Abstract
Chromogranin A (CgA), the major soluble protein in chromaffin granules, is proteolytically processed to generate biologically active peptides including the catecholamine release inhibitory peptide catestatin. Here we sought to determine whether cysteine protease cathepsin L (CTSL), a novel enzyme for proteolytic processing of neuropeptides, acts like the well-established serine proteases [prohormone convertase (PC)1/3 or PC2] to generate catestatin by proteolytic processing of CgA. We found that endogenous CTSL colocalizes with CgA in the secretory vesicles of primary rat chromaffin cells. Transfection of PC12 cells with an expression plasmid encoding CTSL directed expression of CTSL toward secretory vesicles. Deconvolution fluorescence microscopy suggested greater colocalization of CTSL with CgA than the lysosomal marker LGP110. The overexpression of CTSL in PC12 cells caused cleavage of full-length CgA. CTSL also cleaved CgA in vitro, in time- and dose-dependent fashion, and specificity of the process was documented through E64 (thiol reagent) inhibition. Mass spectrometry on CTSL-digested recombinant CgA identified a catestatin-region peptide, corresponding to CgA360–373. The pool of peptides generated from the CTSL cleavage of CgA inhibited nicotine-induced catecholamine secretion from PC12 cells. CTSL processing in the catestatin region was diminished by naturally occurring catestatin variants, especially Pro370Leu and Gly364Ser. Among the CTSL-generated peptides, a subset matched those found in the catestatin region in vivo. These findings indicate that CgA can be a substrate for the cysteine protease CTSL both in vitro and in cella, and their colocalization within chromaffin granules in cella suggests the likelihood of an enzyme/substrate relationship in vivo.
Chromogranin A protein can be a substrate for the cysteine protease Cathepsin L both in vitro and in cella, and their co-localization within chromaffin granules in cella suggests the likelihood of an enzyme/substrate relationship in vivo.
Chromogranin A (CgA), a 48-kDa acidic glycoprotein, is the major soluble protein in the catecholamine storage vesicles and widely distributed in the endocrine cells and neurons (1,2,3). It is costored and coreleased with catecholamines, neuropeptides, and ATP by exocytosis from chromaffin cells of the adrenal medulla as well as sympathetic neurons (4,5). It controls biogenesis of secretory granules by targeting secretory proteins to the granules of regulated pathway (6,7,8,9). This glycoprotein also serves as a precursor of several biologically active peptides that are released on exocytotic secretion, such as pancreastatin (human CgA250–301) (10,11), vasostatin (human CgA1–76) (12), and catecholamine-release inhibitory peptide catestatin (human CgA352–372) (13,14,15,16). During stress, the latter regulates catecholamine secretion through the autocrine-paracrine-negative feedback mechanism.
Although CgA is overexpressed in human essential (hereditary) hypertension, the plasma concentration of catestatin decreased in not only the established cases of hypertension but also normotensive subjects with a family history of hypertension (17,18). Consistent with human findings, we found that mouse chromogranin A gene (Chga) knockout mice are hypertensive and hyperadrenergic (19). Thus, a better understanding of the processing of catestatin fragment from CgA may provide clues to the development of hypertension through adrenergic pathway.
Like the other prohormones, CgA contains eight to 10 dibasic residue sites, which are considered as potential sites for proteolytic cleavage (1). Several groups have examined the role of the serine prohormone convertase (PC) in the processing of chromogranin/secretogranin family of proteins and neuropeptides for generation of bioactive peptides (20,21,22,23,24,25,26,27). Besides the well-established subtilisin-like serine proteases, we reported previously that in vitro digestion of recombinant CgA with another serine protease plasmin also generated a 14-amino acid catestatin region-specific peptide (28,29,30) with an IC50 of about 2–3 μm. However, the protease(s) responsible for generation of the full-length 21-mer are yet to be established. The cysteine protease cathepsin L (CTSL) has recently been implicated as a novel enzyme for proteolytic processing of neuropeptides (31,32,33). To this end, CTSL has been identified as the cysteine protease component of purified prohormone thiol protease in the secretory vesicle (31,32,33,34). The function of CTSL in the secretory vesicles differs from that in the lysosomes for protein degradation (35). Several groups reported various roles of CTSL in the secretory vesicles (36,37,38,39,40). CTSL has also been implicated in multiple physiological and pathological processes (41,42,43,44,45,46). In the present study, we characterized the in vitro processing of catestatin region from CgA by CTSL. The findings indicate that CTSL is localized within chromaffin granules/vesicles and can generate active catestatin-region fragments by proteolytic cleavage of CgA. The results may have implications for the development of hypertension through modulation of catecholamine secretion.
Materials and Methods
Overexpression and purification of recombinant proteins
Recombinant human CgA isoforms, including naturally occurring nonsynonymous amino acid replacement variants (Gly364Ser, Pro370Leu, Arg374Gln), were expressed in Escherichia coli and purified as described previously using a C-terminal 6-His affinity tag (30).
Immunoblot analysis
The proteins were separated in a 10% SDS-PAGE (Novex precast gel; Invitrogen, San Diego, CA) or a 10–20% tricine gel. Rabbit polyclonal antihuman catestatin [1:3000 in tris-buffered saline with 0.1% Tween 20 (TBS-T)], goat polyclonal anti-CTSL (1:500 dilution, SC-6498; Santa Cruz Biotechnology, Santa Cruz, CA), and goat polyclonal antiactin (1:500, SC-1615; Santa Cruz Biotechnology) were used. The membrane was incubated with secondary antibody, horseradish peroxidase-conjugated donkey antirabbit (1:3000 in TBS-T), and donkey antigoat (1:3000 in TBS-T) for 1 h. The membrane was then developed by the Supersignal west pico chemiluminescent substrate (Thermo Scientific, Rockford, IL) according to the manufacturer’s protocol.
Digestion of recombinant CgA by CTSL
Purified CgA wild-type and variant proteins (10 μm final) were incubated with CTSL (human liver, specific activity: 1.54 U/mg of protein; lot no. 035K2741; catalog no. C6854; Sigma-Aldrich, St. Louis, MO) in 400 mm sodium acetate (pH 5.5) with 4 mm EDTA and 8 mm dithiothreitol at 30 C water bath for various periods of time. The reaction was stopped by the addition of the epoxide (sulfhydryl protease) inhibitor E64 (10 μm).
Matrix-assisted laser desorption (MALDI)/ionization time-of-flight (TOF) mass spectrometry
The CTSL digests were acidified in 0.1% trifluoroacetic acid and were purified by using Zip Tip (Millipore, Billencia, MA). Samples were analyzed by mass spectrometry as described previously (30).
Liquid chromatography (LC) and tandem mass spectrometry (MS/MS) analysis
LC-MALDI was acquired on a Tempo LC MALDI spotter (Applied Biosystems/MDS Sciex, Foster City, CA) using a Chromolith CapRod monolithic capillary column (150 × 0.1 mm) (EMD; VWR International, West Chester, PA). LC-MALDI spots were acquired at a rate of 4 sec/spot from 8 to 16 min. A plate model and default calibration were performed for reflector-positive mode as well as a default calibration for 2 kV-positive modes on the 4800 MALDI TOF/TOF analyzer (Applied Biosystems). Reflector-positive spectra were obtained by acquiring 500 shots from mass to charge ratio (m/z) 1000–4000 amu (atomic mass units). MS/MS spectra were acquired in 2 kV-positive modes with collision-induced dissociation (CID) on at resolution 200 full width at half maximum; for 3000 shots. The MS/MS data were analyzed by Global Proteome server 3.0 (Applied Biosystems) and subjected to database search using Mascot 2.1.1 (www.matrixscience.com). Data were searched against the Swissprot database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/) containing 237,168 sequences, in which had been inserted the recombinant sequences of interest. The search results indicated that individual ion scores greater than 42 indicate identity or extensive homology (P < 0.05). Only significant scores for the peptides defined by a Mascot probability analysis 99% or greater confidence interval were listed.
Peptide synthesis
Peptides were synthesized at a 10- to 100-μmol scale using 9-fluorenylmethoxycarbonyl protection chemistry and purified to greater than 80% homogeneity by reverse-phase HPLC on a C18 column.
Secretagogue-stimulated catecholamine release
Norepinephrine secretion was assayed in [3H]norepinephrine-labeled PC12 cells as described previously (47). The cells were treated with nicotine (60 μm) in secretion medium, either alone or in combination with recombinant CgA (1.5 μm), full length, and digested with CTSL. Digestion reaction (1 h) contained CgA (10 μm) and CTSL (0.3 μm). After digestion, peptides were purified by C18 cartridge (Waters, Milford, MA), lyophilized, and suspended in water. For a dose response, five ascending doses (0.075, 0.15, 0.3, 0.9, and 1.5 μm), equivalent of digested CgA, were used. 3[H]norepinephrine in the supernatants and cell lysates were measured by liquid scintillation counting, and the results were expressed as percent secretion: [amount released/(amount released + amount in cell lysate)] × 100. Net secretion is secretagogue-stimulated release minus basal release.
Cell culture and transient transfection
Rat pheochromocytoma PC12 cells were grown as described previously (47). Cells were transfected with pcDNA3.1 (vector) or pcDNA3.1-preproCTSL (a kind gift from Dr. Vivian Hook, Department of Medicine, University of California, San Diego) by using SuperFect transfection reagent as per the manufacturer’s protocol (QIAGEN, Valencia, CA). Transfected cells were cultured on poly-l-lysine-coated coverslips. Cells were fixed at 48 h for immunofluorescence and were harvested at 72 h of posttransfection for protein analysis.
Primary culture of rat chromaffin cells
Young female rats were killed to collect adrenal medulla. The medullary tissues were digested following a published protocol (http://www. natureprotocols.com) in Hanks’ balanced salt solution containing collagenase type I, BSA, deoxyribonuclease I, and hyaluronidase I-S. Cells were grown in DMEM with 10% fetal bovine serum and cultured for 3 d for immunocytochemistry.
Immunofluorescence for subcellular localization
Cells were washed with PBS and fixed with 2.5% paraformaldehyde for 20 min at room temperature. Fixed cells were permeabilized for 10 min with 0.5% Triton X-100 in PBS, incubated for 5 min with 150 mm glycine in PBS, and exposed for 30 min to 5% BSA in PBS. Cells were then incubated for 30 min at room temperature with either goat polyclonal anti-CTSL (1:100; Santa Cruz Biotechnology) and rabbit polyclonal anti-CgA (1:1500) (48) or goat polyclonal anti-CTSL (1:100; Santa Cruz Biotechnology) and rabbit polyclonal LGP110 (1:100) (9) in PBS containing 2% BSA. Cells were subsequently washed and incubated for 30 min with a Alexa Fluor 594-conjugated donkey antigoat IgG (1:350; Invitrogen) and Alexa Fluor 488-conjugated donkey antirabbit at 1:250; (Invitrogen) together with 1 μg/ml Hoechst 33342 in PBS containing 1% BSA. Coverslips were washed with PBS, mounted in SlowFade antifade kit (Invitrogen). Three-dimensional images were captured on a DeltaVision deconvolution microscopy system operated by SoftWoRx software (Applied Precision, Issaquah, WA), using oil immersion objectives (×60, numerical aperture 1.4) as described previously (49).
Statistical analysis
Biochemical data are expressed as the mean ± sem. Curve fitting was accomplished in the program Kaleidagraph (Synergy Software, Reading, PA). Multiple comparisons were made using one-way ANOVA followed by Bonferroni’s post hoc test. Statistical significance was concluded at P < 0.05.
Results
Colocalization in vivo of endogenous CTSL with CgA in primary rat chromaffin cells
To determine the functional interactions between CgA and CTSL, the intracellular distribution of these two proteins were examined in primary cultures of rat chromaffin cells (Fig. 1A). Consistent with the localization of secretory vesicle, deconvolution microscopy revealed colocalization of CgA with CTSL as evidenced by discrete and punctuate immunostaining (Fig. 1A).
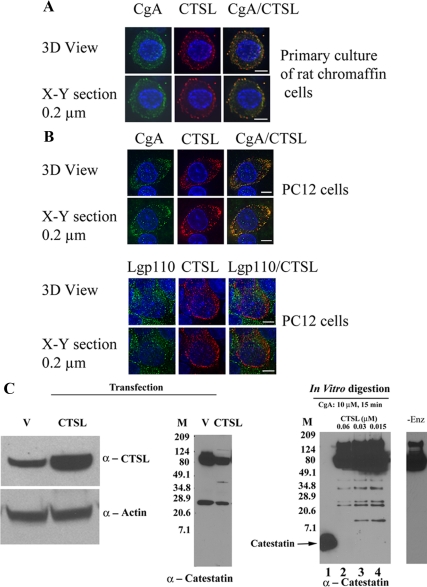
Figure 1.
In vivo role of CTSL for the generation of catestatin-related peptide. A, Subcellular localization of endogenous CTSL and CgA in chromaffin granules or secretory vesicles of primary rat chromaffin cells. Cells were examined by deconvolution microscopy. A series of xy optical sections along the z-axis was acquired with increments of 0.2 μm using ×60 oil immersion objectives. The data set was processed to generate three-dimensional (3D) or representative xy section. Colocalization of CgA (green) and CTSL (red) was shown by overlay of the images, which is illustrated by yellow fluorescence. B, Intracellular distribution of CgA and the endosomal/lysosomal marker LGP110. PC12 cells were transfected with pcDNA3.1-CTSL and processed for immunocytochemistry using rabbit polyclonal anti-CgA primary and antirabbit IgG-Alexa Fluor 488-conjugated secondary (green fluorescence). CTSL was detected with goat polyclonal anti-CTSL and antigoat IgG-Alexa Fluor 594-conjugated secondary (red fluorescence; upper panel). In the lower panel, cells were stained with rabbit polyclonal anti-Lgp110 (green) and goat polyclonal anti-CTSL (red). The nuclei were visualized with Hoechst 33342 (blue). Scale bar, 5 μm. C, Overexpression of CTSL and processing of CgA in cella. PC12 cells were transfected with pcDNA3.1 (lane marked V) or pcDNA3.1-CTSL (lane marked CTSL). Three days after transfection, cells were harvested, lysed, and subjected to 10–20% tricine gel. Proteins were then transferred to polyvinyl difluoride membrane and analyzed by Western blot using anticatestatin antibody. Catestatin immunoreactive bands were shown in the middle panel. A parallel blot was stained with anti-CTSL and antiactin antibody (left panel). In the right panel, purified human recombinant CgA (10 μm) was digested with 0.06, 0.03, and 0.015 μm of CTSL and subjected to immunoblot with anticatestatin antibody. In lane 1 of right panel, synthetic catestatin (CgA352–372) was loaded. Molecular size markers (Bio-Rad prestained broad range) were used in lane marked M. The lane with -Enz represents zero enzyme control.
Overexpression of CTSL in PC12 cells and its effect on proteolytic processing of CgA
The subcellular localization of CTSL was examined in PC12 cells transfected with an expression plasmid encoding prepro-CTSL (Fig. 1B). The cells were stained with either CgA and CTSL (Fig. 1B, upper panel) or CTSL and lysosomal type 1 integral membrane glycoprotein LGP110 (Fig. 1B, lower panel). CTSL showed colocalization with CgA, similar results in primary cultures of rat chromaffin cells. LGP110, as seen by green fluorescence, showed very little overlap with CTSL. The estimated colocalization (by Pearson’s correlation coefficient, scaled from −1 to +1) of CgA with CTSL in primary cultures of rat chromaffin cells (Fig. 1A) and PC12 cells (Fig. 1B, upper panel) were 0.7690 and 0.5972, respectively. The overlap distribution between CTSL (red) and LGP110 (green) was substantially lower, with Pearson’s correlation coefficient of 0.1089 (Fig. 1B, lower panel). PC12 cells were transfected with either the vector pcDNA3 or pcDNA3-prepro-CTSL to test the involvement of CTSL in CgA processing. Overexpression of CTSL (Fig. 1C, left panel) reduced the levels of both full-length CgA and a 24-kDa fragment (Fig. 1B, middle panel). A minor 40-kDa catestatin-positive fragment appeared in CTSL-overexpressing cells, compared with the control (Fig. 1B, middle panel). In parallel, purified recombinant CgA (10 μm) was digested with various concentrations of CTSL (0.06, 0.03, and 0.015 μm) and subjected to immunoblot with anticatestatin antibody for identification of catestatin region-specific peptides (Fig. 1C, right panel). In the lane marked -Enz, recombinant CgA was incubated without the enzyme, and immunoblotting clearly showed the position of undigested full-length CgA; with prolonged exposure, lower-molecular-weight CgA fragments were not observed in the absence of enzyme (results not shown). On the other hand, CTSL-digested CgA samples showed five catestatin-positive fragments, besides undigested CgA (Fig. 1C, lanes 2–4). CgA underwent less digestion on exposure to low protease concentration, as evidenced by the presence of more full-length protein, shown in lanes 3 and 4, compared with lane 2 (Fig. 1C). Catestatin antibody recognized a series of peptides ranging from 10 to 50 kDa (Fig. 1C, right panel). The 11-kDa fragment was observed only at lower enzyme concentrations (0.03 and 0.015 μm; lanes 3 and 4, right panel), suggesting its further processing at higher protease concentrations. The other fragments of 22, 24, 34, and 40 kDa were prominent throughout all conditions. The fragments of 24 and 40 kDa were noticed in the transfected cells as well as in the in vitro digestions (Fig. 1C, middle and right panel). Thus, CTSL generated catestatin immunopositive peptides in CTSL overexpressed PC12 cells that resemble the in vitro digestion of recombinant CgA.
Dose- and time-dependent, specific cleavage of CgA in vitro
Purified recombinant CgA protein (10 μm) was incubated with varying concentrations of CTSL (Fig. 2A, upper panel) to assess the degree of proteolytic processing. CTSL readily digested the recombinant CgA, and the extent of digestion was dose dependent. The major products produced were having apparent molecular masses ranging from 8 to 10 kDa to 65 kDa. At 0.03 and 0.015 μm CTSL, digestion was partial as evidenced by the presence of full-length CgA running as a 70-kDa protein (Fig. 2A, lanes 6 and 7, upper panel). When recombinant CgA (10 μm) was digested with CTSL (0.15 μm) for various periods of time (0–60 min), it suggests a time-dependent degradation of CgA by CTSL (Fig. 2A, lower panel). The sequentially produced major products have apparent molecular masses of 10, 11, 12.5, 16, 18, 20, 27, 34, 43, 50, and 55 kDa as detected by SDS-PAGE. Among the generated peptides, the fragments with 10–11 kDa were prominent at all the time points. CgA was not processed to smaller peptides in either the absence of CTSL (Fig. 2A, lower panel, lane 9) or presence of a general cysteine protease inhibitor E64 (Fig. 2A, lower panel, lane 8), documenting the specificity of CTSL toward processing of CgA.
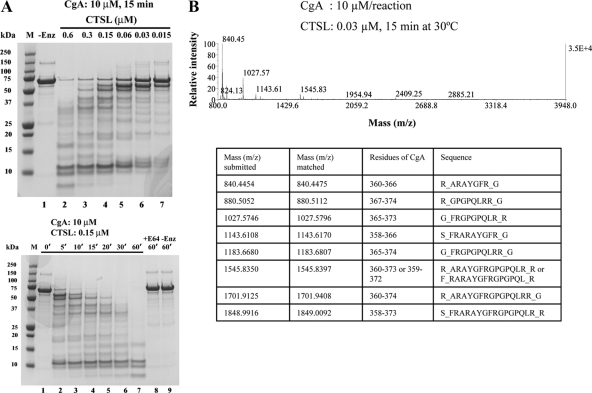
Figure 2.
Proteolysis of purified recombinant CgA by CTSL. A, Dose dependence (upper panel). Recombinant human CgA (10 μm) was incubated with different doses (0.6, 0.3, 0.15, 0.06, 0.03, 0.015 μm; lanes 2–7, upper panel) of CTSL for 15 min at 30 C. Lower panel, Time dependence. CgA (10 μm) was incubated with 0.15 μm of CTSL and aliquots were taken at different time intervals. The reaction products were subjected to SDS-PAGE on a 10% gel and stained with Coomassie blue. Molecular size markers (precision plus protein unstained standards) were used in lane marked M. E64 (CTSL inhibitor) specificity controls are also shown. The lane with-Enz represents zero enzyme control. B, Analysis of peptides generated by CTSL. CgA (10 μm/reaction) was digested with 0.03 μm of CTSL for 15 min at 30 C. The reaction product was subjected to MALDI-TOF. Potential fragments generating from the catestatin region as identified by protein prospector program were listed in the table below.
Identification of different catestatin region-specific peptides generated from CTSL digestion of CgA
We have shown previously that the recombinant wild-type (WT) CgA is cleaved easily by plasmin, generating a very stable 14-mer peptide (CgA360–373) (30). In the present study, we digested recombinant CgA protein (10 μm) with CTSL (0.3 μm for 15 min) to verify whether CTSL can generate the catestatin region-specific peptide. The digestion products were subjected to LC-MS/MS analyses and the peptides generated in the catestatin region, with greater than 99% confidence interval, are listed in supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://endo. endojournals.org. Thirteen peptides were generated from the N-terminal part of the catestatin region, ranging from CgA332–359, with a common stretch of sequence, viz., QEEEEDNRDSSMK (supplemental Table 1). In addition, the following peptides were generated by CTSL: nine peptides in the pancreastatin domain, 11 peptides in the vasostatin region, nine peptides in the WE14 domain, four peptides in the GE25 domain, and four peptides in the chromacin region (supplemental Table 2, A–E).
Digestion of CgA was also attempted at a lower dose of CTSL (0.03 μm). MALDI-TOF analysis of the digests revealed the generation of internal catestatin fragments, CgA360–366, CgA367–374, CgA365–373, CgA358–366, CgA365–374, CgA360–373, CgA360–374, and CgA358–373 (Fig. 2B). Among those, peptides with mass 840, 1545 and 1701 were also generated from plasmin digestion of CgA (30). We have shown previously that the internal peptides CgA360–373 and CgA360–374 act as nicotinic antagonist to catecholamine secretion with IC50 of 2.4 and 3.0 μm, respectively (30). The generation of these active peptides in CTSL digestion implicates CgA as a substrate of CTSL. The peptides with mass 1027 (CgA365–373), 1143 (CgA358–366), 1183 (CgA365–374), and 1848 (CgA358–373) were generated exclusively with CTSL digestion, suggesting additional cleavage specificity for CTSL. Two peptides in the C-terminal part (CgA374–394, mass 2409, and CgA374–393, mass 2253) and one peptide in the N-terminal part (CgA336–359, mass 2883) of catestatin were also generated. In the control experiments, when CTSL or CgA alone were incubated for 15 min at 30 C, no detectable peaks in m/z range 800-4000 were produced (results not shown).
Cleavage of synthetic catestatin-region peptides (CgA360–380 or CgA333–379) by CTSL
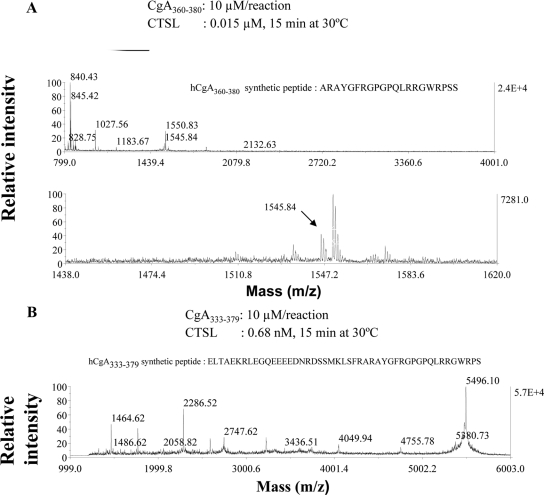
To explore the catestatin-region digestion pattern within CgA by CTSL, we used two synthetic peptides as substrates: a short 21-amino acid synthetic peptide, CgA360–380, with one dibasic site (Arg373Arg374) and a longer 47-amino acid synthetic peptide, CgA333–379, which spans the entire catestatin region including its two dibasic sites (Lys338Arg339 and Arg373Arg374). Each peptide (10 μm) was digested with CTSL and then subjected to MALDI-TOF analysis. Analysis of the mass spectrometric data revealed generation of the following eight peptides CgA360–366 (ARAYGFR, m/z 840), CgA374–380 (RGWRPSS, m/z 845), CgA367–374 (GPGPQLRR, m/z 880), CgA365–373 (FRGPGPQLR, m/z 1027), CgA365–374 (FRGPGPQLRR, m/z 1183), CgA360–373 (ARAYGFRGPGPQLR, m/z 1545), and CgA367–380 (GPGPQLRRGWRPSS, m/z 1550) and CgA365–380 (FRGPGPQLRRGWRPSS, m/z 1853) from the short monodibasic catestatin synthetic peptide (Fig. 3A and Table 1). The peptides with m/z 840, 880, 1027, 1183, and 1545 were also generated from the full-length recombinant protein, thus reinforcing the cleavage specificity of CTSL. The digestion of the long catestatin peptide (CgA333–379) peptide with the flanking dibasic residues was attempted with very low concentration of CTSL (0.7 nm) and for better sensitivity the MALDI-TOF analysis was carried out in linear mode from 1000 to 6000 Da range (Fig. 3B). The presence of undigested peptide of (m/z 5496) was evident and indicated the partial cleavage of the synthetic substrate. The following peptides were deduced from the mass analysis: CgA367–379 (m/z 1464), CgA360–373 (m/z 1546), CgA340–354 (m/z 1768), CgA360–379 (m/z 2286), CgA357–378/CgA358–379 (m/z 2590), CgA341–363 (m/z 2748), CgA333–359 (m/z 3227), CgA333–366 (m/z 4049), and CgA333–373 (m/z 4755) (Table 2).
Figure 3.
Digestion of catestatin-region synthetic peptides by CTSL. Catestatin region synthetic peptides, CgA360–380 (A) or CgA333–379 (B) (10 μm/reaction), were digested with 0.015 μm and 0.68 nm CTSL, respectively, for 15 min at 30 C, and the reaction products were analyzed by MALDI-TOF in reflectron mode for A and linear mode for B.
Table 1.
Digestion of catestatin-region synthetic peptide CgA360–380 by CTSL
| Mass (m/z) submitted | Mass (m/z) matched | Sequence |
|---|---|---|
| 840.4344 | 840.4475 | _ARAYGFR_G |
| 845.4232 | 845.4377 | R_RGWRPSS_ |
| 880.5095 | 880.5112 | F_RGPGPQLR_R or R_GPGPQLRR_G or G_PGPQLRRG_W |
| 1027.5590 | 1027.5796 | G_FRGPGPQLR_R |
| 1183.6744 | 1183.6807 | G_FRGPGPQLRR_G |
| 1545.8408 | 1545.8397 | _ARAYGFRGPGPQLR_R |
| 1550.8342 | 1550.8299 | R_GPGPQLRRGWRPSS_ |
| 1853.9360 | 1853.9994 | G_FRGPGPQLRRGWRPSS_ |
Catestatin region synthetic peptide was digested with CTSL for 15 min at 30 C, and the reaction products were analyzed by MALDI-TOF. Possible fragments generated in the catestatin region are listed in the table. CgA360–380 was 10 μm/reaction, and CTSL was 0.015 μm. hCgA360–380 synthetic peptide: ARAYGFRGPGPQLRRGWRPSS.
Table 2.
Digestion of catestatin-region synthetic peptide CgA333–379 by CTSL
| Mass (m/z) Average | Sequence | Residues of CgA |
|---|---|---|
| 1464.6 | R_GPGPQLRRGWRPS_ | 367–379 |
| 1546.4 | R_ARAYGFRGPGPQLR_R or F_RARAYGFRGPGPQL_R | 360–373 or 359–372 |
| 1767.9 | R_LEGQEEEEDNRDSSM_K | 340–354 |
| 2286.5 | R_ARAYGFRGPGPQLRRGWRPS_ | 360–379 |
| 2590.1 | L_SFRARAYGFRGPGPQLRRGWRP_S or S_FRARAYGFRGPGPQLRRGWRPS_ | 357–378 or 358–379 |
| 2747.6 | E_EEDNRDSSMKLSFRARAYGFRGPG_P or L_EGQEEEEDNRDSSMKLSFRARAY_G | 346–369 or 341–363 |
| 3227.6 | _ELTAEKRLEGQEEEEDNRDSSMKLSFR_A | 333–359 |
| 4049.9 | _ELTAEKRLEGQEEEEDNRDSSMKLSFRARAYGFR_G | 333–366 |
| 4755.7 | _ELTAEKRLEGQEEEEDNRDSSMKLSFRARAYGFRGPGPQLR_R | 333–373 |
| 5496.1 | _ELTAEKRLEGQEEEEDNRDSSMKLSFRARAYGFRGPGPQLRRGWRPS_ | 333–379 |
Catestatin region synthetic peptide was digested with CTSL for 15 min at 30 C, and the reaction products were analyzed by MALDI-TOF. Possible fragments generated in the catestatin region are listed in the table. CgA333–379 was 10 μm/reaction, and CTSL was 0.68 nm. hCgA333–379 synthetic peptide: ELTAEKRLEGQEEEEDNRDSSMKLSFRARAYGFRGPGPQLRRGWRPS.
Active catestatin fragment CgA360–373 was generated by CTSL digestion from both full-length CgA and the long catestatin region-spanning synthetic peptide CgA333–379, with m/z about 1546 in each case; thus, CTSL’s cleavage preference within the catestatin region was similar in both CgA and a synthetic local region (Figs. 2B and 3).
Suppression of catecholamine release by CTSL-generated CgA fragments
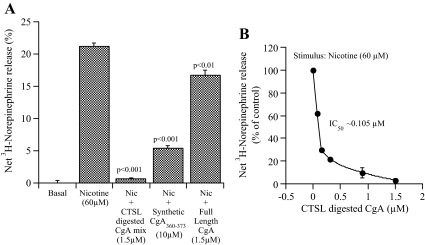
We have shown previously that plasmin generated CgA peptides inhibited nicotine-stimulated catecholamine release (30). To determine the potency of the peptides generated by the CTSL digestion, we performed norepinephrine secretion assay in 3H-norepinephrine-loaded PC12 cells. Recombinant CgA (10 μm) was digested with CTSL (0.3 μm), and the resulting peptides were purified with a C18 cartridge. The nicotine-stimulated norepinephrine secretion was then tested in the presence of undigested CgA, digested CgA, or synthetic CgA360–373 (Fig. 4A, left panel), which caused differential inhibition of nicotine-evoked catecholamine release: 20.87% by undigested CgA, 96.79% by CTSL-generated peptide mixture, and 74.32% by the synthetic catestatin peptide (CgA360–373). The CTSL-generated peptide mixtures abolished nicotine-evoked catecholamine secretion with an IC50 of 0.105 μm (Fig. 4B, right panel).
Figure 4.
Effect of CTSL-digested peptides on secretagogue-stimulated norepinephrine secretion by PC12 cells. A, PC12 cells were stimulated with 60 μm nicotine (Nic) in the absence (basal) or presence of undigested CgA (1.5 μm), CTSL-digested CgA (1.5 μm), or synthetic catestatin peptide (CgA360–373; 10 μm). After 30 min incubation, the cells and media were analyzed for norepinephrine secretion. In control, the net norepinephrine release (100%) was measured in the presence of 60 μm nicotine alone. B, Dose-dependent inhibition of CTSL digests on secretion assay was shown.
Digestion of CgA containing naturally occurring catestatin variants by CTSL
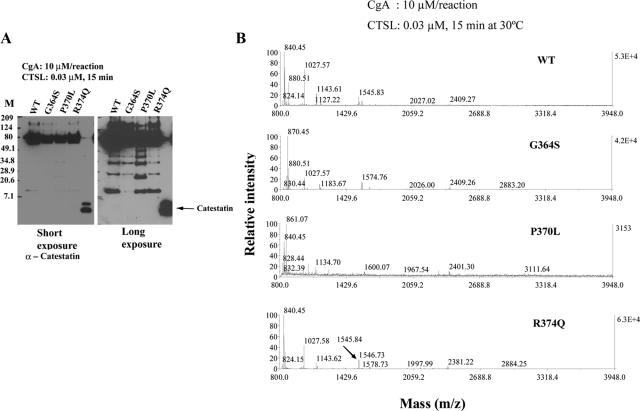
In our previous communication, we have shown differential cleavage of human catestatin variants by plasmin (30). In the present study, we sought to determine whether CTSL is also capable of displaying differential cleavage pattern to human catestatin variants. Therefore, the following purified proteins CgA-WT, CgA-Gly364Ser, CgA-Pro370Leu, and CgA-Arg374Gln at 10 μm were subjected to proteolysis by CTSL (0.03 μm) and the resulting peptides were analyzed by SDS-PAGE (Fig. 5A) and MALDI-mass spectrometry (Fig. 5B).
Figure 5.
Digestion of CgA with catestatin variants by CTSL. A, Immunoblots. WT and mutant CgA (10 μm/reaction) containing the catestatin variants was digested with 0.03 μm CTSL for 15 min and subjected to separation on a 10–20% tricine gel. Proteins were then transferred to polyvinyl difluoride membrane, and immunoblotted with anticatestatin antibody. In the last lane, synthetic catestatin (CgA352–372) was loaded. Molecular size markers (Biorad prestained broad range) were used in lane marked M. B, MALDI-TOF. CTSL digests (same as A) were subjected to MALDI-TOF analysis.
On SDS-PAGE, the digestion pattern, as evidenced by the immunoblot with catestatin antibody, appeared qualitatively comparable and quantitatively different for the four proteins. Several catestatin-positive fragments were detected with mass 11, 26, 34, and 40 kDa (Fig. 5A). In all four proteins, the detected lowest-molecular-mass catestatin-positive fragment was about 11 kDa that differed quantitatively: Gly364Ser (9.67% compared with WT), Pro370Leu (70.5% compared with WT), and Arg374Gln (80.6% compared with WT) (Fig. 5A).
MALDI-TOF revealed the generation of short internal fragments with m/z 840 (CgA360–366), 880 (CgA367–374), 1027 (CgA365–373), 1143 (CgA358–366), 1183 (CgA365–374), 1318 (CgA362–373), 1545 (CgA360–373), 1567 (CgA349–361), and 1848 (CgA358–373) from the wild-type CgA (Fig. 5B). Digestion of CgA-Gly364Ser did not generate the small catestatin peptide (CgA360–373; expected m/z ∼1576); instead, two internal fragments (CgA360–366 and CgA367–374) within the functional catestatin domain were generated (Fig. 5B). Consistent with our plasmin digestion, CTSL digestion of CgA-Pro370Leu also failed to generate functional catestatin peptide (CgA360–373; expected m/z ∼1561) (Fig. 5B). Despite having a mutation within the dibasic site (Arg373Arg374), the CTSL digestion pattern of CgA-Arg374Gln was comparable with WT-CgA (Fig. 5B).
Thus, CTSL displayed differential proteolytic digestion pattern in human variants of catestatin, and the generation of small catestatin fragments (such as CgA360–373) was impaired in the Gly364Ser and Pro370Leu versions of CgA.
Discussion
Overview
The biosynthesis of the physiologically active catestatin peptide (CgA352–372) requires a proteolytic processing of its CgA precursor within the chromaffin granules/vesicles of adrenal medulla or large dense core vesicles of endocrine or neuroendocrine cells. But the mechanism and the specific proteases involved in the processing of the prohormone precursors have so far been incompletely understood. We have shown previously that plasmin, a major fibrinolytic enzyme, cleaved CgA in vitro to produce a catestatin fragment that inhibited nicotine-stimulated norepinephrine secretion from PC12 cells (30). The involvement of the serine proteases PC1/PC3 and PC2 in the proteolytic degradation of CgA have already been demonstrated (20,21,22,23,24,25,26,27). Recent findings revealed that CTSL is present in the secretory vesicles and is responsible for the processing of the neuropeptide enkephalin and neuropeptide Y (31,32,33). Overexpression of CTSL in PC12 cells reduced the abundance of full-length CgA, indicating substrate-like behavior of the latter (Fig. 1C). In addition, the generated catestatin immunopositive peptides in CTSL-overexpressing PC12 cells resembled in vitro digestion products of recombinant CgA (Fig. 1C). Subcellular colocalization of CgA with CTSL in secretory granules also implicated the functional relationship between these two proteins (Fig. 1, A and B). Cathepsin B, D, and K are secreted from thyroid epithelial cells in a regulated pathway and involved in extracellular prohormone processing (50,51,52). Although CTSL is described as lysosomal protease, our immunofluorescence data in sympathochromaffin PC12 cells indicated substantial colocalization of CTSL with the secretory protein CgA. This is consistent with the observation in mouse pituitary, in which CTSL showed partial colocalization with the lysosomal membrane-associated protein 1 but more extensive colocalization with adrenocorticotropic hormone, β-endorphin, and α-melanocyte stimulating hormone (33). Previously we demonstrated very little overlap in localization between CgA and the late endosome/lysosome marker protein LGP110 in the A35C cells (9).
Cleavage in vitro
In the present communication, we undertook biochemical approaches coupled with mass spectrometry to characterize the proteolytic digestion of CgA with CTSL. At the outset, we used a three-step purification procedure to obtain highly purified proteins, and the in vitro proteolysis study showed that the human recombinant CgA proteins were processed readily in a time- and dose-dependent fashion (Fig. 2A). At high protease concentrations, a series of N-terminal catestatin fragments were generated (supplemental Table 1). Earlier we identified a minimal active core of bovine catestatin, consisting of the N-terminal 15-amino acid residues (bovine CgA344–358) within the catestatin (bovine CgA344–364) region (15) using synthetic N- and C-terminal and bidirectional deletion peptides.
Correspondence of CTSL cleavage sites to previously established sites in CgA in vivo
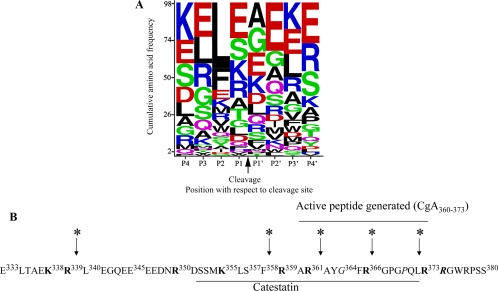
In the catestatin region, previous analyses in bovine chromaffin granule peptides identified cleavage at R331↓L332 of bovine CgA (53); we found cleavage at the corresponding human site R339↓L340 (Table 2 and Fig. 6B). Likewise, previously determined cleavage at bovine CgA R358↓G359 corresponds to our human CgA R366↓G367, and bovine CgA R353↓G354 corresponds to our human CgA R361↓A362. In previously determined sites of human CgA cleavage in pheochromocytoma (54), site R361↓A362 is also found in our CTSL digests (Fig. 6B), whereas L372↓R373 would be expected after carboxypeptidase digestion of the peptide resulting from our R373↓R374 cleavage (Fig. 6B).
Figure 6.
CTSL cleavage sites in CgA. A, Bioinformatic analysis. Recombinant CgA (10 μm) was digested with CTSL (0.3 μm) for 15 min and subjected to LC-MS/MS analyses. The 98 resulting cleavage sites (P1 and P1′ positions) and the flanking sequences (supplemental Tables 1 and 2) were combined into a 20 × 8 amino acid frequency matrix. WebLogo was used to generate a cumulative frequency sequence from this matrix. Amino acids are arranged from top to bottom in order of descending frequency, with the height of each letter proportional to its frequency. B, Depiction of preferred cleavage sites of CTSL within the catestatin region of human CgA. Catestatin region within human CgA was shown as underlined CgA352–372. All the monobasic and paired dibasic sites were shown in bold. The predicted cleavage sites were shown by arrow. The catestatin variant positions were shown in italics.
We obtained additional fragments from in vitro digestion, which were not described previously. It is conceivable that posttranslational modifications of the purified protein are not the same as native protein, in which case different conformations may influence accessibility to proteases. We also used a wide range of CTSL concentrations to digest CgA; some of those conditions may not replicate the in vivo substrate to enzyme ratio and might have resulted in generation of additional fragments. Additional in vivo proteomic analyses along with the in vitro digestion data will be necessary to clarify this point.
Previous studies on mice with targeted ablation of the CTSL gene demonstrated decreased processing of ACTH, β-endorphin, α-MSH, and proneuropeptide Y in brain (33,55).
Consensus CTSL cleavage site in CgA
The WebLogo evaluation (56) of CTSL-generated peptides from CgA indicates that this protease does not have a strong, unequivocal preference for basic residues at the cleavage site (P1-P1′), although it does prefer leucine at the P2 subsite (Fig. 6A). Whereas a preference of CTSL for aromatic and hydrophobic residues upstream of the cleavage site has been reported previously (57,58,59), we did observe such a preference in CgA. In the absence of Leu at the P2 or P3 position, CTSL did show a propensity toward positively charged residues lysine and arginine at the P1 site, which is in congruence with typical prohormone processing. Based on the frequencies, the proposed consensus or most frequent cleavage sequence (P4 to P4′) for CTSL at CgA is KELE↓AEKE. This consensus seems to be unique among the cathepsins (60), but the abolition of cleavage by the specific CTSL inhibitor E64 (Fig. 2A) reinforces the weight of our findings.
Biological activity of CTSL-generated catestatin fragments
Of note, CgA360–373, formed at lower enzyme concentrations, was found to act as a nicotinic antagonist (Figs. 2B and 5B). The same peptide was also detected as the major proteolytic product in the plasmin-catalyzed digestion of CgA (30). CTSL also yielded CgA360–373 active peptide from a catestatin-spanning synthetic peptide, establishing the specificity of CTSL to the digestion of CgA (Fig. 3).
The CTSL-digested CgA peptides almost completely abolished nicotine-evoked catecholamine secretion (Fig. 4). Our previous studies demonstrated that only the catestatin region of CgA has the intrinsic property to inhibit nicotine-stimulated catecholamine release (13). Therefore, our present studies inevitably support for the generation of catestatin region-specific peptide from CTSL-catalyzed digestion of CgA. The observed IC50 value of 0.105 μm was even lower than that of catestatin itself (0.8 μm) (61). The enhanced inhibitory effect could result from multiple active peptides in the digestion reaction. Besides the known active peptides CgA360–373 and CgA360–374, peptide fragments with mass 880 (CgA367–374), 1027 (CgA365–373), 1143 (CgA358–366), 1183 (CgA365–374), and 1848 (CgA358–373) may have contributed to the enhanced inhibitory effect of the CTSL digestion mixture. The activity of these individual peptides as nicotinic antagonists has not been tested yet.
Naturally occurring amino acid replacement variants of catestatin
The catestatin variants are associated with human hypertension (62) and also exert differential potencies as nicotinic antagonists to catecholamine secretion from PC12 cells (61). Hence, it was imperative to understand the processing of the matured proteins containing these variants. These four proteins furnished similar fragment size as evidenced by the catestatin-specific Western blot (Fig. 5A). Among the three amino acid replacement single-nucleotide polymorphisms discovered in the catestatin region at the human CHGA locus, there was a change in the dibasic processing site in CgA-Arg374Gln that might alter its processing. In fact, the digestion pattern of CgA-Arg374Gln with CTSL was very similar to the wild type, suggesting the preferential protease cleavage site for CTSL was before Arg374; hence, any mutation of the latter overall did not change the mass of the generated peptides (Fig. 5B). Also, CgA-Pro370Leu did not produce any detectable amounts of CgA360–373, indicating its Arg373-Arg374 bond cleavage was inefficient (Fig. 5B). In particular, both plasmin and CTSL proteases were unable to generate the catestatin fragment (CgA360–373) from CgA-Pro370Leu variant. In other words, CgA-Pro370Leu variant displayed resistance to cleavage by both plasmin and CTSL. The digestion pattern of CgA-Gly364Ser by CTSL was different from that of the wild-type protein (Fig. 5B), in which the formation of catestatin-positive 11-kDa fragment and CgA360–373 short peptide was less efficient (Fig. 5, A and B). The processing sites at Arg359↓ and Arg373↓ in CgA-Gly364Ser were similar to that of wild-type, as evidenced from the generation of internal peptides CgA360–366, CgA365–373, and CgA365–374, but the generation of 14-amino acid active fragment (CgA360–373) could not be substantiated. It is possible that change of glycine to serine induced more internal cleavage, generating short fragments. The digestion patterns of the full-length protein and synthetic peptides delineated the preferred cleavage site of CTSL are in and around the catestatin region, as summarized in Fig. 6B. Unlike plasmin, the digestion of CgA with a broad range of CTSL concentrations did not offer preferential generation of any particular peptide. The internal cleavage at Ser357↓, Arg359↓, ↓Gly364, and Arg366↓ were apparent in wild-type and the catestatin mutants. The formation of CgA360–373 within a narrow window of substrate to protease ratio suggested a complex proteolytic cleavage of CgA by CTSL. The concentration of the thiol protease in the secretory vesicles may be quite low (63), and under that substrate to enzyme ratio, in lieu of the internal sites, other sites may be preferably cleaved to generate the longer catestatin fragments. The endogenous inhibitors of general cysteine protease might also play a role to direct the cleavage specificity.
Conclusions
Taken together, our results indicate that CgA could be a substrate of CTSL, and the catestatin nonsynonymous variants showed differential processing toward CTSL digestion. Further experiments will be designed to verify the proteolytic profiling of the proteins using a different protease and/or a combination of proteases to better understand the development of human hypertension.
Supplementary Material
Acknowledgments
We are grateful to Professor Elizabeth A. Komives (Department of Chemistry and Biochemistry, University of California, San Diego) for helping us in MALDI-TOF and MS/MS experiments.
Footnotes
This work was supported by National Institutes of Health Grants R29 DA011311 and R01 DA011311 (to S.K.M.), DK 60702 (to D.T.O.), P01 HL58120 and U01 HL69758 (to S.K.M. and D.T.O.); and grants from the Department of Veterans Affairs (to S.K.M. and D.T.O.).
Disclosure Summary: The authors have nothing to disclose.
First Published Online April 16, 2009
Abbreviations: CgA, Chromogranin A protein; CTSL, cathepsin L; LC, liquid chromatography; MALDI, matrix-assisted laser desorption ionization; MS/MS, tandem mass spectrometry; m/z, mass to charge ratio; PC, prohormone convertase; TBS-T, tris-buffered saline with 0.1% Tween 20; TOF, time of flight; WT, wild type.
References
- Winkler H, Fischer-Colbrie R 1992 The chromogranins A and B: the first 25 years and future perspectives. Neuroscience 49:497–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupenot L, Harper KL, O'Connor DT 2003 Mechanisms of disease: the chromogranin-secretogranin family. N Engl J Med 348:1134–1149 [DOI] [PubMed] [Google Scholar]
- Montero-Hadjadje M, Vaingankar S, Elias S, Tostivint H, Mahata SK, Anouar Y 2008 Chromogranins A and B and secretogranin II: evolutionary and functional aspects. Acta Physiol (Oxf) 192:309–324 [DOI] [PubMed] [Google Scholar]
- Takiyyuddin MA, Cervenka JH, Hsiao RJ, Barbosa JA, Parmer RJ, O'Connor DT 1990 Chromogranin A. Storage and release in hypertension. Hypertension 15:237–246 [DOI] [PubMed] [Google Scholar]
- Takiyyuddin MA, Cervenka JH, Sullivan PA, Pandian MR, Parmer RJ, Barbosa JA, O'Connor DT 1990 Is physiologic sympathoadrenal catecholamine release exocytotic in humans? Circulation 81:185–195 [DOI] [PubMed] [Google Scholar]
- Kelly RB 1985 Pathways of protein secretion in eukaryotes. Science 230:25–32 [DOI] [PubMed] [Google Scholar]
- Kim T, Tao-Cheng JH, Eiden LE, Loh YP 2001 Chromogranin A, an “on/off” switch controlling dense-core secretory granule biogenesis. Cell 106:499–509 [DOI] [PubMed] [Google Scholar]
- Kim T, Zhang CF, Sun Z, Wu H, Loh YP 2005 Chromogranin A deficiency in transgenic mice leads to aberrant chromaffin granule biogenesis. J Neurosci 25:6958–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courel M, Rodemer C, Nguyen ST, Pance A, Jackson AP, O'connor DT, Taupenot L 2006 Secretory granule biogenesis in sympathoadrenal cells: identification of a granulogenic determinant in the secretory prohormone chromogranin A. J Biol Chem 281:38038–38051 [DOI] [PubMed] [Google Scholar]
- Tatemoto K, Efendiæ S, Mutt V, Makk G, Feistner GJ, Barchas JD 1986 Pancreastatin, a novel pancreatic peptide that inhibits insulin secretion. Nature 324:476–478 [DOI] [PubMed] [Google Scholar]
- Cadman PE, Rao F, Mahata SK, O'Connor DT 2002 Studies of the dysglycemic peptide, pancreastatin, using a human forearm model. Ann NY Acad Sci 971:528–529 [DOI] [PubMed] [Google Scholar]
- Aardal S, Helle KB, Elsayed S, Reed RK, Serck-Hanssen G 1993 Vasostatins, comprising the N-terminal domain of chromogranin A, suppress tension in isolated human blood vessel segments. J Neuroendocrinol 5:405–412 [DOI] [PubMed] [Google Scholar]
- Mahata SK, O'Connor DT, Mahata M, Yoo SH, Taupenot L, Wu H, Gill BM, Parmer RJ 1997 Novel autocrine feedback control of catecholamine release. A discrete chromogranin A fragment is a noncompetitive nicotinic cholinergic antagonist. J Clin Invest 100:1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahata SK, Mahata M, Parmer RJ, O'Connor DT 1999 Desensitization of catecholamine release: the novel catecholamine release-inhibitory peptide catestatin (chromogranin A344–364) acts at the receptor to prevent nicotinic cholinergic tolerance. J Biol Chem 274:2920–2928 [DOI] [PubMed] [Google Scholar]
- Mahata SK, Mahata M, Wakade AR, O'Connor DT 2000 Primary structure and function of the catecholamine release inhibitory peptide catestatin (chromogranin A344–364): Identification of amino acid residues crucial for activity. Mol Endocrinol 14:1525–1535 [DOI] [PubMed] [Google Scholar]
- Mahata SK 2004 Catestatin—the catecholamine release inhibitory peptide: a structural and functional overview. Curr Med Chem Immun Endocr Metab Agents 4:221–234 [Google Scholar]
- Takiyyuddin MA, Parmer RJ, Kailasam MT, Cervenka JH, Kennedy B, Ziegler MG, Lin MC, Li J, Grim CE, Wright FA, et al 1995 Chromogranin A in human hypertension. Influence of heredity. Hypertension 26:213–220 [DOI] [PubMed] [Google Scholar]
- O'Connor DT, Kailasam MT, Kennedy BP, Ziegler MG, Yanaihara N, Parmer RJ 2002 Early decline in the catecholamine release-inhibitory peptide catestatin in humans at genetic risk of hypertension. J Hypertens 20:1335–1345 [DOI] [PubMed] [Google Scholar]
- Mahapatra NR, O'Connor DT, Vaingankar SM, Hikim AP, Mahata M, Ray S, Staite E, Wu H, Gu Y, Dalton N, Kennedy BP, Ziegler MG, Ross J, Mahata SK 2005 Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J Clin Invest 115:1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjannet S, Leduc R, Adrouche N, Falgueyret JP, Marcinkiewicz M, Seidah NG, Mbikay M, Lazure C, Chretien M 1987 Chromogranin B (secretogranin I), a putative precursor of two novel pituitary peptides through processing at paired basic residues. FEBS Lett 224:142–148 [DOI] [PubMed] [Google Scholar]
- Hoflehner J, Eder U, Laslop A, Seidah NG, Fischer-Colbrie R, Winkler H 1995 Processing of secretogranin II by prohormone convertases: importance of PC1 in generation of secretoneurin. FEBS Lett 360:294–298 [DOI] [PubMed] [Google Scholar]
- Eskeland NL, Zhou A, Dinh TQ, Wu H, Parmer RJ, Mains RE, O'Connor DT 1996 Chromogranin A processing and secretion: specific role of endogenous and exogenous prohormone convertases in the regulated secretory pathway. J Clin Invest 98:148–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Day R, Marcinkiewicz M, Chrétien M 1998 Precursor convertases: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann NY Acad Sci 839:9–24 [DOI] [PubMed] [Google Scholar]
- Laslop A, Weiss C, Savaria D, Eiter C, Tooze SA, Seidah NG, Winkler H 1998 Proteolytic processing of chromogranin B and secretogranin II by prohormone convertases. J Neurochem 70:374–383 [DOI] [PubMed] [Google Scholar]
- Seidah NG, Prat A 2002 Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem 38:79–94 [DOI] [PubMed] [Google Scholar]
- Doblinger A, Becker A, Seidah NG, Laslop A 2003 Proteolytic processing of chromogranin A by the prohormone convertase PC2. Regul Pept 111:111–116 [DOI] [PubMed] [Google Scholar]
- Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM 2006 Proprotein convertases: lessons from knockouts. FASEB J 20:1954–1963 [DOI] [PubMed] [Google Scholar]
- Parmer RJ, Mahata M, Gong Y, Mahata SK, Jiang Q, O'Connor DT, Xi XP, Miles LA 2000 Processing of chromogranin A by plasmin provides a novel mechanism for regulating catecholamine secretion. J Clin Invest 106:907–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Taupenot L, Mahata SK, Mahata M, O'Connor DT, Miles LA, Parmer RJ 2001 Proteolytic cleavage of chromogranin A (CgA) by plasmin: selective liberation of a specific bioactive CgA fragment that regulates catecholamine release. J Biol Chem 276:25022–25029 [DOI] [PubMed] [Google Scholar]
- Biswas N, Vaingankar SM, Mahata M, Das M, Gayen JR, Taupenot L, Torpey JW, O'Connor DT, Mahata SK 2008 Proteolytic cleavage of human chromogranin a containing naturally occurring catestatin variants: differential processing at catestatin region by plasmin. Endocrinology 149:749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V 2003 Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci USA 100:9590–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SR, Garza C, Mosier C, Toneff T, Wunderlich E, Goldsmith P, Hook V 2007 Cathepsin L expression is directed to secretory vesicles for enkephalin neuropeptide biosynthesis and secretion. J Biol Chem 282:9556–9563 [DOI] [PubMed] [Google Scholar]
- Funkelstein L, Toneff T, Hwang SR, Reinheckel T, Peters C, Hook V 2008 Cathepsin L participates in the production of neuropeptide Y in secretory vesicles, demonstrated by protease gene knockout and expression. J Neurochem 106:384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR 2008 Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol 48:393–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke H, Barrett AJ, Rawlings ND 1995 Proteinases 1: lysosomal cysteine proteinases. Protein Profile 2:1581–1643 [PubMed] [Google Scholar]
- Docherty K, Carroll R, Steiner DF 1983 Identification of a 31,500 molecular weight islet cell protease as cathepsin B. Proc Natl Acad Sci USA 80:3245–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty K, Hutton JC, Steiner DF 1984 Cathepsin B-related proteases in the insulin secretory granule. J Biol Chem 259:6041–6044 [PubMed] [Google Scholar]
- Kukor Z, Mayerle J, Krüger B, Tóth M, Steed PM, Halangk W, Lerch MM, Sahin-Tóth M 2002 Presence of cathepsin B in the human pancreatic secretory pathway and its role in trypsinogen activation during hereditary pancreatitis. J Biol Chem 277:21389–21396 [DOI] [PubMed] [Google Scholar]
- Neves FA, Duncan KG, Baxter JD 1996 Cathepsin B is a prorenin processing enzyme. Hypertension 27:514–517 [DOI] [PubMed] [Google Scholar]
- Jutras I, Reudelhuber TL 1999 Prorenin processing by cathepsin B in vitro and in transfected cells. FEBS Lett 443:48–52 [DOI] [PubMed] [Google Scholar]
- McCoy K, Gal S, Schwartz RH, Gottesman MM 1988 An acid protease secreted by transformed cells interferes with antigen processing. J Cell Biol 106:1879–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretzki L, Schmitt M, Mann K, Calvete J, Chucholowski N, Kramer M, Günzler WA, Jänicke F, Graeff H 1992 Effective activation of the proenzyme form of the urokinase-type plasminogen activator (pro-uPA) by the cysteine protease cathepsin L. FEBS Lett 297:112–118 [DOI] [PubMed] [Google Scholar]
- Kasai M, Shirasawa T, Kitamura M, Ishido K, Kominami E, Hirokawa K 1993 Proenzyme from of cathepsin L produced by thymic epithelial cells promotes proliferation of immature thymocytes in the presence of IL-1, IL-7, and anti-CD3 antibody. Cell Immunol 150:124–136 [DOI] [PubMed] [Google Scholar]
- Stypmann J, Gläser K, Roth W, Tobin DJ, Petermann I, Matthias R, Mönnig G, Haverkamp W, Breithardt G, Schmahl W, Peters C, Reinheckel T 2002 Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci USA 99:6234–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Zhang Y, Pan J, Sun J, Liu J, Libby P, Sukhova GK, Doria A, Katunuma N, Peroni OD, Guerre-Millo M, Kahn BB, Clement K, Shi GP 2007 Cathepsin L activity controls adipogenesis and glucose tolerance. Nat Cell Biol 9:970–977 [DOI] [PubMed] [Google Scholar]
- Li W, Kornmark L, Jonasson L, Forssell C, Yuan XM 2009 Cathepsin L is significantly associated with apoptosis and plaque destabilization in human atherosclerosis. Atherosclerosis 202:92–102 [DOI] [PubMed] [Google Scholar]
- Mahata M, Mahata SK, Parmer RJ, O'Connor DT 1996 Vesicular monoamine transport inhibitors. Novel action at calcium channels to prevent catecholamine secretion. Hypertension 28:414–420 [DOI] [PubMed] [Google Scholar]
- Taupenot L, Remacle JE, Helle KB, Aunis D, Bader MF 1995 Recombinant human chromogranin A: expression, purification and characterization of the N-terminal derived peptides. Regul Pept 56:71–88 [DOI] [PubMed] [Google Scholar]
- Courel M, Vasquez MS, Hook VY, Mahata SK, Taupenot L 2008 Sorting of the neuroendocrine secretory protein secretogranin II into the regulated secretory pathway: role of N- and C-terminal α-helical domains. J Biol Chem 283:11807–11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix K, Linke M, Tepel C, Herzog V 2001 Cysteine proteinases mediate extracellular prohormone processing in the thyroid. Biol Chem 382:717–725 [DOI] [PubMed] [Google Scholar]
- Colombo B, Longhi R, Marinzi C, Magni F, Cattaneo A, Yoo SH, Curnis F, Corti A 2002 Cleavage of chromogranin A N-terminal domain by plasmin provides a new mechanism for regulating cell adhesion. J Biol Chem 277:45911–45919 [DOI] [PubMed] [Google Scholar]
- Tepel C, Brömme D, Herzog V, Brix K 2000 Cathepsin K in thyroid epithelial cells: sequence, localization and possible function in extracellular proteolysis of thyroglobulin. J Cell Sci 113 Pt 24:4487–4498 [DOI] [PubMed] [Google Scholar]
- Metz-Boutigue MH, Garcia-Sablone P, Hogue-Angeletti R, Aunis D 1993 Intracellular and extracellular processing of chromogranin A. Determination of cleavage sites. Eur J Biochem 217:247–257 [DOI] [PubMed] [Google Scholar]
- Orr DF, Chen T, Johnsen AH, Chalk R, Buchanan KD, Sloan JM, Rao P, Shaw C 2002 The spectrum of endogenous human chromogranin A-derived peptides identified using a modified proteomic strategy. Proteomics 2:1586–1600 [DOI] [PubMed] [Google Scholar]
- Funkelstein L, Toneff T, Mosier C, Hwang SR, Beuschlein F, Lichtenauer UD, Reinheckel T, Peters C, Hook V 2008 Major role of cathepsin L for producing the peptide hormones ACTH, β-endorphin, and α-MSH, illustrated by protease gene knockout and expression. J Biol Chem 283:35652–35659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004 WebLogo: a sequence logo generator. Genome Res 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasothornsrikul S, Aaron W, Toneff T, Hook VY 1999 Evidence for the proenkephalin processing enzyme prohormone thiol protease (PTP) as a multicatalytic cysteine protease complex: activation by glutathione localized to secretory vesicles. Biochemistry 38:7421–7430 [DOI] [PubMed] [Google Scholar]
- Puzer L, Cotrin SS, Alves MF, Egborge T, Araújo MS, Juliano MA, Juliano L, Brömme D, Carmona AK 2004 Comparative substrate specificity analysis of recombinant human cathepsin V and cathepsin L. Arch Biochem Biophys 430:274–283 [DOI] [PubMed] [Google Scholar]
- Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, Vlodavsky I 2008 Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem 283:18167–18176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon RJ, Bond JS 1989 Proteolytic enzymes: a practical approach. New York: IRL Press at Oxford University Press [Google Scholar]
- Mahata SK, Mahata M, Wen G, Wong WB, Mahapatra NR, Hamilton BA, O'Connor DT 2004 The catecholamine release-inhibitory “catestatin” fragment of chromogranin A: naturally occurring human variants with different potencies for multiple chromaffin cell nicotinic cholinergic responses. Mol Pharmacol 66:1180–1191 [DOI] [PubMed] [Google Scholar]
- Rao F, Wen G, Gayen JR, Das M, Vaingankar SM, Rana BK, Mahata M, Kennedy BP, Salem RM, Stridsberg M, Abel K, Smith DW, Eskin E, Schork NJ, Hamilton BA, Ziegler MG, Mahata SK, O'Connor DT 2007 Catecholamine release-inhibitory peptide catestatin [chromogranin A(352–372)]: naturally occurring amino acid variant Gly364Ser causes profound changes in human autonomic activity and alters risk for hypertension. Circulation 115:2271–2281 [DOI] [PubMed] [Google Scholar]
- Krieger TJ, Hook VY 1991 Purification and characterization of a novel thiol protease involved in processing the enkephalin precursor. J Biol Chem 266:8376–8383 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.