al. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zang H, et al. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 20.Nair DT, et al. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat. Struct. Mol. Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 21.Zang H, et al. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 22.Bauer J, et al. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling H, et al. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu GW, et al. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J. Biol. Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 25.Hsu GW, et al. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. doi: 10.1038/nature02908. [DOI] [PubMed] [Google Scholar]
- 26.Eoff RL, et al. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 27.Broyde S, et al. DNA adduct structure-function relationships: Comparing solution with polymerase structures. Chem. Res. Toxicol. 2008;21:45–52. doi: 10.1021/tx700193x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong I, et al. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 29.Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 30.Franklin MC, et al. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 31.Kiefer JR, et al. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 32.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLucia AM, et al. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry. 2007;46:10790–10803. doi: 10.1021/bi7006756. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, et al. A water-mediated and substrate-assisted catalytic mechanism for Sulfolobus solfataricus DNA polymerase IV. J. Am. Chem. Soc. 2007;129:4731–4737. doi: 10.1021/ja068821c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David SS, et al. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu P, et al. Following an environmental carcinogen N2-dG adduct through replication: elucidating blockage and bypass in a high-fidelity DNA polymerase. Nucleic Acids Res. 2007;35:4275–4288. doi: 10.1093/nar/gkm416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlow-Poehnelt RA, et al. The spacious active site of a Y-Family DNA polymerase facilitates promiscuous nucleotide incorporation opposite a bulky carcinogen-DNA adduct: elucidating the structure-function relationship through experimental and computational approaches. J. Biol. Chem. 2004;279:36951–36961. doi: 10.1074/jbc.M404332200. [DOI] [PubMed] [Google Scholar]
- 38.Jarosz DF, et al. Proficient and accurate bypass of persistent DNA lesions by DinB DNA polymerases. Cell Cycle. 2007;6:817–822. doi: 10.4161/cc.6.7.4065. [DOI] [PubMed] [Google Scholar]
- 39.Avkin S, et al. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase κ. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki N, et al. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 41.Rechkoblit O, et al. Translesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 2002;277:30488–30494. doi: 10.1074/jbc.M201167200. [DOI] [PubMed] [Google Scholar]
- 42.Cheng SC, et al. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem. Res. Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 43.Zhang L, et al. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): modeling and dynamics studies. Nucleic Acids Res. 2006;34:3326–3337. doi: 10.1093/nar/gkl425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandani S, Loechler EL. Molecular modeling benzo[a]pyrene N2-dG adducts in the two overlapping active sites of the Y-family DNA polymerase Dpo4. J. Mol. Graph. Model. 2007;25:658–670. doi: 10.1016/j.jmgm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Meneni S, et al. Examination of the long-range effects of aminofluorene-induced conformational heterogeneity and its relevance to the mechanism of translesional DNA synthesis. J. Mol. Biol. 2007;366:1387–1400. doi: 10.1016/j.jmb.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lone S, et al. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 47.Nair DT, et al. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 48.Trincao J, et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 49.Nair DT, et al. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 50.Alt A, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 51.Perlow RA, et al. DNA adducts from a tumorigenic metabolite of benzo[a]pyrene block human RNA polymerase II elongation in a sequence- and stereochemistry-dependent manner. J. Mol. Biol. 2002;321:29–47. doi: 10.1016/s0022-2836(02)00593-4. [DOI] [PubMed] [Google Scholar]
- 52.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 53.Scicchitano DA. Transcription past DNA adducts derived from polycyclic aromatic hydrocarbons. Mutat. Res. 2005;577:146–154. doi: 10.1016/j.mrfmmm.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 54.Svejstrup JQ. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem. Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Livneh Z. Keeping mammalian mutation load in check: regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5:1918–1922. doi: 10.4161/cc.5.17.3193. [DOI] [PubMed] [Google Scholar]
- 56.Schmutz V, et al. Requirements for PCNA monoubiquitination in human cell-free extracts. DNA Repair (Amst.) 2007;6:1726–1731. doi: 10.1016/j.dnarep.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Fujii S, Fuchs RP. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J. Mol. Biol. 2007;372:883–893. doi: 10.1016/j.jmb.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 58.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 59.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kokoska RJ, et al. Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J. Biol. Chem. 2002;277:19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 61.Boudsocq F, et al. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic polη. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiefer JR, et al. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 Å resolution. Structure. 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, et al. Fidelity of eucaryotic DNA polymerase δ holoenzyme from Schizosaccharomyces pombe. J. Biol. Chem. 2000;275:17677–17682. doi: 10.1074/jbc.M910278199. [DOI] [PubMed] [Google Scholar]
- 64.Tabor S, et al. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 65.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 66.Lehmann AR. New functions for Y family polymerases. Mol. Cell. 2006;24:493–495. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 67.Evans MD, et al. Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 68.Brieba LG, et al. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dizdaroglu M. Chemistry of free radical damage to DNA and nucleoprotein. In: Halliwell B, Aruoma OI, editors. DNA and Free Radicals. Ellis Horwood; 1993. pp. 19–39. [Google Scholar]
- 70.Park JH, et al. Polycyclic aromatic hydrocarbon (PAH) o-quinones produced by the aldo-keto-reductases (AKRs) generate abasic sites, oxidized pyrimidines, and 8-oxo-dGuo via reactive oxygen species. Chem. Res. Toxicol. 2006;19:719–728. doi: 10.1021/tx0600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 72.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 73.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol. Appl. Pharmacol. 2005;206:73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 74.Geacintov NE, et al. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 75.Fernandes A, et al. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2-2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 76.Yin J, et al. A role for DNA polymerase V in G→T mutations from the major benzo[a]pyrene N2-dG adduct when studied in a 5′-TGT sequence E. coli. DNA Repair (Amst.) 2004;3:323–334. doi: 10.1016/j.dnarep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 77.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 78.Zhuang P, et al. Base sequence dependence of in vitro translesional DNA replication past a bulky lesion catalyzed by the exo− Klenow fragment of Pol I. Biochemistry. 2001;40:6660–6669. doi: 10.1021/bi010005o. [DOI] [PubMed] [Google Scholar]
- 79.Lipinski LJ, et al. Effect of single benzo[a]pyrene diol epoxide-deoxyguanosine adducts on the action of DNA polymerases in vitro. Int. J. Oncol. 1998;13:269–273. doi: 10.3892/ijo.13.2.269. [DOI] [PubMed] [Google Scholar]
- 80.Alekseyev YO, et al. Effects of benzo[a]pyrene DNA adducts on Escherichia coli DNA polymerase I (Klenow fragment) primer-template interactions: evidence for inhibition of the catalytically active ternary complex formation. Biochemistry. 2001;40:2282–2290. doi: 10.1021/bi002245u. [DOI] [PubMed] [Google Scholar]
- 81.Mizukami S, et al. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]