Abstract
The B cell translocation gene (BTG) family regulates gene transcription and cellular differentiation and inhibits proliferation. The present study investigated the spatiotemporal expression pattern of BTG members and their potential role in the rat ovary during the periovulatory period. Immature female rats (22–23 d old) were injected with pregnant mare serum gonadotropin to stimulate follicular development. Ovaries or granulosa cells were collected at various times after hCG administration (n = 3 per time point). Real-time PCR analysis revealed that mRNA for Btg1, Btg2, and Btg3 were highly induced both in intact ovaries and granulosa cells by 4–8 h after hCG treatment, although their temporal expression patterns differed. In situ hybridization analysis demonstrated that Btg1 mRNA expression was highly induced in theca cells at 4 h after hCG, primarily localized to granulosa cells at 8 h, and decreased at 24 h. Btg2 and Btg3 mRNA was also induced in granulosa cells; however, Btg2 mRNA was observed in newly forming corpora lutea. Inhibition of progesterone action and the epidermal growth factor pathway did not change Btg1 and Btg2 mRNA expression, whereas inhibition of prostaglandin synthesis or RUNX activity diminished Btg2 mRNA levels. Overexpression of BTG1 or BTG2 arrested granulosa cells at the G0/G1 phase of the cell cycle and decreased cell apoptosis. In summary, hCG induced Btg1, Btg2, and Btg3 mRNA expression predominantly in the granulosa cell compartment. Our findings suggest that the induction of the BTG family may be important for theca and granulosa cell differentiation into luteal cells by arresting cell cycle progression.
The BTG family is induced by hCG and may regulate theca and granulosa cell differentiation into luteal cells by arresting cell cycle progression
The LH surge initiates a series of changes in the granulosa cells of the preovulatory follicle culminating in their transition to luteal cells. The hallmarks of this transition from granulosa cells to luteal cells include a conversion from a proliferative, estrogen-producing granulosa cell to a differentiated, progestin-secreting luteal cell (1). One key feature regulating cell proliferation and differentiation lies in control of the cell cycle. There are numerous signals controlling cell progression or arrest (2). One gene family that negatively regulates cell proliferation is the B cell translocation gene (BTG) family. Btg1 was originally described as a chromosomal translocation associated with B-cell chronic lymphocytic leukemia (3). Since its original discovery, more than 20 members of this family have been described in several species (4,5,6,7). However, several of the members of the family have been classified with different names for the same gene. Currently, six distinct proteins of this family have been identified in the human (BTG1, BTG2/TIS21/PC3, BTG3, BTG4/PC3B, Transducer of ErbB-2, and TOB2) (8,9,10).
Evidence for the BTG family inhibiting cellular proliferation and regulating differentiation is found in a variety of model systems (10). For example, NIH 3T3 cells overexpressing Btg genes inhibited cell proliferation (3,11,12). Similarly, PC12 cells overexpressing Btg2 (PC3) demonstrated an inhibition of the transition from G1 to the S phase of cell cycle and a diminution of cell division (13). In the testis, all the members of the BTG family are expressed (5,11,14,15,16), but of interest is the finding that BTG1 is highly expressed only in round spermatids that have completed meiosis and are undergoing the differentiative process of spermiogenesis (15). The presence of BTG1 at this stage of seminiferous epithelial maturation may reflect the role of BTG1 in the irreversible exit from the cell cycle leading toward differentiation (15).
Members of the PC3/BTG/TOB family have been reported in the oocyte and ovary mainly from gene screening approaches (5,16,17,18,19). Gene screening between dominant and subordinate bovine follicles revealed that Btg3 mRNA was elevated in the subordinate follicles (18). Early reports on the cloning of PC3B (BTG4) found it was highly expressed in the murine testis and oocyte. Its expression decreased in the morula and blastocyst, which led to the suggestion that PC3B/BTG4 may have a role in gametogenesis (4). Likewise, Btg4 mRNA was identified as one of the top five oocyte-specific genes expressed in bovine oocytes compared with somatic tissues (17).
Based upon the paucity of data regarding the members of the BTG family in the ovary and their potential role in regulating cellular proliferation, the present study was undertaken to characterize the expression patterns during the periovulatory period in the rat. We hypothesized that the LH surge would induce the BTG family members and that their induction may play a role in the transition of granulosa cells to luteal cells by regulating cell cycle kinetics.
Materials and Methods
Materials and reagents
Unless otherwise noted, all chemicals and reagents were purchased from Sigma Chemical Co. (St. Louis, MO). Molecular biological enzymes, molecular size markers, oligonucleotide primers, pCRII-TOPO vector, culture media, and Trizol were purchased from Invitrogen Life Technologies, Inc. (Carlsbad, CA).
Tissue collection
All animal procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee. Immature 15-d-old female Sprague Dawley rats were purchased and shipped with the mother from Harlan Sprague Dawley Inc. (Indianapolis, IN). Animals were maintained on a 12-h light, 12-h dark cycle and provided water and rat chow ad libitum. On the morning of d 22–23, rats (approximately 58.3 ± 7.6 g) were injected with pregnant mare serum gonadotropin (PMSG, 10 IU) sc. Forty-eight hours later, animals were injected with human chorionic gonadotropin (hCG, 5 IU) sc. Ovulation in this model occurs 12–16 h after hCG (personal observation, about 52 ova per rat released). Ovaries were collected at 0 h (i.e. time of hCG administration) and 4, 8, 12, or 24 h after hCG administration (n = 3–4 animals per time point). Ovaries were stored at −70 C for later isolation of total RNA or protein or placed in Tissue-Tek OCT compound (VWR Scientific, Atlanta, GA), snap frozen, and stored at −70 C until sectioned and processed for in situ hybridization analyses.
Rat granulosa cell culture
To isolate granulosa cells, ovaries were collected 48 h after PMSG administration and processed as described previously (20). Briefly, follicles were punctured and cells gently expressed, pooled, filtered, and pelleted by centrifugation. Cells were resuspended in defined medium consisting of Opti-MEM medium supplemented with 0.05 mg/ml gentamicin and 1× ITS (insulin, transferrin, and selenium). The cells were cultured in the absence or presence of various reagents for time points outlined below for each experiment at 37 C in a humidified atmosphere of 5% CO2. When reagents were dissolved in dimethylsulfoxide (DMSO), the same concentrations of DMSO were added to medium for the control cells. The final concentration of DMSO was less than 0.1%. For the indomethacin experiments, rats were injected with indomethacin (1 mg/animal, ip; Sigma) 48 h after PMSG priming. Rats were then injected with hCG 1 h later, and granulosa cells were collected from ovaries 4 h after hCG administration. At the end of each culture period or collection, cells were collected and snap-frozen for later isolation of total RNA or protein.
Quantification of mRNA for BTG genes
Total RNA was isolated from ovaries or granulosa cells using Trizol™ reagent according to the manufacturer’s protocol. Real-time PCR was used to measure expression of Btg1, Btg2, and Btg3 mRNA in vitro and in vivo. Oligonucleotide primers corresponding to cDNA for rat L32 (forward, 5′-GAA-GCC-CAA-GAT-CGT-CAA-AA-3′; reverse, 5′-AGG-ATC-TGG-CCC-TGG-CCC-TTG-AAT CT-3′), rat Btg1 (forward, 5′-CGT-TGT-ATT-CGC-ATC-AAC-C-3′; reverse, 5′-AGC-CAT-CCT-CTC-CAA-TCC-3′), rat Btg2 (forward, 5′-TCA-AAG-CTC-CAG-GGA-ACT-CC-3′; reverse, 5′-CTA-AAA-CCC-ACC-AGG-AAT-CAG-G-3′), and rat Btg3 (forward, 5′-TTC-AGA-ACT-GAT-ATT-CCC-ACC-3′; reverse, 5′-TGA-TTC-CGA-TCA-CAA-TGC-3′) were designed using PRIMER3 software (21). The specificity for each primer set was confirmed by both electrophoresis of the PCR products on a 2.0% agarose gel and analyzing the melting (dissociation) curve in the MxPro real-time PCR analysis program (Stratagene, La Jolla, CA) after each real-time PCR. The PCR products were sequenced before using. The real-time PCR contained 10% of the RT reaction product, 0.4 μm forward and reverse primers, 0.3 μl 1:10 diluted ROX reference dye (provided with SYBR Green ER qPCR SuperMix Universal kit, Invitrogen), and SYBR Green SuperMix. PCR was performed on Mx3000P QPCR System (Stratagene). The thermal cycling steps included 2 min at 50 C to permit optimal AmpErase uracil-N-glycosylase activity, 10 min at 95 C for initial denaturation, and then each cycle of 15 sec at 95 C, 30 sec at 55 C, and 45 sec at 72 C for 40 cycles, followed by 1 min at 95 C, 30 sec at 58 C, and then 30 sec at 95 C for ramp dissociation. The relative amount of each Btg gene transcript was calculated using the 2−ΔΔCT method (22) and normalized to the endogenous L32 reference gene.
Western blot analysis
Intact ovarian tissues were homogenized in RIPA buffer/protease inhibitor cocktail. Granulosa cells were resuspended in RIPA buffer/protease inhibitor cocktail for 30 min on ice. Tissue and cell lysates were then centrifuged at 14,000 × g for 10 min. The supernatants were stored at −70 C until use. Twenty micrograms of protein, measured by the Lowry method (23), were separated on a 15% SDS-PAGE gel and transferred to a nitrocellulose membrane (Whatman, Sanford, ME). Western blotting was performed by blocking nonspecific binding with 5% dry milk in Tris-buffered saline buffer containing 0.1% Tween 20 for 1 h. Blots were incubated with the primary antibody for BTG1 (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), BTG2 (1:1000; Abcam, Cambridge, MA), or β-actin (1:2000; Cell Signaling Technology, Danvers, MA) overnight at 4 C on a rocking platform. Blots were washed four times with PBS plus 0.1% Tween 20 and incubated with the respective secondary antibodies linked to horseradish peroxidase for 1 h. After extensive washing, blots were analyzed using an enhanced chemiluminescence detection system (GE Healthcare, Piscataway, NJ) and exposed to x-ray film. BTG3 protein expression was not explored because we were unable to get a distinct band of the appropriate size using anti-BTG3 antibodies from different commercial vendors.
Generation of recombinant adenovirus vector and granulosa cell infection
Adenovirus vectors were created to overexpress BTG1 and BTG2 to explore the function of these BTG family members. The Btg1 and Btg2 cDNA were amplified by PCR using total RNA isolated from rat granulosa cells. The specific primers for rat Btg1 (forward, 5′-TTA-GTC-GAC-CAC-CAT-GCA-TCC-CTT-CTA-CAC-TC-3′; reverse, 5′-TTA-AAG-CTT-TTA-ACC-TGA-TAC-AGT-CAT-CAT-A-3′) and rat Btg2 (forward, 5′-TTA-GTC-GAC-CAC-CAT-GAG-CCA-CGG-GAA-GAG- AA-3′; reverse, 5′-TTA-AAG-CTT-CTA-GCT-GGA-GAC-AGT-CAT-CAC-G-3′) contained SalI and HindIII sites. The PCR product was subcloned into the pShuttle-CMV vector (Stratagene). AdEasy XL Adenovirus vector system was used for generation of the recombinant Ad-BTG1 and Ad-BTG2 vectors. The resultant plasmids encoded the Btg1 or Btg2 gene under the control of a CMV promoter. These plasmids were linearized using Pme1 and then cotransformed into electro-competent BJ5183 bacteria with pAdEasy-1, which contains the viral backbone. The recombinant plasmid was selected on kanamycin LB plates. These plasmids were amplified and purified using a plasmid maxiprep system (QIAGEN, Valencia, CA). The recombinant adenovectors were linearized using PacI and transfected to Ad293 cells where viral particles were further amplified. The adenovectors were purified and then titered using an AdEasy Viral Titer Kit (Stratagene).
Rat granulosa cells were collected at 48 h after PMSG priming as detailed above and cultured on six-well plates in Opti-MEM medium supplemented with 0.05 mg/ml gentamicin and 1× ITS (insulin, transferrin, and selenium) for 4 h before addition of the adenovirus vectors containing Btg1 (Ad-BTG1), Btg2 (Ad-BTG2), or a control GFP adenovirus vector (Ad-GFP). The granulosa cells were exposed to Ad-BTG1, Ad-BTG2, or Ad-GFP at a multiplicity of infection of 100 plaque-forming units (pfu)/cell for 2 h. We routinely observe approximately a 70% infection efficiency of GFP-adenovirus in granulosa cells. Medium was replaced with fresh Opti-MEM medium. At 48 h after adenovirus exposure, granulosa cells were collected for total RNA isolation, protein extraction, or flow cytometric analysis.
To examine the regulation of BTG2 by the nuclear transcription factor RUNX, granulosa cells collected at 48 h after PMSG priming were infected with or without an adenovirus vector containing a dominant-negative (DN) inhibitor for RUNX (Ad-DNRUNX) or Ad-GFP at a multiplicity of infection of 10 pfu/cell. The construction of Ad-DNRUNX was described previously (Liu, J., et al., submitted for publication). Two hours later, the media was replaced with fresh Opti-MEM medium containing forskolin (10 μm) and/or phorbol 12-myristate 13-acetate (PMA, 20 nm) to mimic the actions of hCG. The cells were cultured for an additional 6 h. Subsequently, granulosa cells were collected for total RNA or protein isolation.
Flow cytometric analysis of granulosa cells
To determine the impact of overexpression of BTG1 or BTG2 on cell cycle kinetics, granulosa cells were collected at 48 h after adenovirus infection as described above. At the end of culture, granulosa cells were suspended using 3% trypsin and then stained for DNA content (24). Briefly, ribonuclease A (final concentration of 0.1 mg/ml) was added to 1 × 106 cells and incubated at 37 C for 30 min. Afterward, granulosa cells were resuspended in 50 μg/ml propidium iodide and incubated for 1 h in the dark at 4 C. The cell cycle distribution along with the percentage of cells with degraded DNA were determined at an excitation wavelength of 488 nm using a FACS Calibur flow cytometer (Becton Dickinson, San Jose, CA) at the core flow cytometry laboratory at the University of Kentucky. Cell cycle histograms were obtained from three determinations, each with a total of 100,000 cells per treatment.
Statistical analyses
All data are presented as means ± sem. One-way ANOVA was used to test differences in Btg1, Btg2, and Btg3 mRNA expression across time of culture or among treatments in vitro. If ANOVA revealed significant effects of time of tissue collection, time of culture, or treatment, the means were compared by Duncan’s test, with P < 0.05 considered significant.
Results
hCG induced BTG1, BTG2, and BTG3 expression in periovulatory rat ovaries and granulosa cells
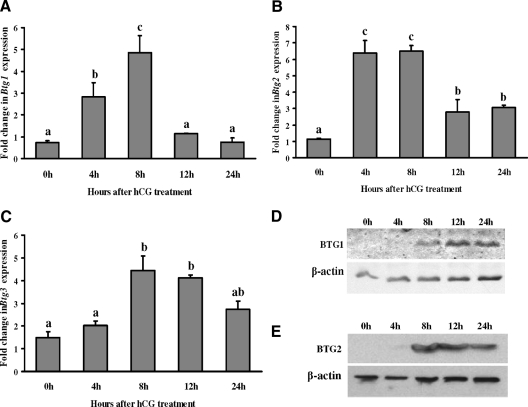
To determine whether hCG induces ovarian Btg1, Btg2, and Btg3 mRNA and protein, expression patterns were analyzed from ovaries collected at different times after hCG administration. The mRNA levels for all three of the Btg genes increased at 8 h after hCG, although the pattern of expression differed among the family members (Figs. 1, A–C). Btg1 mRNA expression increased between 4 and 8 h after hCG to levels approximately 6-fold higher than that of the 0-h time point (i.e. control) before decreasing to control levels by 12 and 24 h (Fig. 1A). In contrast, Btg2 mRNA expression increased 6.5-fold at 4 and 8 h after hCG but did not decrease to control levels by the 24-h time point (Fig. 1B). Btg3 mRNA levels increased 3-fold at 8 h after hCG and remained elevated throughout the periovulatory period (Fig. 1C). Western blot results showed that both BTG1 (Fig. 1D) and BTG2 (Fig. 1E) protein levels were highly induced at 8 h after hCG treatment and remained elevated at 12 and 24 h after treatment.
Figure 1.
Stimulation of BTG1, BTG2, and BTG3 expression by hCG in rat ovary. Real-time PCR analysis shows the expression of Btg1 (A), Btg2 (B), and Btg3 (C) mRNA in preovulatory ovaries after hCG administration. Rats were injected with PMSG for 48 h and treated with hCG, and ovaries were collected at 0, 4, 8, 12, or 24 h after treatment. Relative levels of mRNA for Btg1, Btg2, and Btg3 were normalized to the L32 band in each sample (mean ± sem; n = 3 independent culture experiments). Western blot analysis shows BTG1 (D) and BTG2 (E) protein levels at different time points after hCG treatment in rat ovary. Bars with no common superscripts are significantly different (P < 0.05).
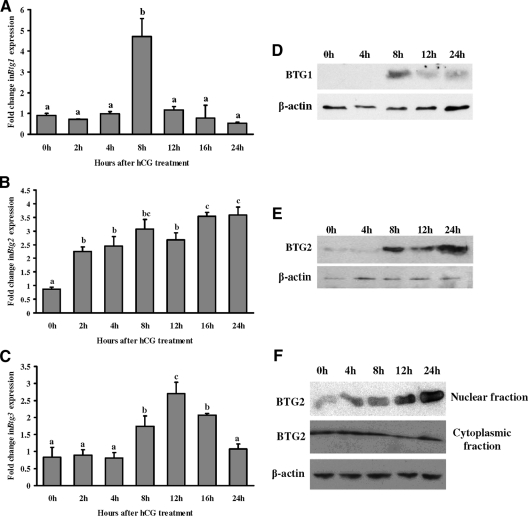
To determine the relative expression of the BTG family members in the granulosa cell compartment, granulosa cells were collected at various times after hCG (i.e. in vivo). The levels of Btg1 mRNA were highest at 8 h after hCG (Fig. 2A), similar to the peak levels observed in the whole ovary. Btg2 mRNA expression had a slightly different pattern than in intact ovaries with an elevation in Btg2 mRNA up to 24 h after hCG (Fig. 2B). Btg3 mRNA levels in granulosa cells mimicked the pattern observed in intact ovaries (Fig. 2C). Both BTG1 (Fig. 2D) and BTG2 (Fig. 2E) protein levels were highly induced at 8 h after hCG treatment. BTG1 protein level decreased gradually until 24 h after hCG treatment, whereas BTG2 protein remained elevated until 24 h after hCG treatment.
Figure 2.
Induction of BTG1, BTG2, and BTG3 expression by hCG in rat granulosa cells in vivo. Real-time PCR analysis shows the expression of Btg1 (A), Btg2 (B), and Btg3 (C) mRNA in rat granulosa/luteal cells obtained from PMSG-primed rat preovulatory ovaries collected at 0, 2, 4, 8, 12, 16, or 24 h after hCG administration. Relative levels of mRNA for Btg1, Btg2, and Btg3 were normalized to the L32 band in each sample (mean ± sem; n = 3 independent culture experiments). Western blot analysis shows BTG1 (D) and BTG2 (E) protein levels at different time points after hCG treatment of rat granulosa/luteal cells collected in vivo. Western blot demonstrates that BTG2 expression in the nuclear and cytoplasmic compartments of rat granulosa/luteal cells obtained from preovulatory ovaries (48 h after PMSG) and then treated with hCG (1 IU/ml) for 0, 4, 8, 12, or 24 h (F). Bars with no common superscripts are significantly different (P < 0.05).
Subcellular location of BTG2 was reported to be important for its function (25). Therefore, we assessed the subcellular location of the BTG2 protein by Western blot analysis. Granulosa cell cytoplasmic and nuclear fractions were extracted and analyzed. Western blot results showed that BTG2 expression in the cytoplasm was relatively unchanged after hCG, whereas its expression in the nucleus was markedly induced through 24 h after hCG (Fig. 2F).
Localization of Btg1 and Btg2 mRNA in the ovary
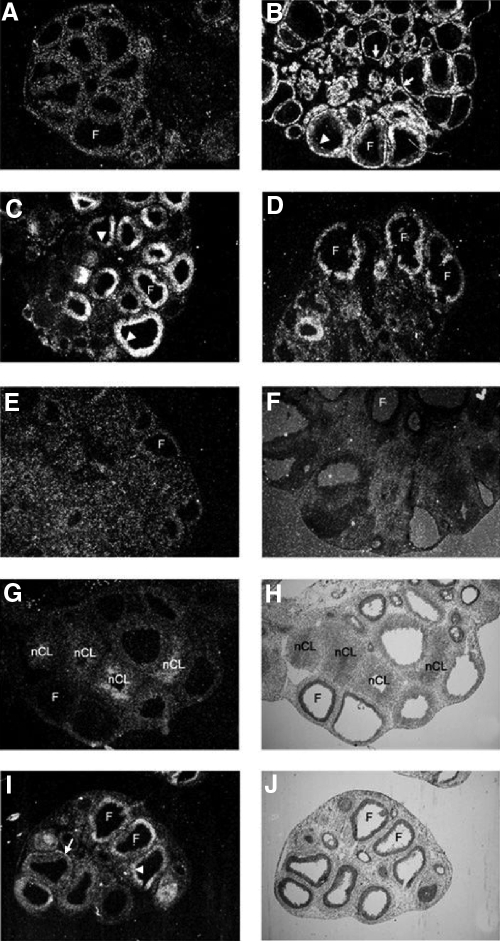
In situ hybridization of Btg1 mRNA revealed that the expression of this gene was low and primarily in the theca cells at 0 h (Fig. 3A) but was induced in the theca and granulosa cells of certain follicles at 4 h after hCG injection (Fig. 3B). Strikingly, there was a switch in the expression from the theca to the granulosa cell compartment at 8 and 12 h after hCG (Fig. 3, C and D). After ovulation, Btg1 mRNA expression was similar to the 0-h ovary with little detectable signal in the forming corpus luteum (Fig. 3E). There was no detected signal on the sections that were hybridized to the sense probe of Btg1 (Fig. 3F). Btg2 mRNA expression was very low at 0 h and gradually increased in the granulosa cells, and at 24 h, Btg2 mRNA was found in the central portion of the newly formed corpus luteum. A representative panel of Btg2 expression from an ovary collected at 24 h after hCG is illustrated (Fig. 3, G and H). Btg3 mRNA expression was induced at 8 h in both granulosa cells and theca cells (Fig. 3, I and J), and its expression gradually decreased and was low at the 24-h time point (data not shown).
Figure 3.
Cellular localization of Btg1, Btg2, and Btg3 mRNA in periovulatory rat ovaries. Sections of rat ovaries obtained at 0 h (48 h after PMSG) (panel A), 4 h (panel B), 8 h (panels C, I, and J), 12 h (panel D), or 24 h (panels E–H) after hCG injection were hybridized with the appropriate antisense probes for Btg1 (panels A–E), Btg2 (panels G and H), or Btg3 (panels I and J) mRNA or sense probe of Btg1 (panel F). Representative bright-field (panels H and J) and corresponding dark-field (panels G and I) photomicrographs are depicted. Arrows indicate the theca layer. Arrowheads indicate the granulosa cell layer. F, Follicle; nCL, newly formed corpus luteum. Original magnification of all slides, ×20.
Effects of hCG on granulosa cell expression of Btg1 and Btg2 mRNA in vitro
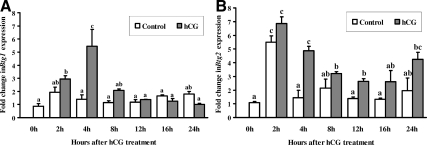
Because the most striking changes were observed with Btg1 and Btg2 mRNA expression, studies were conducted to determine whether the increase in Btg1 and Btg2 mRNA in vivo can be mimicked in vitro by the action of hCG. Real-time PCR analysis revealed that hCG induced a similar pattern of Btg1 mRNA expression in cultured granulosa cells (Fig. 4A) compared with cells collected in vivo (Fig. 2A) with the exception that the peak expression was observed at 4 h after hCG in vitro. Btg2 mRNA expression increased at 4 h, declined at 12 h, and then was elevated at 24 h after hCG (Fig. 4B). Of interest was the induction of Btg2 mRNA immediately after the initiation of culture (i.e. 2 h) in the untreated cells (Fig. 4B), which may be a response to the stress of cell isolation. For example, Btg2 mRNA is increased within 30–60 min in response to the stress of hypoxia and ischemia and was proposed to control the progression of apoptosis (26,27).
Figure 4.
Stimulation of Btg1 (A) and Btg2 (B) mRNA expression by hCG in rat granulosa cells in vitro. Real-time PCR analysis shows the expression of Btg1 and Btg2 mRNA in granulosa cells obtained from rat preovulatory ovaries (48 h after PMSG) and cultured in medium alone (control) or with hCG (1 IU/ml) for 0, 2, 4, 8, 12, 16, or 24 h. Relative levels of mRNA for Btg1 and Btg2 were normalized to the L32 band in each sample (mean ± sem; n = 3 independent culture experiments). Bars with no common superscripts are significantly different (P < 0.05).
Regulation of Btg1 and Btg2 mRNA expression
Both the protein kinase A and protein kinase C signaling pathways are known to be activated by hCG in preovulatory granulosa cells (28). To determine which signaling pathway is involved in the up-regulation of Btg1 and Btg2 mRNA expression in response to hCG stimulation, granulosa cells from rat preovulatory ovaries (48 h after PMSG) were cultured with hCG, forskolin (FSK), which is an activator of adenylate cyclase, or an activator of protein kinase C, PMA, for 4 h. FSK mimicked the action of hCG and induced both Btg1 and Btg2 mRNA expression (Fig. 5, A and B). PMA alone stimulated Btg1 mRNA expression slightly, had no effect on Btg2 mRNA expression, and did not enhance the action of FSK (Fig. 5, A and B). These results suggest that Btg1 and Btg2 mRNA expression was mainly mediated by the hCG-induced activation of the protein kinase A pathway.
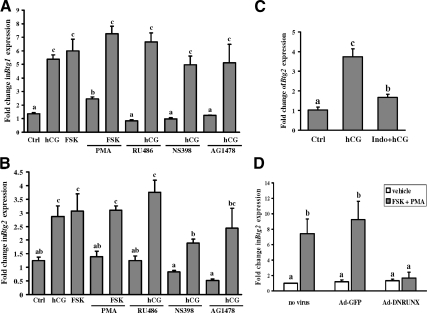
Figure 5.
Regulation of Btg1 and Btg2 mRNA expression by activators or inhibitors of intracellular signaling pathways in granulosa cells in vitro. A and B, Real-time PCR shows mRNA expression for Btg1 (A) and Btg2 (B) in granulosa cells from rat preovulatory ovaries (48 h after PMSG) cultured in medium alone (Ctrl) or with hCG (1 IU/ml), FSK1(0 μm), or PMA (20 nm) for 4 h. Granulosa cells were also cultured with hCG in the absence or presence of the progesterone receptor antagonist RU486 (1 μm), the PTGS2 inhibitor (NS-398, 1 μm), or the EGF receptor tyrosine kinase selective inhibitor (AG1478, 1 μm) for 4 h. C, PMSG-primed rats were treated with or without indomethacin (1 mg/animal) 1 h before hCG injection. Granulosa cells were collected either from the ovaries at 0 h hCG treatment (control) or after 4 h hCG treatment to measure Btg2 expression. Ctrl, Control; Indo, indomethacin. D, Granulosa cells obtained from rat preovulatory ovaries (48 h after PMSG) were untreated, infected with a control adenovirus vector (Ad-GFP), or infected with a DN inhibitor of RUNX (Ad-DNRUNX) adenovirus vector at 10 pfu/cell for 48 h. Cells were stimulated with FSK and PMA for 6 h. Relative levels of mRNA for Btg1 and Btg2 were normalized to the L32 band in each sample (mean ± sem; n = 3 independent culture experiments). Bars with no common superscripts are significantly different (P < 0.05).
hCG also sets in motion a number of steps that are crucial for follicular rupture and oocyte release including activation of epidermal growth factor (EGF) signaling, induction of progesterone receptors, as well as stimulation of prostaglandin-endoperoxide synthase 2 (PTGS2) (29). We tested whether the up-regulation of Btg1 and Btg2 mRNA is mediated by the hCG-induced activation of these signaling pathways using RU486 to block progesterone receptor action, NS398 to inhibit PTGS2, and AG1478 to prevent EGF signaling. NS398 was able to reduce the hCG-stimulated increase in Btg2 mRNA expression in vitro (Fig. 5B), and the indomethacin treatment could block the hCG-induced expression of Btg2 mRNA in vivo (Fig. 5C).
Reduction of Btg2 mRNA expression by DNRUNX in preovulatory granulosa cells
Members of the RUNX family of nuclear transcription factors are induced by the LH surge and known to regulate gene expression in periovulatory granulosa cells (20). Because there are five RUNX binding sites in the promoter of the rat Btg2 gene but none reported in the promoter of the Btg1 gene, we determined whether Btg2 mRNA expression is regulated by the RUNX transcription factors. Granulosa cells isolated from PMSG-primed immature rats were infected with either a control GFP adenovirus vector (Ad-GFP) or a DN inhibitor for RUNX (Ad-DNRUNX). Treatment with Ad-DNRUNX inhibited Btg2 mRNA expression stimulated by FSK plus PMA treatment (Fig. 5D), which suggests that RUNX is involved in Btg2 transcriptional regulation.
Cell cycle analysis of granulosa cells after overexpression of rat BTG1 and BTG2
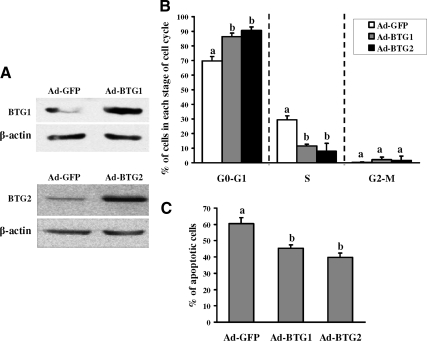
To investigate the potential function of BTG1 and BTG2, granulosa cells from preovulatory ovaries were infected with either a recombinant Ad-BTG1 or Ad-BTG2 adenovirus. Granulosa cells infected with Ad-GFP were used as a control. We examined BTG1 and BTG2 because their expression was most markedly induced by hCG in intact ovaries and granulosa cells, and we are able to monitor the protein in our overexpression studies. Western blot results confirmed that both BTG1 and BTG2 were highly expressed in the granulosa cells (Fig. 6A). Overexpression of BTG1 or BTG2 resulted in an alteration in granulosa cells entering the cell cycle (Fig. 6B) and a decrease in cell apoptosis (Fig. 6C). The percentage of cells in the G0/G1 phase increased markedly in granulosa cells expressing either BTG1 or BTG2, whereas the percentage of cells in the S phase decreased (Fig. 6B). The apoptosis rate in the granulosa cells overexpressing BTG1 and BTG2 also decreased compared with cells infected with the control GFP vector (Fig. 6C). The experimental paradigm employed in the current study used serum-starved cells, which increased the rate of apoptosis and magnified the impact of overexpression of BTG1 and BTG2 on cell survival. However, under conditions in which the cells were not stressed (i.e. in the presence of serum), fewer cells were observed undergoing apoptosis, but overexpression of BTG1 and BTG2 still decreased apoptosis, although the magnitude of this protective effect was diminished (data not shown). Irrespective, these findings on the cell cycle and apoptosis suggest that both BTG1 and BTG2 can block granulosa cell proliferation while maintaining the health of the cell.
Figure 6.
Cell cycle analysis of granulosa cells after overexpression of rat BTG1 and BTG2. A, Western blot shows that both BTG1 and BTG2 are highly expressed in granulosa cells that were infected with the Ad-BTG1 or Ad-BTG2 adenovirus vector for 48 h. B and C, Granulosa cells infected with the Ad-BTG1, Ad-BTG2, or Ad-GFP (control) adenovirus vector for 48 h were analyzed for cell cycle kinetics (B) and apoptosis (C). The cells at each stage of cell cycle, G0–G1, S, and G2-M, are depicted. Values represent the mean ± sem; n = 3 independent culture experiments. Bars with no common superscripts are significantly different (P < 0.05).
Discussion
In response to the LH surge, granulosa cells of preovulatory follicles cease dividing (30) and initiate a program of terminal differentiation to become luteal cells (31). It is well established that cellular proliferation and differentiation are controlled by altering the balance of positive and negative regulators of the checkpoints of the cell cycle. Members of the BTG family can regulate these checkpoints by modulating cyclin activity and thereby control cell cycle kinetics resulting in an inhibition of cellular proliferation (32,33). In this study, we demonstrate that the BTGs are induced before ovulation and that overexpression of the BTGs impacts the cell cycle kinetics and health of rat granulosa cells.
There are limited reports on the expression and localization of the BTG family in the ovary. In the human, BTG2 (TIS21/PC3) was found in granulosa cells by expression screening of BTG2 in a variety of normal human tissues (34). The BTG family has also been associated with folliculogenesis in the rat (19) as well as follicular dominance in cattle (18). However, a detailed analysis of the expression patterns and regulation of the BTG family has not been performed. In the current study, mRNA expression for Btg1, Btg2, and Btg3 was highly induced both in intact ovaries and granulosa cells by hCG. Expression of mRNA for all of the BTG family members was increased within 4–8 h after hCG, although the temporal patterns of mRNA expression differed between the BTG family members. Using in situ hybridization analysis, this stimulation of Btg mRNA was apparent throughout the ovary with an induction of all three Btg members in the granulosa cells. However, unlike Btg2 and Btg3, Btg1 mRNA expression was initially observed in the theca cells and then switched to the granulosa cell compartment of antral follicles by 8 and 12 h after hCG. This pattern of expression where Btg1 mRNA switches from the theca to the granulosa cells is very unique. Although the physiological significance of this switch is unclear, we would speculate that this reflects the rate at which theca and granulosa cells respond to hCG to induce differentiation. Expression of Btg2 mRNA was unique in that it was present in the forming corpus luteum, suggesting a potential role in luteal function as discussed below.
The expression of the three BTG family members (BTG1, BTG2, and BTG3) in the granulosa cell compartment led us to hypothesize that they may be involved in the process of luteinization, specifically the transition from a proliferative granulosa cell to a differentiated luteal cell. It is well established that nearly half the granulosa cell population before an ovulatory stimulus is proliferative, whereas within 36 h after hCG treatment, less than 10% of the cells are proliferating. This reduction in granulosa cell proliferation is not steroid dependent but rather is initiated by the ovulatory gonadotropin surge, which is associated with a complex and diverse set of changes in various components of the cell-cycle machinery such as the cyclins (35). The basis for our postulate that the BTG family may play a role in the switch from proliferation to differentiation is the body of literature that the BTGs are antiproliferative and modulate the cell cycle. Btg1 was reported to be maximally expressed in the G0/G1 phases of the cell cycle and that it negatively regulated cell proliferation (3,36,37). Subsequently, both Btg2 (12) and Btg3 (38) have been reported to exhibit antiproliferative properties. For example, BTG2 (PC3) was able to inhibit proliferation in chromaffin PC12 cells and nonneuronal cells by arresting G1 to S progression (12,13). These reported actions of BTG have led to the theory that the BTG family is active or induced at the transition between cell differentiation and commitment to terminal differentiation (3). To determine whether the BTG family impacted cell cycle kinetics and potentially the granulosa-luteal transition, we overexpressed BTG1 or BTG2 in granulosa cells and observed an increase in the number of cells in the G0/G1 stage of the cell cycle, suggesting a role in cell cycle control for these members of the BTG family. Overexpression of BTG1 and BTG2 also decreased the number of cells undergoing apoptosis. This effect of BTG1 and BTG2 on cell survival has been observed in other systems where BTG2 expression promotes neuronal differentiation and is required for survival of terminally differentiated cells (39). Of interest was our observation that Btg2 mRNA persisted and was highly expressed in the developing CL. These findings, along with previous reports, support the hypothesis for a potential role of the BTG members in promoting survival of the differentiated luteal cells.
In the present study, expression of mRNA for Btg1, Btg2, and Btg3 was induced by hCG, which was used to mimic the preovulatory LH surge. The hormonal induction of the BTG family members was further explored using granulosa cells in vitro. The PKA pathway appears to be the dominant signaling pathway regulating Btg1 and Btg2 mRNA because FSK but not PMA mimicked hCG induction of the Btg genes. Because the LH/hCG pathway sets in motion a series of downstream events that are crucial for follicular rupture (40), we further examined the impact of progesterone, prostaglandins, and the EGF signaling pathway on the regulation of Btg1 and Btg2 mRNA. There was no effect of the progesterone receptor antagonist RU486 or the EGF receptor tyrosine kinase inhibitor AG1478 on hCG-induced expression of Btg1 and Btg2 mRNA. The lack of an effect by RU486 at 4 h after hCG is not unexpected because this is immediately before maximal induction of the progesterone receptor (41). The early induction of Btg2 mRNA occurs at a time concordant with the increase in Ptgs2 mRNA within 3–4 h after hCG (42) and the initial stimulation of PGF2α and PGE production within 4 h after hCG (43). The observation that inhibition of prostaglandin synthesis by indomethacin (in vivo) or NS398 (in vitro) blocked hCG-induced Btg2 mRNA expression suggests that prostaglandins play an important role in regulating Btg2 mRNA.
In addition to progesterone, prostaglandins, and the EGF signaling pathway, the LH/hCG pathway stimulates dramatic changes in the expression patterns of a myriad of genes (44) including the RUNX family of transcription factors (20,45), which plays an essential role in differentiation of various cells (46,47,48). In the ovary, RUNX1 and RUNX2 are highly induced by hCG and involved in regulating preovulatory gene expression and progesterone production (20,45). Because hCG induces both BTG and RUNX expression and there are five RUNX binding sites on the Btg2 promoter, we examined whether RUNX could regulate Btg2 expression. This was accomplished by inhibiting RUNX expression using a DN inhibitor for RUNX (DNRUNX). Our findings that the inhibition of Btg2 mRNA expression by the DNRUNX suggest that RUNX1 or RUNX2 was involved in regulating BTG2. Further evidence for RUNX regulation of BTG2 is forthcoming from chromatin immunoprecipitation assays where we have observed that RUNX interacts with the Btg2 promoter (Park, E.-S., unpublished observation).
A predominantly nuclear localization of BTG2 was observed in granulosa cells by Western analysis. We observed that nuclear expression of BTG2 increased after hCG treatment, whereas the expression level of BTG2 in the cytoplasmic compartments did not change. These findings are in agreement with previous reports that BTG2 has a predominantly nuclear localization in epithelial cells (49). The presence of members of the BTG family in the nucleus is consistent with the observation that BTG1 and BTG2 act as transcriptional cofactors of the Hoxb9 protein (50) as well as estrogen receptor α (ERα) (51). The presence of two copies of an LXXLL motif known as the nuclear receptor (NR) box within BTG1 and BTG2, which is essential for the interaction of coactivators with nuclear receptors (52), led Prévôt and co-workers (51) to examine the regulation of ERα by BTG1 and BTG2. Interestingly, transfection studies suggest that BTG1 and BTG2 can function either as a coactivator or corepressor of ERα depending on the promoter context. These findings suggest that BTG1 and BTG2 may regulate ERα or other members of the Hox gene family (53,54) to impact ovarian follicular dynamics.
In summary, the present findings demonstrate an induction of Btg1, Btg2, and Btg3 expression in granulosa cells after hCG administration. A unique pattern of expression is observed with Btg1 mRNA appearing in the theca and then being expressed in the granulosa cell compartment. BTG2 is found predominantly in the nucleus. Our findings suggest BTG family members may regulate the exit of granulosa cells from the cell cycle and promote their survival, which would direct their differentiation into luteal cells.
Acknowledgments
We thank Dr. Susan Kraner for the assistance with the construction of the BTG1 and BTG2 adenoviruses.
Footnotes
This work was supported by National Institutes of Health Grants P20 RR 15592 (T.E.C.) and HD051727-01 (M.J.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online April 9, 2009
Abbreviations: BTG, B cell translocation gene; DMSO, dimethylsulfoxide; DN, dominant negative; EGF, epidermal growth factor; ERα, estrogen receptor α; FSK, forskolin; hCG, human chorionic gonadotropin; pfu, plaque-forming units; PMA, phorbol 12-myristate 13-acetate; PMSG, pregnant mare serum gonadotropin; PTGS2, prostaglandin-endoperoxide synthase 2.
References
- Niswender GD, Nett TM 2000 The corpus luteum and its control in infraprimate species. In: Knobil E, Neill J, eds. The physiology of reproduction. 2nd ed. New York: Raven Press; 781–816 [Google Scholar]
- Schafer KA 1998 The cell cycle: a review. Vet Pathol 35:461–478 [DOI] [PubMed] [Google Scholar]
- Rouault JP, Rimokh R, Tessa C, Paranhos G, Ffrench M, Duret L, Garoccio M, Germain D, Samarut J, Magaud JP 1992 BTG1, a member of a new family of antiproliferative genes. EMBO J 11:1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buanne P, Corrente G, Micheli L, Palena A, Lavia P, Spadafora C, Lakshmana MK, Rinaldi A, Banfi S, Quarto M, Bulfone A, Tirone F 2000 Cloning of PC3B, a novel member of the PC3/BTG/TOB family of growth inhibitory genes, highly expressed in the olfactory epithelium. Genomics 68:253–263 [DOI] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T 1999 Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 18:7432–7441 [DOI] [PubMed] [Google Scholar]
- Reiss K, Wang JY, Romano G, Furnari FB, Cavenee WK, Morrione A, Tu X, Baserga R 2000 IGF-I receptor signaling in a prostatic cancer cell line with a PTEN mutation. Oncogene 19:2687–2694 [DOI] [PubMed] [Google Scholar]
- Ajima R, Ikematsu N, Ohsugi M, Yoshida Y, Yamamoto T 2000 Cloning and characterization of the mouse tob2 gene. Gene 253:215–220 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Rouault J, Magaud J, Berthet C 2001 In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett 497:67–72 [DOI] [PubMed] [Google Scholar]
- Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A 2004 BTG2, its family and its tutor. Bull Cancer 91:E242–E253 [PubMed] [Google Scholar]
- Tirone F 2001 The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J Cell Physiol 187:155–165 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Kawamura-Tsuzuku J, Ohsugi M, Yoshida M, Emi M, Nakamura Y, Onda M, Yoshida Y, Nishiyama A, Yamamoto T 1996 Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene 12:705–713 [PubMed] [Google Scholar]
- Rouault JP, Falette N, Guéhenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud JP, Ozturk M, Samarut C, Puisieux A 1996 Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet 14:482–486 [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Guardavaccaro D, Starace G, Tirone F 1996 Overexpression of the nerve growth factor-inducible PC3 immediate early gene is associated with growth inhibition. Cell Growth Differ 7:1327–1336 [PubMed] [Google Scholar]
- Tippetts MT, Varnum BC, Lim RW, Herschman HR 1988 Tumor promoter-inducible genes are differentially expressed in the developing mouse. Mol Cell Biol 8:4570–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raburn DJ, Hamil KG, Tsuruta JK, O'Brien DA, Hall SH 1995 Stage-specific expression of B cell translocation gene 1 in rat testis. Endocrinology 136:5769–5777 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Matsuda S, Ikematsu N, Kawamura-Tsuzuku J, Inazawa J, Umemori H, Yamamoto T 1998 ANA, a novel member of Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene 16:2687–2693 [DOI] [PubMed] [Google Scholar]
- Vallée M, Gravel C, Palin MF, Reghenas H, Stothard P, Wishart DS, Sirard MA 2005 Identification of novel and known oocyte-specific genes using complementary DNA subtraction and microarray analysis in three different species. Biol Reprod 73:63–71 [DOI] [PubMed] [Google Scholar]
- Evans AC, Ireland JL, Winn ME, Lonergan P, Smith GW, Coussens PM, Ireland JJ 2004 Identification of genes involved in apoptosis and dominant follicle development during follicular waves in cattle. Biol Reprod 70:1475–1484 [DOI] [PubMed] [Google Scholar]
- Schmidt J, de Avila J, McLean D 2006 Regulation of protein tyrosine phosphatase 4a1, B-cell translocation gene 2, nuclear receptor subfamily 4a1 and diacylglycerol O-acyltransferase 1 by follicle stimulating hormone in the rat ovary. Reprod Fertil Dev 18:757–765 [DOI] [PubMed] [Google Scholar]
- Jo M, Curry Jr TE 2006 Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol 20:2156–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000 Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ 1951 Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275 [PubMed] [Google Scholar]
- Vindeløv LL, Christensen IJ, Nissen NI 1983 A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 3:323–327 [DOI] [PubMed] [Google Scholar]
- Kawamura-Tsuzuku J, Suzuki T, Yoshida Y, Yamamoto T 2004 Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene 23:6630–6638 [DOI] [PubMed] [Google Scholar]
- Fiedler F, Mallo GV, Bödeker H, Keim V, Dagorn JC, Iovanna JL 1998 Overexpression of the PC3/TIS21/BTG2 mRNA is part of the stress response induced by acute pancreatitis in rats. Biochem Biophys Res Commun 249:562–565 [DOI] [PubMed] [Google Scholar]
- Gubits RM, Burke RE, Casey-McIntosh G, Bandele A, Munell F 1993 Immediate early gene induction after neonatal hypoxia-ischemia. Brain Res Mol Brain Res 18:228–238 [DOI] [PubMed] [Google Scholar]
- Jo M, Curry Jr TE 2004 Regulation of matrix metalloproteinase-19 messenger RNA expression in the rat ovary. Biol Reprod 71:1796–1806 [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Ochsner S, Hsieh M, Doyle KH, Falender AE, Lo YK, Sharma SC 2002 Novel signaling pathways that control ovarian follicular development, ovulation and luteinization. Recent Prog Horm Res 57:195–220 [DOI] [PubMed] [Google Scholar]
- Hirshfield AN 1991 Development of follicles in the mammalian ovary. Int Rev Cytol 124:43–101 [DOI] [PubMed] [Google Scholar]
- Richards JS 1994 Hormonal control of gene expression in the ovary. Endocr Rev 15:725–751 [DOI] [PubMed] [Google Scholar]
- Lim IK 2006 TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J Cancer Res Clin Oncol 132:417–426 [DOI] [PubMed] [Google Scholar]
- Donato LJ, Suh JH, Noy N 2007 Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res 67:609–615 [DOI] [PubMed] [Google Scholar]
- Melamed J, Kernizan S, Walden PD 2002 Expression of B-cell translocation gene 2 protein in normal human tissues. Tissue Cell 34:28–32 [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Schwinof KM, Stouffer RL 2001 Gonadotropin and steroid control of granulosa cell proliferation during the periovulatory interval in rhesus monkeys. Biol Reprod 65:755–762 [DOI] [PubMed] [Google Scholar]
- Marchal S, Cassar-Malek I, Magaud JP, Rouault JP, Wrutniak C, Cabello G 1995 Stimulation of avian myoblast differentiation by triiodothyronine: possible involvement of the cAMP pathway. Exp Cell Res 220:1–10 [DOI] [PubMed] [Google Scholar]
- Rodier A, Marchal-Victorion S, Rochard P, Casas F, Cassar-Malek I, Rouault JP, Magaud JP, Mason DY, Wrutniak C, Cabello G 1999 BTG1: a triiodothyronine target involved in the myogenic influence of the hormone. Exp Cell Res 249:337–348 [DOI] [PubMed] [Google Scholar]
- Guehenneux F, Duret L, Callanan MB, Bouhas R, Hayette S, Berthet C, Samarut C, Rimokh R, Birot AM, Wang Q, Magaud JP, Rouault JP 1997 Cloning of the mouse BTG3 gene and definition of a new gene family (the BTG family) involved in the negative control of the cell cycle. Leukemia 11:370–375 [DOI] [PubMed] [Google Scholar]
- el-Ghissassi F, Valsesia-Wittmann S, Falette N, Duriez C, Walden PD, Puisieux A 2002 BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene 21:6772–6778 [DOI] [PubMed] [Google Scholar]
- Richards JS 2005 Ovulation: new factors that prepare the oocyte for fertilization. Mol Cell Endocrinol 234:75–79 [DOI] [PubMed] [Google Scholar]
- Park OK, Mayo KE 1991 Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol 5:967–978 [DOI] [PubMed] [Google Scholar]
- Sirois J, Simmons DL, Richards JS 1992 Hormonal regulation of messenger ribonucleic acid encoding a novel isoform of prostaglandin endoperoxide H synthase in rat preovulatory follicles. Induction in vivo and in vitro. J Biol Chem 267:11586–11592 [PubMed] [Google Scholar]
- Espey LL, Norris C, Forman J, Siler-Khodr T 1989 Effect of indomethacin, cycloheximide, and aminoglutethimide on ovarian steroid and prostanoid levels during ovulation in the gonadotropin-primed immature rat. Prostaglandins 38:531–539 [DOI] [PubMed] [Google Scholar]
- Leo CP, Pisarska MD, Hsueh AJ 2001 DNA array analysis of changes in preovulatory gene expression in the rat ovary. Biol Reprod 65:269–276 [DOI] [PubMed] [Google Scholar]
- Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, Richards JS 2006 Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: does this expand their role in the ovulation process? Mol Endocrinol 20:1300–1321 [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H 2004 AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med 10:299–304 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR 2002 Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111:621–633 [DOI] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT 2005 Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem 280:30689–30696 [DOI] [PubMed] [Google Scholar]
- Kawakubo H, Brachtel E, Hayashida T, Yeo G, Kish J, Muzikansky A, Walden PD, Maheswaran S 2006 Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res 66:7075–7082 [DOI] [PubMed] [Google Scholar]
- Prévôt D, Voeltzel T, Birot AM, Morel AP, Rostan MC, Magaud JP, Corbo L 2000 The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem 275:147–153 [DOI] [PubMed] [Google Scholar]
- Prévôt D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L 2001 Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor α signaling pathway. J Biol Chem 276:9640–9648 [DOI] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG 1997 A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733–736 [DOI] [PubMed] [Google Scholar]
- Villaescusa JC, Verrotti AC, Ferretti E, Farookhi R, Blasi F 2004 Expression of Hox cofactor genes during mouse ovarian follicular development and oocyte maturation. Gene 330:1–7 [DOI] [PubMed] [Google Scholar]
- Ota T, Choi KB, Gilks CB, Leung PC, Auersperg N 2006 Cell type- and stage-specific changes in HOXA7 protein expression in human ovarian folliculogenesis: possible role of GDF-9. Differentiation 74:1–10 [DOI] [PubMed] [Google Scholar]