Abstract
FSH acts through the FSH receptor (FSHR) to modulate cell processes that are required to support developing spermatozoa. Within the testis, only Sertoli cells possess receptors for FSH and are the major targets for this regulator of spermatogenesis. FSH stimulation of Sertoli cells for 24–48 h is known to induce Fshr mRNA expression through an E-box motif (CACGTG) located 25 bp upstream of the transcription start site. In contrast, FSH stimulation for 8 h inhibits Fshr transcription. DNA-protein binding studies performed using nuclear extracts from Sertoli cells show that protein binding to the Fshr promoter E-box was reduced 68% after 6 h of FSH stimulation but increased 191% over basal levels after 48 h of stimulation. The proteins binding to the Fshr E-box were identified as upstream stimulatory factor (USF)-1 and -2. FSH stimulation transiently decreased USF1 levels and increased the expression of the inhibitor of DNA binding/differentiation (ID)-2 repressor protein with the same kinetics as the decreased USF/E-box interactions. Overexpression of ID2 resulted in a dose-dependent decrease in USF-driven Fshr promoter activity in the MSC-1 Sertoli cell line, and ID2 inhibited USF binding to the Fshr E-box. Together, these studies suggest that stimulation of Sertoli cells with FSH transiently decreases expression of the USF1 activator and induces accumulation of the ID2 repressor, to block USF binding to the Fshr promoter and delay activation of Fshr transcription. This FSH-regulated mechanism may explain the cyclical changes in Fshr expression that occurs in Sertoli cells in vivo.
FSH stimulation of Sertoli cells transiently decreases the expression of the USF1 transcriptional activator and induces the expression of the ID2 repressor to block USF binding to the Fshr promoter and delay Fshr transcription.
The process of spermatogenesis is regulated by a complex interplay of endocrine and paracrine signals. FSH and LH are the major components of the hypothalamic-pituitary-gonadal axis that regulate reproductive function and ultimately fertility. In the testis, LH acts on the Leydig cells to cause the production of testosterone. Both FSH and testosterone bind to receptors on the surface or in the cytoplasm of somatic Sertoli cells, respectively. In response to FSH or testosterone, the Sertoli cells supply developing germ cells with growth factors and nutrients and control the biochemical environment in which germ cells develop into mature sperm (1).
In adult male rats, serum FSH levels remain relatively constant. However, the expression of FSH receptor (FSHR) on Sertoli cells varies more than 3-fold in a cyclical manner throughout the cycle of the seminiferous epithelium resulting in the cyclical activation of FSH-mediated signaling pathways (2). FSH binding to FSHR and subsequent receptor coupling with Gs is responsible for FSH-mediated increases in the levels of the intracellular signaling factor cAMP and the activation of protein kinase A (3). FSH stimulation of Sertoli cells also results in calcium influx as well as the activation of the phosphatidylinositol 3-kinase, p38 MAPK and ERK MAPK intracellular signaling pathways. A feedback mechanism controls FSHR expression because FSH has been shown to down-regulate Fshr mRNA expression (4,5), but the mechanism by which expression is down-regulated is not well understood.
The promoter region regulating expression of the Fshr gene has been characterized. Transient transfection analysis of deletion and block replacement mutants indicated that an E-box element (CACGTG), located 120 bp upstream of the translation start site and 25 bp upstream of the major transcriptional start site, is a critical control site for Fshr transcription (6). Members of the basic helix-loop-helix (HLH) family of proteins called E-box proteins bind as dimers to E-box motifs that have the consensus sequence CANNTG. The ubiquitously expressed E-box proteins upstream stimulatory factor (USF)-1 and USF2 (7,8) have been shown to bind to the Fshr E-box and are known to regulate the expression of the Fshr promoter within Sertoli cells (6,9,10,11). However, there are no reported studies regarding FSH regulation of USF proteins and potential FSH regulation of USF binding to the Fshr promoter E-box.
Inhibitors of DNA binding/differentiation (ID) protein are HLH proteins that lack the DNA binding region but are able to dimerize with and inhibit the binding of E-box proteins to DNA and inhibit E-box-mediated transcription. Four ID proteins have been characterized in mammalian cells (ID1-4) (12,13,14,15). In Sertoli cells, the ID1 protein was shown to repress Fshr promoter activity, but it is not known whether ID1 directly interferes with USF protein activity (9). ID-2 is another potential negative regulator of USF-mediated activation of the Fshr promoter because Id2 mRNA is transiently induced by FSH stimulation of Sertoli cells (16). Thus far, it is not known whether FSH-mediated increases in ID2 levels feed back to block USF actions and Fshr promoter activity. However, FSH stimulation was shown to transiently decrease Fshr mRNA levels (4) concurrent with increased Id2 mRNA expression (16). These results raise the possibility that ID2 may act as a repressor of FSH-mediated gene transcription in Sertoli cells by interfering with E-box protein binding to the Fshr promoter. We now demonstrate FSH or agents that elevate cAMP levels transiently decrease USF protein binding to the E-box. FSH also was found to transiently decrease USF1 expression and elevate ID2 levels corresponding with the decrease in binding to the Fshr promoter E-box. Furthermore, we show that USF binding to the Fshr promoter E-box and USF-mediated stimulation of Fshr expression is inhibited by ID2 in a dose-dependent manner.
Materials and Methods
Isolation of primary Sertoli cells and cell culture
Sertoli cells were isolated from 15-d-old Sprague Dawley rats and cultured in serum-free media as previously described (16).
The mouse-derived Sertoli cell line MSC-1, which lacks FSHRs (17), was cultured at 33 C in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine. COS7 cells were cultured under the same conditions as MSC-1 cells at 37 C.
Animals used in these studies were maintained and killed according to the principles and procedures described in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. These studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
DNA-protein binding studies
32P-radiolabeled DNA probes were generated by annealing 26–27 nucleotide templates containing protein binding motifs plus flanking promoter sequences to 10-nucleotide primers that are complementary to the 5′ end of the templates. The overhangs were filled in with Klenow in the presence of [α-32P] dATP and 5 mm each dCTP, dGTP, and dTTP.The probes used and their coding strand template sequences included the Fshr E-box, 5′-GATCGGTGGGTCACGTGACTTTGC-3′ (the E-box motif is underlined), Fshr mt E-box,-5′ GATCGGTGGGTACCGTGACTTTGC-3′ and a consensus cAMP response element-binding protein (CREB) binding site, 5′-GATCCGGCTGACGTCATCAAGCTAGAA-3′. EMSAs were performed as described (18). For supershift assays, antiserum was added to the mixture for 15 min at room temperature before the addition of the probe. All antisera were obtained from Santa Cruz Biotechnologies (Santa Cruz Biotechnologies, Santa Cruz, CA): α-USF1 (sc-229), α-USF2 (sc-862), α-MYC (sc-40), α-E47 (sc-763). For comparing protein-binding affinities of various probes, labeled probes with similar specific activities (±20%) were used in simultaneous reactions. DNA-protein complex formation was visualized by autoradiography and quantified using Image J software (version 1.37; National Institutes of Health, Bethesda, MD) (19).
For the EMSA experiments investigating the interaction of ID and USF proteins, whole-cell extracts were made from COS7 or primary Sertoli cells overexpressing ID2 and USF proteins using enhanced lysis buffer as previously described (20). The extracts were mixed to have a total of 30 μg (COS7) or 10 μg (Sertoli) of protein. For COS7 cells, constant levels of extract from cells transfected with USF expression vectors (5 μg) were mixed with increasing levels of extracts from cells overexpressing ID2 (2.5, 5, 10 μg). Whole-cell extracts from untransfected COS7 cells without overexpressed proteins were added to keep the protein concentration in the mixes equal. For Sertoli cells, extracts from cells transfected with empty expression vector were mixed with increasing levels of extracts from cells transfected with an ID2 expression vector. The extracts were mixed for 30 min at 4 C and 0.9 μg (COS7) or 1.2 μg (Sertoli) of the mixtures were added to either the Fshr E-box or CREB probes in EMSA reactions.
RNA isolation and cDNA preparation
RNA was obtained from cultured Sertoli cells using the method of Chomczynski and Sacchi (21). After digestion with ribonuclease-free deoxyribonuclease, the RNA was subjected to reverse transcription using random hexamers (22). For reverse transcription, 250 ng of RNA was incubated with 100 μl of reaction mix containing 7.5 mm MgCl2, 400 μm deoxynucleotide triphosphates (Promega, Madison, WI), 10× PCR II Buffer (Applied Biosystems), 40 U RNasin ribonuclease inhibitor (Promega, Madison, WI), 2.25 μm random hexamers (Integrated DNA Technologies, Coralville, IA), 250 U SuperScript reverse transcriptase II (Invitrogen), and nuclease-free water (Ambion, Austin, TX). Parallel reactions were performed without reverse transcriptase to control for the presence of contaminant DNA. The samples were incubated at 25 C for 10 min, 48 C for 30 min, and 95 C for 5 min followed by 4 C for 5 min.
Quantitative real-time PCR (qPCR) analysis of gene expression
qPCR amplifications were performed in a 96-well plate in the ABI Prism 7900HT sequence detection system version 2.3 (Applied Biosystems, Foster City, CA) in a total volume of 20 μl, which included 2 μl of cDNA, 10 μl of Perfecta SYBR Green FastMix ROX (Quanta Biosciences, Inc., Gaithersburg, MD), and 600 μm of each primer. Primers used for each gene are listed in Table 1 and were independently validated for use in the ΔΔCt method of gene expression analysis. Ppia (peptidylprolyl isomerase A, commonly known as cyclophillin) was used as an endogenous control. The qPCR analysis initiated with melting of cDNA at 95 C for 15 min, followed by 40 amplification cycles (15 sec at 95 C and 1 min at 60 C). A dissociation curve was performed immediately after amplification to ensure there was only one (gene specific) amplification peak. Cycle threshold (Ct) values were recorded and analyzed via the ΔΔCt method (23). The means (±sd) of three individual experiments were determined for each treatment group for each gene of interest.
Table 1.
Primers for qPCR
| Gene | Accession no. | Sequence (5′–3′) | Region amplified | Referencea |
|---|---|---|---|---|
| Fshr | NM_199237.1 | Forward, ACGCCATTTTCACCAAGAAC | 1944–2022 | |
| Reverse, TGGGCTTGCATTTCATAACA | ||||
| Usf1 | NM_031777 | Forward, AAGTCAGAGGCTCCCAGGA | 693–761 | (42) |
| Reverse, CGGCGCTCCACTTCGTTAT | ||||
| Usf2 | NM_031139 | Forward, AGACCAACCAGCGTATGCAG | 1386–1473 | (42) |
| Reverse, GCTCCTCGGATCTGCTGCCT | ||||
| Ppia | M19533.1 | Forward, atggtcaaccccaccgtgt | 43–143 | |
| Reverse, tctgctgtctttggaactttgtct |
Primers without a reference were designed for use in this study with the aid of Primer Express software (version 3.0; Applied Biosystems).
Immunofluorescence assays
Immunostaining was performed on primary cultures of rat Sertoli cells grown on glass coverslips. The cells were incubated with vehicle or FSH (100 ng/ml) + 3-isobutyl-1-methylxanthine (IBMX; 0.5 mm) for 2–48 h and then fixed in 2% paraformaldehyde for 30 min, washed in PBS, permeabilized in 0.05% Triton-X-100 for 5 min on ice, washed with PBS, and blocked in goat serum. The cells were probed with either USF1 antiserum (USF1 C-20, SC-229; Santa Cruz Biotechnology) or ID2 antiserum made in rabbit against a His-tagged-full-length ID2 protein that was purified by Ni-agarose affinity chromatography followed by fractionation by SDS-PAGE (obtained from E. Prochownik, University of Pittsburgh) (24). All cells were costained with antiserum against Sertoli cell-specific vimentin (V 6630; Sigma-Aldrich, St. Louis, MO). Immunoreactivity was observed using Alexa 488 conjugated fluorescent secondary goat antirabbit antiserum (USF1 and ID2) or Cy3 conjugated fluorescent secondary goat antirabbit antiserum (vimentin). Nuclei were stained with 4′,6′-diamino-2-phenylindole to determine that comparable numbers of cells were viewed in images.
Reporter plasmid constructs, transient transfections, and luciferase assays
The FSHR(−220/+123)luciferase (Luc) reporter plasmid contains the Fshr promoter region extending from −220 to +123 (6). The luciferase reporter driven by a consensus κB-enhancer (κBLUC) reporter plasmid contains the −105 to −79 region of the HIV promoter including two κB-enhancer elements inserted upstream of the luciferase gene in pGL2basic. MSC-1 Sertoli cells were transfected with 0.5 μg of reporter plasmid in the presence or absence of 0.5–2.0 μg of empty expression vector or vectors expressing USF1, USF2, or ID2 (pSVUSF1, pSVUSF2, pRCCMVID2) (24,25) using FUGENE reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. Two days after transfection the MSC-1 cells were stimulated with vehicle or forskolin (10 μm) plus IBMX (0.5 mm) for 8 h after which the cells were collected and total cellular proteins were extracted using reporter lysis buffer (Promega). Luciferase assays were performed using a Victor2 luminometer (PerkinElmer, Waltham, MA). Luciferase activities were normalized for total protein as determined by Bradford assay.
Statistical analysis
Results were analyzed by ANOVA with Newman-Keuls protected least significant difference at a 5% significance level using GraphPad Prism 4.3 (GraphPad Software, San Diego, CA).
Results
Elevating FSH or cAMP levels for 48 h results in increased USF protein binding to the Fshr promoter
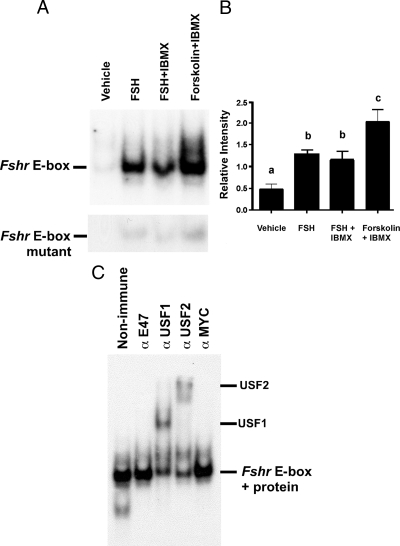
To characterize the effects of FSH and elevated cAMP levels on protein interactions with the E-box motif within the Fshr promoter, we performed EMSA DNA-protein binding studies using nuclear extracts of cultured Sertoli cells isolated from 15-d-old rats and a probe containing the Fshr promoter E-box and flanking sequences. Stimulation of the cultured Sertoli cells with FSH for 48 h resulted in a dramatic increase in binding to the Fshr E-box probe. After treatment with FSH and the phosphodiesterase inhibitor IBMX or the potent cAMP inducer forskolin plus IBMX, binding activity was as high or higher than FSH stimulation alone (Fig. 1, A and B). A probe containing a 2-bp mutation of the E-box (Fshr E-box mt) was not an effective binder of proteins from the Sertoli nuclear extracts, indicating that protein binding requires the presence of the E-box motif. These data suggest that protein binding to the E-box within the Fshr promoter is enhanced after long-term elevation of cAMP levels.
Figure 1.
FSH and cAMP induce USF binding to the Fshr E-box. A, EMSA studies were performed using probes containing either the E-box region of the Fshr promoter (upper panel) or the same probe with a mutated E-box (lower panel) and nuclear extracts from primary Sertoli cells that were stimulated with vehicle, FSH (100 ng/ml), FSH + IBMX (0.5 mm), or forskolin (10 μm) + IBMX as indicated. The DNA-binding complexes are indicated on the left. B, For studies performed as in A, the mean (±se) of protein binding (relative intensity) to the wild-type E-box region of the Fshr promoter is shown (n = 3). Values with different lower-case letters differ significantly (P < 0.05). C, EMSA DNA-protein binding studies were performed with the Fshr E-box probe and extracts from Sertoli cells stimulated with FSH and IBMX for 48 h. The DNA-protein reactions were incubated with nonimmune serum or antisera against E47, USF1, USF2, or MYC. The supershifted complexes containing USF1 or USF2 proteins are indicated on the right. DNA-protein complexes were visualized by autoradiography. The unbound radiolabeled probe was run off of the gel for all studies. The data shown are representative of more than three independent experiments.
Supershift EMSA studies were performed to identify the proteins binding to the E-box in the Fshr promoter. Extracts from Sertoli cells stimulated with FSH and IBMX for 48 h were incubated with nonimmune sera and antisera against E-box proteins. Antisera against E47 or MYC did not alter the migration of the DNA-protein complexes; however, antisera against USF1 or USF2 proteins supershifted the complexes (Fig. 1C). These studies are consistent with those of Heckert and colleagues (11) that demonstrated USF binding to the Fshr promoter in vivo. Furthermore, these results indicate that the levels of USF1 and USF2 binding to the Fshr E-box increase after stimulation with FSH for 48 h.
FSH stimulation causes time-dependent changes in protein binding to the Fshr promoter E-box
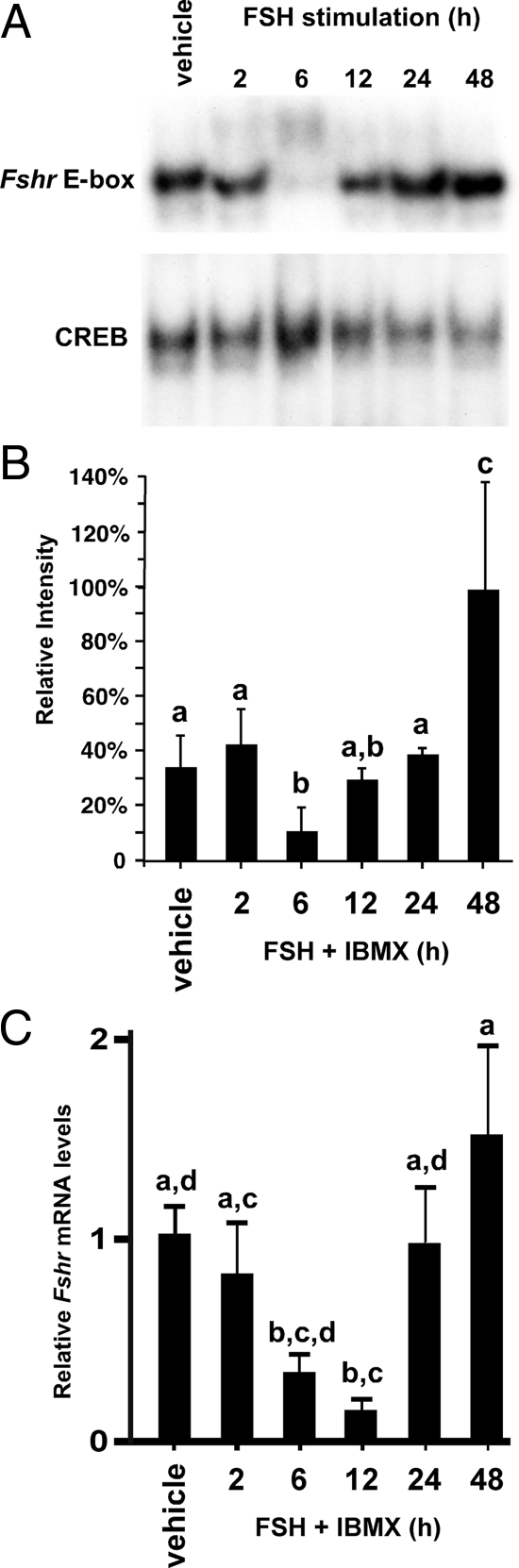
To determine the kinetics of FSH-mediated enhancement of binding to the Fshr E-box motif, binding activity was assayed after Sertoli cells were stimulated with FSH + IBMX for 2–48 h. Instead of a linear increase in FSH-induced DNA-protein interactions, we observed that protein binding to the Fshr E-box was essentially unchanged after 2 h, but binding decreased precipitously by 68% after 6 h before gradually increasing to 191% greater than basal levels after 48 h of FSH stimulation (Fig. 2, A and B).
Figure 2.
Transient down-regulation of USF binding to the Fshr E-box corresponds with decreased Fshr expression. A, FSH stimulation transiently decreases USF binding to the Fshr E-box. Representative EMSA images are shown for assays using probes containing either the Fshr E-box probe or a CREB binding site and nuclear extracts from Sertoli cells stimulated with vehicle or FSH (100 ng/ml) + IBMX (0.5 mm) for 2, 6, 12, 24, and 48 h. B, The mean (±se) of the relative signal intensities of E-box binding activity normalized to CREB binding from four independent experiments are displayed. The binding intensities were arbitrarily made relative to that of 48 h stimulation (100%). C, qPCR assays were performed on RNA isolated from cultured Sertoli cells treated with vehicle or FSH (100 ng/ml) + IBMX (0.5 mm) for 2–24 h. The relative mRNA levels for Fshr were normalized to Ppia (cyclophillin). The means (±se) of three experiments are presented. Values with different lower-case letters differ significantly (P < 0.05).
To determine how changes in protein binding to the E-box corresponded with Fshr mRNA expression, qPCR analysis was performed using RNA from vehicle-treated Sertoli cells and cells treated with FSH + IBMX for 2–24 h. These studies revealed that Fshr mRNA levels decreased significantly after 6 h, reached a nadir at 12 h, but started to return to initial levels after 24 h of stimulation and were elevated over baseline levels by 48 h (Fig. 2C). The kinetics of the transiently decreasing Fshr mRNA levels in response to FSH is consistent with previous reports (4,5,26). Furthermore, the pattern of mRNA expression also closely followed that of protein binding to the Fshr E-box, although the relative timing of the mRNA decrease was delayed by 4–6 h.
FSH stimulation transiently decreases USF expression
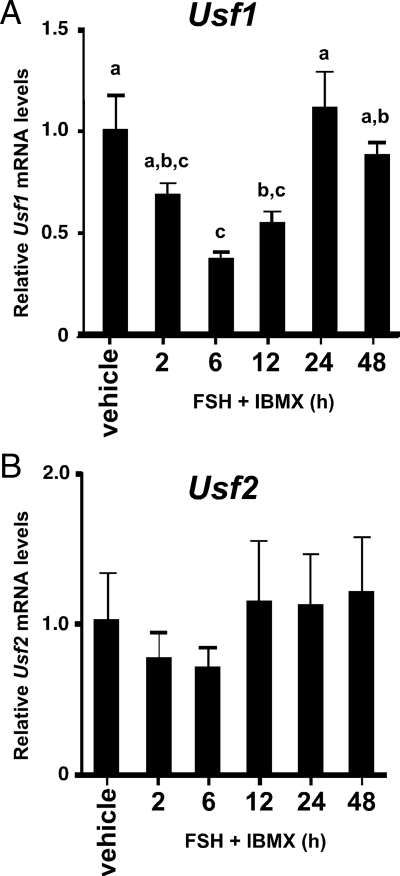
One explanation for decreased protein binding to the Fshr promoter E-box in response to FSH treatment would be that FSH stimulation decreases the expression of the proteins binding to the Fshr promoter E-box. Because USF proteins were shown by supershift analysis to be the predominant protein binding to the Fshr E-box probe, the expression of the genes encoding USF1 and USF2 were assayed during stimulation with FSH + IBMX. qPCR analysis of Usf1 and Usf2 mRNA levels in cultured Sertoli cells determined that Usf1 mRNA expression transiently decreases after 2–12 h of FSH + IBMX stimulation (Fig. 3A). The lowest levels of Usf1 expression occurred after 6 h when mRNA levels declined 67%. Usf2 mRNA expression followed a similar trend but with more limited decreases that did not reach statistical significance when compared with vehicle-treated cells (Fig. 3B).
Figure 3.
FSH stimulation transiently decreases Usf1 mRNA levels. qPCR assays for Usf1 (A) and Usf2 (B) were performed as described in Fig. 2C. The means (±se) of three experiments are presented. Values with different lower-case letters differ significantly (P < 0.05).
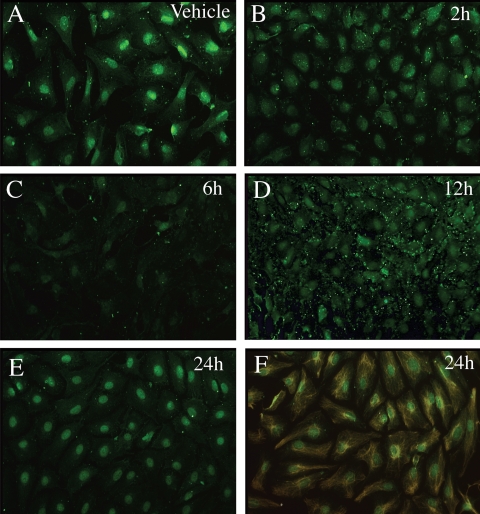
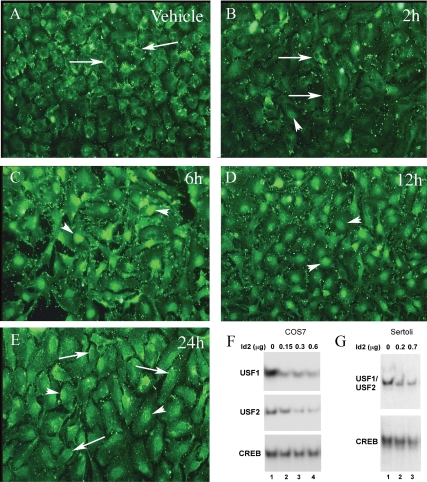
For Usf1, which showed the greater response to FSH, additional protein expression studies were performed to determine whether the decrease in Usf1 mRNA translated to decreased protein levels. Previously we found that Western analysis of endogenous USF protein expression in cultured Sertoli cells is possible but requires large amounts of Sertoli cell extract (Wood, M. A., and W. H. Walker, unpublished results). Therefore, to assay USF1 levels at multiple time points, we used the USF antiserum validated in our Western blot assays for immunofluorescence studies. This study revealed that USF1 protein was present predominantly in the nuclei of vehicle-treated Sertoli cells but that USF1 immunostaining decreased after 2–12 h of FSH + IBMX stimulation with the lowest levels of USF1 being present after 6 h of treatment (Fig. 4, A–E). Nuclear USF1 immunostaining returned to near vehicle levels after 24 h of FSH + IBMX stimulation. The highly enriched nature of the Sertoli cell cultures was confirmed after probing with antiserum against the Sertoli cell-specific intermediate filament protein vimentin (27) (Fig. 4F). Together the mRNA and protein expression results suggest that FSH-induced decreases in USF1 expression contribute to the decrease in protein binding to the Fshr promoter E-box.
Figure 4.
USF1 expression in the nuclei of Sertoli cells is transiently repressed by FSH stimulation. Cultures of primary Sertoli cells were treated with vehicle or FSH (100 ng/ml) + IBMX (0.5 mm) for 2–24 h as indicated and probed with antiserum against USF1 (green, A–E) or vimentin (orange) + USF1 after 24 h of stimulation (F). All images contained similar numbers of cells as determined by 4′,6′-diamino-2-phenylindole staining (not shown). Results are representative of three independent experiments.
FSH stimulation transiently increases ID2 expression
A second potential mechanism to decrease protein binding to the Fshr promoter E-box is the increased expression of an inhibitor of protein binding to E-boxes such as ID2. We noted that the timing of the FSH-mediated decrease in E-box binding activity and Fshr mRNA expression corresponded to the previously demonstrated induction of Id2 mRNA that occurs 2–12 h after FSH stimulation (16). Therefore, ID2 protein levels were assayed in immunofluorescence studies to determine whether ID2 levels corresponded with the known Id2 mRNA expression patterns. Analysis of ID2 protein expression revealed that Sertoli cells treated with vehicle or FSH + IBMX for 2 h had low overall levels of ID2 immunostaining that was nuclear excluded (Fig. 5, A and B). The ID2 staining that could be detected was predominantly perinuclear. ID2 immunostaining increased in Sertoli cells after 6 h of stimulation with most of the ID2 staining being detected within nuclei (Fig. 5C). ID2 levels then diminished slightly at 12 h and were more dramatically reduced after 24 (Fig. 5, D and E). The kinetics of ID2 protein expression in response to FSH + IBMX stimulation was consistent with the results of the earlier mRNA analysis (16) and the hypothesis that induction of ID2 protein expression contributes to decreasing the E-box binding activity of USF proteins in Sertoli cells.
Figure 5.
FSH transiently increases ID2 levels in Sertoli cells and ID2 blocks USF protein binding to the Fshr promoter E-box. A–E, Cultured Sertoli cells were treated with vehicle or FSH (100 ng/ml) + IBMX (0.5 mm) for 2–24 h as indicated and probed with antiserum against ID2 (green). Arrows indicate perinuclear ID2 staining and nuclear exclusion of ID2. The arrowheads indicate nuclear staining of ID2. Results are representative of three independent experiments. Similar results were observed using two other ID2 antiserum (data not shown). F and G, EMSA studies were performed using either the Fshr E-box or consensus CREB binding site probe incubated with mixtures of whole-cell extracts from COS7 cells (F) or Sertoli cells (G) that were transfected with empty, USF, or ID2 expression plasmids. A constant level of extract (0.3 μg) from COS7 cells transfected with USF1 or USF2 expression plasmids were mixed with extracts from cells transfected with empty vector (lane 1) or 0.15, 0.3, or 0.6 μg of extract from cells transfected with ID2 expression vector. Extracts from cells transfected with empty vector were added to provide a total of 0.9 μg protein for each EMSA reaction (lanes 2–4). Binding of USF1 and USF2 to the Fshr E-box probe are shown in the top and middle panels, respectively. The binding of endogenous CREB proteins to the CREB probe is shown in the bottom panel. Whole-cell extracts from Sertoli cells transfected with either empty vector or ID2 expression vector were mixed and incubated with Fshr E-box or CREB probes in EMSA reactions. A total of 1.2 μg of extract was used for each DNA-protein binding reaction and consisted of 0, 0.2, or 0.7 μg from Sertoli cells transfected with ID2 expression vector. The relative binding of endogenous USF1/USF2 and CREB proteins to their respective probes are shown. The results displayed are representative of three (F) and two (G) independent experiments. Unbound probes were run off of the gels.
ID2 inhibits binding to the Fshr promoter E-box
The FSH-mediated increase in ID2 expression in Sertoli cells raised the possibility that ID2 can interfere with the binding of USF proteins to the Fshr E-box. To test this concept, whole-cell extracts prepared from COS7 cells transfected with ID2 or USF expression vectors were mixed and subjected to EMSA analysis. Constant levels of extracts from cells transfected with USF1 or USF2 expression plasmids were mixed with extracts transfected with empty expression vector or increasing levels of extracts from cells transfected with an ID2 expression vector. Equal levels of total protein in the mixtures were maintained by addition of protein from untransfected control cells. Incubation of the extract mixtures with probes containing the Fshr promoter E-box indicated that USF1 and USF2 protein binding to the Fshr promoter E-box probe was inhibited with increasing ID2 levels (Fig. 5F, upper panels). In contrast, interactions of endogenous CREB transcription factors with a probe containing a consensus CREB binding site was not significantly altered by ID2 (Fig. 5F, bottom panel). The results of these DNA-protein binding studies indicate that increasing the levels of ID2 proteins results in blocking of USF binding to the E-box motif within the Fshr promoter. To confirm that elevated ID2 levels block DNA binding of endogenous USF proteins in Sertoli cells, extracts were prepared from Sertoli cells transfected with empty or ID2 expression vectors. EMSA analysis of mixtures of the two extracts revealed that mixtures containing higher percentages of extract from cells transfected with the ID2 expression vector displayed less USF binding to the Fshr E-box probe (Fig. 5G). CREB binding activity was not greatly altered in the various extract mixtures.
ID2 decreases Fshr promoter activity
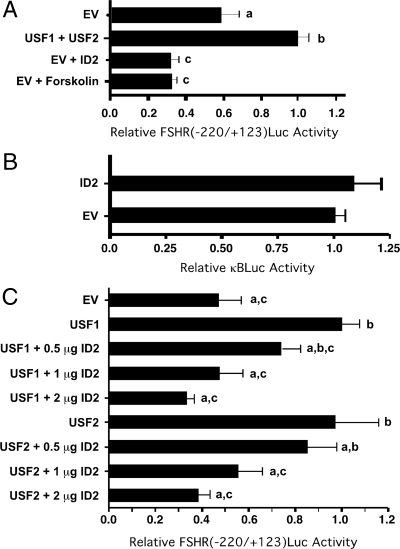
To test the hypothesis that ID proteins interfere with USF-mediated induction of Fshr, promoter activity, the luciferase reporter construct FSHR(−220/+123)Luc containing the Fshr E-box and known to be regulated by USF proteins was used for transient transfection assays (6). Preliminary studies indicated that the FSHR(−220/+123)Luc construct was active in the MSC-1 mouse Sertoli cell line. Because transfection efficiency is relatively high in the readily available MSC-1 cells, we chose to use this cell model to study potential ID2 regulation of USF-mediated transcription. Cotransfection of FSHR(−220/+123)Luc with plasmids expressing USF1 and USF2 induced promoter activity 1.7-fold over basal levels (Fig. 6A). In contrast, cotransfection of the ID2 expression plasmid decreased basal Fshr promoter activity to levels similar to that found after stimulation with forskolin + IBMX for 8 h. These data demonstrate that that overexpression of ID2 inhibits transcription from the Fshr promoter to levels that are consistent with that resulting from forskolin-mediated induction of the ID2 repressor protein. Overexpression of ID2 did not cause a general inhibition of transcription as the activity of a κBLUC was not affected by cotransfection with the ID expression vector (Fig. 6B).
Figure 6.
ID2 down-regulates basal and USF-induced Fshr promoter activity. A, MSC-1 cells were transfected with FSHR(−220/+123)Luc and empty vector (EV) or plasmids expressing USF1, USF2, or ID2 in the absence of forskolin or with forskolin stimulation for 8 h. Relative luciferase activities are expressed relative to the activity resulting from the overexpression of USF1 + USF2 (=1). B, MSC-1 cells were cotransfected with the κBLUC reporter plasmid and either 2 μg of empty expression vector (EV) or ID2 expression vector. Relative luciferase activities were expressed relative to the activity resulting from the cotransfection of EV (=1). C, MSC-1 cells were transfected with FSHR(−220/+123)Luc and expression vectors for USF1 or USF2 (1 μg), empty vector and/or varying levels of ID2 as indicated. Appropriate amounts of empty vector were added to maintain equal total levels (3 μg) of expression vector for each transfection. Relative luciferase activities are expressed relative to that in which USF1 is overexpressed alone (=1). For A–C, the results represent the mean (±se) luciferase activity for four (A and B) and three (C) independent experiments using two replicates. Values with different lower-case letters differ significantly (P < 0.05).
Additional transient transfection studies were performed to assess the effects of ID2 on USF-mediated Fshr promoter activity in MSC-1 cells. Overexpression of USF1 or USF2 alone increased Fshr promoter activity 2.1- and 2.0-fold, respectively (Fig. 6C). Cotransfection of 0.5–2.0 μg of ID2 expression vector in the presence of constant levels of USF1 or USF2 expression vectors resulted in a dose-dependent decrease in USF-inducible promoter activity.
Discussion
FSH is capable of activating several intracellular pathways in Sertoli cells including the stimulation of adenylate cyclase and subsequent elevation of cAMP levels, causing the activation of protein kinase A and resulting downstream effects including the production of factors required by germ cells. We found that one result of FSH stimulation is a transient decrease in USF protein binding to an E-box motif in the Fshr gene promoter. This finding is based on EMSA studies that revealed a decrease in protein binding to the E-box of Fshr gene promoter after stimulation with FSH. Supershift assays determined that USF proteins were the predominant proteins present in the DNA-protein complexes. To our knowledge there has been only one other report of cAMP-mediated inhibition of USF protein binding to an E-box motif, and thus far the mechanism is not understood (28). Other cis-acting elements that have been identified in the regulation of Fshr expression include binding sites for activator protein-1, E2F, and GATA factors. In addition two binding sites for steroidogenic factor-1 (SF-1) have been identified (6,9,29,30). However, mutation of the E-box has the greatest impact on rat Fshr promoter activity (6). In the rat gene, the E-box (5′-CACGTG-3′) is located at position −120 relative to the translation start site and is conserved across species except for the human gene, which has a single G-to-A change at the first guanosine of the core sequence (5′-CACGTG-3′ to 5′-CACATG-3′).
FSH-mediated repression of Fshr mRNA has been shown in previous studies (4,5,26). Griswold et al. (26) showed that Fshr mRNA levels decreased by 50% after 5 h of FSH stimulation. Maguire et al. (4) showed that FSH stimulation of Sertoli cells decreased Fshr mRNA levels and that mRNA levels were minimal after 8 h of stimulation followed by recovery of Fshr mRNA expression by 24 and 48 h. The timing of the transient repression and recovery of Fshr mRNA expression that we and previous investigators observed closely follows the altered USF protein binding to the Fshr promoter E-box detected by EMSA analysis. Interestingly, the decrease in USF mRNA levels is delayed somewhat compared with the decrease in protein binding. This result suggests that Fshr mRNA levels do not decrease immediately after transcriptional activators are removed and is consistent with the relatively long (6 h) half-life of Fshr mRNA (31).
One mechanism for inhibiting USF binding to the E-box within the Fshr promoter would be decreasing the expression of the proteins that bind to the site. Our results indicate that of the two major proteins that bind to the Fshr promoter E-box (USF1 and USF2), one factor (USF1) has its mRNA expression transiently reduced at least 67% 6 h after FSH stimulation. The decrease in Usf1 mRNA as well as the observed decrease in protein expression directly corresponds with the FSH-mediated decrease in protein binding to the Fshr promoter E-box. Thus, the decrease in USF1 expression likely contributes to the transient decrease in protein binding to the Fshr promoter E-box observed after stimulation with FSH. However, because USF proteins bind as homo- and heterodimers and the relative activities of the USF1/USF2 dimer combinations and the levels of the two proteins in the Sertoli cells are not yet known, the significance of a dramatic decrease in one USF factor is not easily modeled. Nevertheless, if USF1 and USF2 levels are relatively similar and the activities of the dimers are not dramatically different in Sertoli cells, then a 67% decrease in USF1 expression would be expected to decrease binding to Fshr promoter E-box and Fshr mRNA expression.
A second mechanism to inhibit USF from interacting with the Fshr promoter E-box would be to induce the expression of ID2 repressor protein. ID proteins inhibit E-box-mediated gene expression by interacting with and blocking the DNA binding activity of basic HLH proteins (32,33). Previously we showed that Id2 mRNA expression was transiently induced for 2–12 h during FSH stimulation of Sertoli cells (16). In this study, the results of immunofluorescence analysis showed that ID2 protein expression is induced in Sertoli cell nuclei with similar kinetics.
The significance of increased expression of ID2 proteins was demonstrated in EMSA studies. These studies also provided the first indication of potential interactions between ID2 and USF proteins. The mixing of COS7 cell extracts containing steady-state levels of either USF1 or USF2 with increasing levels of extracts containing ID2 resulted in decreased USF binding to the Fshr E-box. Specifically, USF1 binding to the E-box within the Fshr promoter was inhibited using a 1:2 ratio of extracts expressing USF1 or ID2, but USF2 binding to the Fshr promoter was not inhibited until equal or greater ratios of ID2 were added. Comparisons of ID2 inhibition of USF1- or USF2-mediated Fshr promoter activity showed similar patterns. These results raise the possibility that ID2 inhibits USF1 and USF2 DNA binding with different efficiencies. Nevertheless, the finding that increasing levels of ID2 inhibit USF-Fshr E-box interactions suggests that FSH-mediated repression of Fshr promoter activity is established via ID2 blocking USF binding to the Fshr E-box. Additional studies using extracts from Sertoli cells transfected with either an empty expression vector or an ID2 expression vector confirmed that ID2 can block endogenous USF proteins from binding to the Fshr E-box in Sertoli cells.
The results of transient transfection studies using the FSHR(−220/+123)Luc reporter plasmid and the MSC-1 Sertoli cell line suggest that ID2 proteins repress Fshr expression via inhibiting USF-mediated Fshr promoter activity. We confirmed that USF proteins activate the Fshr promoter as cotransfection of USF1 and USF2 expression vectors increased Fshr promoter activity by 1.7-fold. In contrast, ID2 overexpression decreased Fshr promoter activity below basal levels, suggesting that the activity of endogenous USF proteins was blocked by the overexpressed ID2. ID2-mediated repression of the Fshr promoter was similar to that after 8 h of forskolin stimulation, which is consistent with the induction of Id2 mRNA in MSC-1 cells in response to forskolin that we reported previously (16). Cotransfection of the ID2 expression vector with either the USF1 or USF2 expression vectors also down-regulated Fshr promoter activity in a dose-dependent manner. Transfection of USF1 or USF2 alone resulted in the induction of the Fshr promoter by 2.1- and 2.0-fold, respectively. At the highest concentrations of ID2 expression vector, ID2 inhibited USF-activated Fshr promoter activity to basal levels. These results show that ID2 inhibits USF-mediated activation of the Fshr promoter.
Although statistically significant, the increase in Fshr promoter activity (1.7- to 2.1-fold) after overexpression of various combinations of USF proteins in MSC-1 cells is somewhat limited and may reflect endogenous USF levels that approach saturation. In this regard, Heckert et al. (10) were unable to elicit increased Fshr promoter activity in MSC-1 cells after overexpression of USF proteins except when using a construct having a mutated E-box with lower affinity for USF proteins. In our studies, heterodimer and homodimer combinations of USF1/USF2 proteins were found to be nearly equally effective in activating the Fshr promoter. These results provide biochemical support for the findings from USF knockout mice, demonstrating that only one of the USF proteins is sufficient for Fshr expression in Sertoli cells. In contrast, both USF1 and USF2 are required for Fshr expression in the ovary (34). Interestingly, other transient transfection studies using Chinese hamster ovary cells demonstrated that overexpression of USF1 or USF2 inhibited Fshr promoter activity via the E-box by 55–67%, respectively (35). Together, these findings suggest the possibility that USF proteins may regulate the Fshr expression differently in the ovary than in Sertoli cells.
Previously it was proposed that FSH-mediated repression of the Fshr promoter might be due to FSH-induced expression of the inducible cAMP early repressor (ICER) repressor protein, which can bind to a cAMP response element-like sequence (TGACTCA) located at position −561 to −568 of the rat Fshr promoter (4,36). However, the cAMP response element-like sequence is not conserved in the mouse and human promoters and thus is not a strong candidate for the negative element regulating Fshr expression. Furthermore, Griswold et al. (26) found no evidence for ICER down-regulating the Fshr promoter in Sertoli cells, and ICER was not able to down-regulate a CAT reporter gene driven by the Fshr promoter (36). Nevertheless, it cannot be ruled out that ICER may directly or indirectly interfere with the expression or activity of USF proteins.
It is possible that other ID proteins in addition to ID2 may be induced by FSH in Sertoli cells and contribute to the inhibition of Fshr mRNA expression. In the only other published study of ID protein regulation in Sertoli cells, Skinner and colleagues (37) reported that ID4 protein but not mRNA levels were elevated by FSH or cAMP stimulation of Sertoli cells for 72 h. The ID1 transcriptional repressor also has been shown to inhibit the activity of a Fshr promoter extending 317 bp upstream of the translation start site, but no link was made between ID1-mediated repression and USF protein binding to the E-box (9). However, ID1 was shown to block USF binding to the promoter of the α-glycoprotein hormone subunit gene and repress USF-mediated activation of the promoter (38).
Taken in total, the present findings and our earlier results (16) suggest the following model for the regulation of Fshr expression: FSH stimulation concurrently decreases USF1 expression and increases ID2 expression. As a result, USF binding to the Fshr promoter E-box is blocked, contributing to the transient decrease in endogenous Fshr RNA observed within 6–12 h of FSH stimulation (4,26). After 12–24 h, USF1 and ID2 expression returns to basal levels (16), allowing increased USF protein binding to the Fshr promoter, resulting in the activation of Fshr transcription and elevating the concentration of FSHR and ID2 levels once again. The use of two synergistic mechanisms to regulate the temporal expression of Fshr suggests that maintaining the correct levels of FSHR is highly important for the maintenance of male fertility. In fact, the feedback pattern of USF1- and ID2-mediated regulation in cultured Sertoli cells has physiological relevance in that it could explain the cyclical regulation of FSHR in testicular Sertoli cells during the cycle of the seminiferous epithelium. However, whereas the cycle of FSHR expression in cultured Sertoli cells would occur approximately every 24 to 48 h, the cycle of FSHR expression and cAMP production in vivo occurs over approximately a 13-d period in the rat (39). It is possible that the responses of cultured Sertoli cells in the presence and absence of FSH may be amplified compared with the situation in the testis in which FSH is always present. In vivo, changes in FSHR, USF1, and ID2 protein levels may not be as dramatic, and thus, the cyclical pattern may be prolonged. It is also possible that signals from germ cells may contribute to the regulation of USF1 and ID2. The consequences of cyclical FSHR expression in Sertoli cells in vivo are not yet well understood, although it has been established that FSH signaling is lowest when testosterone signaling is greatest (40,41). Therefore, it is possible that FSH signals must be decreased to permit some testosterone actions. Further studies are required to determine whether USF1 and ID2 levels are altered in a cyclical fashion over the 13-d cycle in vivo. In addition, further investigation of the more complex environment of the testis that includes other somatic cells as well as the developing germ cells is needed to identify potential factors and signals that may slow the cycling of FSHR expression.
Acknowledgments
We thank Leslie Heckert for the FSHR(−220/+123)Luc plasmid and Edward Prochownik for ID2 antiserum. We also thank Steven Reisenweber, Kelly Sopko, Corey Toocheck, Alyssa James, and John Shupe for excellent technical assistance.
Footnotes
This work was supported by National Institute of Child Health and Human Development/National Institutes of Health through cooperative U54 (HD008610) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
Disclosure Summary: The authors have nothing to disclose.
First Published Online May 7, 2009
Abbreviations: κBLUC, Luciferase reporter driven by a consensus κB-enhancer; CREB, cAMP response element-binding protein; Ct, cycle threshold; FSHR, FSH receptor; HLH, helix-loop-helix; IBMX, 3-isobutyl-1-methylxanthine; ICER, inducible cAMP early repressor; ID, inhibitor of DNA binding/differentiation; Luc, luciferase; qPCR, quantitative real-time PCR; USF, upstream stimulatory factor.
References
- Walker WH, Cheng J 2005 FSH and testosterone signaling in Sertoli cells. Reproduction 130:15–28 [DOI] [PubMed] [Google Scholar]
- Kangasniemi M, Kaipia A, Toppari J, Perheentupe A, Huhtaniemi I, Parvinen M 1990 Cellular regulation of follicle-stimulating hormone (FSH) binding in rat seminiferous tubules. J Androl 11:336–343 [PubMed] [Google Scholar]
- Steinberger A, Hintz M, Heindel JJ 1978 Changes in cAMP responses to FSH in isolated rat Sertoli cells during sexual maturation. Biol Reprod 19:566–572 [DOI] [PubMed] [Google Scholar]
- Maguire SM, Tribley WA, Griswold MD 1997 Follicle-stimulating hormone (FSH) regulates the expression of FSH receptor messenger ribonucleic acid in cultured Sertoli cells and in hypophysectomized rat testis. Biol Reprod 56:1106–1111 [DOI] [PubMed] [Google Scholar]
- Themmen AP, Blok LJ, Post M, Baarends WM, Hoogerbrugge JW, Parmentier M, Vassart G, Grootengoed JA 1991 Follitropin receptor down-regulation involves a cAMP-dependent post-transcriptional decrease of receptor mRNA expression. Mol Cell Endocrinol 78:R7–R13 [DOI] [PubMed] [Google Scholar]
- Heckert LL, Daggett MA, Chen J 1998 Multiple promoter elements contribute to activity of the follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Mol Endocrinol 12:1499–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M 1988 Multiple forms of the human gene-specific transcription factor USF. II. DNA binding properties and transcriptional activity of the purified HeLa USF. J Biol Chem 263:11994–12001 [PubMed] [Google Scholar]
- Sawadogo M, Van Dyke MW, Gregor PD, Roeder RG 1988 Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J Biol Chem 263:11985–11993 [PubMed] [Google Scholar]
- Goetz TL, Lloyd TL, Griswold MD 1996 Role of E box and initiator region in the expression of the rat follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. J Biol Chem 271:33317–33324 [DOI] [PubMed] [Google Scholar]
- Heckert LL, Sawadago M, Daggert MA, Chen JK 2000 The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol Endocrinol 14:1836–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Hornbaker K, Rice DA, Sawadogo M, Heckert LL 2008 In vivo regulation of FSH-receptor (Fshr) by the transcription factors USF1 and USF2 is cell specific. Endocrinology 149:5297–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H 1990 The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell 61:49–59 [DOI] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D 1991 Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol 11:5603–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D 1991 An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci USA 88:1815–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechman V, van Crüchten I, Sablitzky F 1994 The expression of Id4, a novel dominant negative helix-loop-helix protein is distinct from Id1, Id2 and Id3. Nucleic Acids Res 22:749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobey MJ, Fix CA, Walker WH 2004 The Id2 transcriptional repressor is induced by follicle-stimulating hormone and cAMP. J Biol Chem 279:16064–16070 [DOI] [PubMed] [Google Scholar]
- McGuinness MP, Linder CC, Morales CR, Heckert LL, Pikus J, Griswold MD 1994 Relationship of a mouse Sertoli cell line (MSC-1) to normal Sertoli cells. Biol Reprod 51:116–124 [DOI] [PubMed] [Google Scholar]
- Delfino FJ, Walker WH 1998 Stage-specific nuclear expression of NF-κB in mammalian testis. Mol Endocrinol 12:1696–1707 [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ 2004 Image processing with Image J. Biophoton Int 11:36–42 [Google Scholar]
- Shell SA, Fix C, Olejniczak D, Gram-Humphrey N, Walker WH 2002 Regulation of cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) expression by Sp1 in the mammalian testis. Biol Reprod 66:659–666 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N 1987 Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Innis MA 1990 PCR protocols: a guide to methods and applications. San Diego: Academic Press [Google Scholar]
- Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF 2006 High-throughput real-time quantitative reverse transcription PCR. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, eds. Current protocols in molecular biology. Chap 15, unit 15.8.1–15.8.28. New York: John Wiley, Sons, Inc. [DOI] [PubMed] [Google Scholar]
- Langlands K, Yin X, Anand G, Prochownik EV 1997 Differential interactions of Id proteins with basic helix-loop-helix transcription factors. J Biol Chem 272:19785–19793 [DOI] [PubMed] [Google Scholar]
- Meier JL, Luo X, Sawadogo M, Straus SE 1994 The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol Cell Biol 14:6896–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold MD, Kim JS, Tribley WA 2001 Mechanisms involved in the homologous down-regulation of transcription of the follicle-stimulating hormone receptor gene in Sertoli cells. Mol Cell Endocrinol 173:95–107 [DOI] [PubMed] [Google Scholar]
- Paranko J, Kallajoki M, Pelliniemi LJ, Lehto VP, Virtanen I 1986 Transient coexpression of cytokeratin and vimentin in differentiating rat Sertoli cells. Dev Biol 117:35–44 [DOI] [PubMed] [Google Scholar]
- Grassadonia A, Tinari N, Fiorentino B, Nakazato M, Chung HK, Giuliani C, Napolitano G, Iacobelli S, Howcroft TK, Singer DS, Kohn LD 2007 Upstream stimulatory factor regulates constitutive expression and hormonal suppression of the 90K (Mac-2BP) protein. Endocrinology 148:3507–3517 [DOI] [PubMed] [Google Scholar]
- Kim JS, Griswold MD 2001 E2F and GATA-1 are required for the Sertoli cell-specific promoter activity of the follicle-stimulating hormone receptor gene. J Androl 22:629–639 [PubMed] [Google Scholar]
- Levallet J, Koskimies P, Rahman N, Huhtaniemi I 2001 The promoter of murine follicle-stimulating hormone receptor: functional characterization and regulation by transcription factor steroidogenic factor 1. Mol Endocrinol 15:80–92 [DOI] [PubMed] [Google Scholar]
- Minegishi T, Tano M, Kishi H, Kameda T, Miyamoto K 1997 Follicle-stimulating hormone regulation on its receptor messenger ribonucleic acid levels in cultured rat granulosa cells. Biochim Biophys Acta 1359:165–173 [DOI] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM 2003 Id proteins in cell growth and tumorigenesis. Cancer Cell 3:525–530 [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C 2000 Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol 20:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Heckert LL 2006 Transcriptional regulation of the FSH receptor: new perspectives. Mol Cell Endocrinol 260–262:100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putowski LT, Schillings WJ, Lee CM, Reddy EP, Jakowicki JA 2004 Human follicle-stimulating hormone receptor (FSH-R) promoter/enhancer activity is inhibited by transcriptional factors, from the upstream stimulating factors family, via E-box and newly identified initiator element (Inr) in FSH-R non-expressing cells. Gynecol Endocrinol 19:9–17 [DOI] [PubMed] [Google Scholar]
- Monoco L, Foulkes NS, Sassone-Corsi P 1995 Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: involvement in long term desensitization of the FSH receptor. Proc Natl Acad Sci USA 92:10673–10677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary J, Johnson J, Kim G, Skinner MK 2001 Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology 142:1727–1736 [DOI] [PubMed] [Google Scholar]
- Jackson SM, Gutiierrez-Hartman A, Hoeffler JP 1995 Upstream stimulatory factor, a basic-helix-loop-helix-zipper protein, regulates the activity of the α-glycoproteins hormone subunit gene in pituitary cells. Mol Endocrinol 9:278–291 [DOI] [PubMed] [Google Scholar]
- Kangasniemi M, Kaipia A, Mali P, Toppari J, Huhtaniemi I, Parvinen M 1990 Modulation of basal and FSH-dependent cyclic AMP production in rat seminiferous tubules staged by an improved transillumination technique. Anat Rec 227:62–76 [DOI] [PubMed] [Google Scholar]
- Bremner WJ, Millar MR, Sharpe RM, Saunders PTK 1994 Immunohistochemical localization of androgen receptors in the rat testis: evidence for stage-dependent expression and regulation by androgens. Endocrinology 135:1227–1234 [DOI] [PubMed] [Google Scholar]
- Parvinen M, Marana R, Robertson DM, Hansson V, Ritzen EM 1980 Functional cycle of rat Sertoli cells: differential binding and action of follicle stimulating hormone at various stages of the spermatogenic cycle. In: Steinberger A, Steinberger E, eds. Testicular development, structure and function. New York: Raven Press; 425–432 [Google Scholar]
- Zhu Y, Casado M, Vaulont S, Sharma K 2005 Role of upstream stimulatory factors in regulation of renal transforming growth factor-β1. Diabetes 54:1976–1984 [DOI] [PubMed] [Google Scholar]