Abstract
Objective To evaluate the performance of a rapid test for chlamydia with first void male urine samples as a potential tool for diagnosis and screening of chlamydial infection in men.
Design Evaluation of test performance in prospective cohort study.
Settings A young people’s sexual health centre (site 1) and a genitourinary medicine clinic (site 2) in the United Kingdom.
Participants 1211 men aged 16-73 attending either of the two sites.
Main outcome measures Sensitivity, specificity, positive predictive value, and negative predictive value of the Chlamydia Rapid Test versus polymerase chain reaction assay. Relation between the visual signal of the Chlamydia Rapid Test and organism load.
Results Detection rates for Chlamydia trachomatis infection with polymerase chain reaction were 4.4% (20/454) at site 1 and 11.9% (90/757) at site 2. Compared with polymerase chain reaction assay, the resolved sensitivity, specificity, positive predictive value, and negative predictive value of the Chlamydia Rapid Test was 82.6% (90/109), 98.5% (1085/1102), 84.1% (90/107), and 98.3% (1085/1104), respectively. The organism load in first void urine samples that were positive for chlamydia ranged from 7.28×102 to 6.93×106 plasmids/ml and correlated significantly with the visual signal of the Chlamydia Rapid Test (r=0.7897, P<0.001).
Conclusions The performance of the new Chlamydia Rapid Test with first void male urine samples indicates that it would be an effective diagnostic tool for chlamydial infection in men. The availability of test results within an hour allows for immediate treatment and contact tracing, potentially reducing the risks of persistent infection and onward transmission. The test could also provide a simple and reliable alternative to nucleic acid amplification assays for testing of male urine in chlamydial screening programmes in high prevalence settings.
Introduction
Chlamydia trachomatis infection is the most common sexually transmitted bacterial infection in the United States and United Kingdom. In the US in 2006, the number of cases of genital C trachomatis infection reported to the Centers for Disease Control and Prevention exceeded one million for the first time.1 This number has increased annually for more than two decades, probably because of a combination of a true increase in prevalence, more widespread screening, the availability of more sensitive tests, and more complete national reporting. In the UK, the number of diagnoses of uncomplicated chlamydial infection in genitourinary medicine clinics has also climbed steadily since the mid-1990s. Between 2005 and 2006, it increased by 4% (from 109 418 to 113 585), with the number remaining stable in women but increasing by 8% in men.2
Chlamydial infection is commonly asymptomatic and remains underdiagnosed. Efforts aimed at promoting safer sexual behaviours and at detecting and treating infected individuals, especially young adult men, are therefore important to reduce transmission to sexual partners.3 Chlamydial infection is most common among sexually active men and women aged under 25.1 4 In 2006, the overall rate of reported chlamydial infection in the US among women was almost three times that among men, reflecting the larger number of women screened.1 With the growing availability of urine testing, however, men are increasingly being tested for chlamydial infection.1 Recent evidence hinting at a causative role for chlamydial infection in male subfertility might also encourage increased uptake of testing in men.5 6 More widespread and systematic male testing could substantially reduce the infective pool, hence preventing transmission to female partners and reducing serious complications such as pelvic inflammatory disease, chronic pelvic pain, infertility, and ectopic pregnancy.
Given that the collection of urethral swabs is uncomfortable for the patient and must be performed by a trained clinician, screening for chlamydial infection in asymptomatic men can be optimised with a test that performs adequately on urine samples or less painful meatal swabs. Furthermore, the availability of a high performance, rapid, and simple to use test would allow the implementation of a test and treat regimen, whereby patients are treated on site during the initial visit. The use of rapid tests can result in the treatment of more infected individuals than the use of nucleic acid amplification tests if the return rate is less than 65%.7 Nucleic acid amplification tests are commonly used for chlamydial diagnosis and are more sensitive than enzyme immunoassay based tests. Nucleic acid amplification tests, however, take hours or days (if re-testing is required) to complete and are costly. While results of laboratory based tests usually take many days to reach the clinics, currently available rapid tests for the detection of chlamydia in male urine lack the required sensitivity, even when used in a test and treat setting.8
We have evaluated the performance of a new Chlamydia Rapid Test, developed at the diagnostics development unit of the University of Cambridge. The assay is used in conjunction with a collection device for male urine (FirstBurst),9 10 specifically designed to collect the first voided urine that contains a higher organism load, and test results are available in less than an hour. This assay was developed to aid in the diagnosis of chlamydial infection and to provide a screening tool for the detection of such infection among young sexually active males.
Methods
Sites
The evaluation sites comprised a young people’s sexual health centre (Brook in Birmingham, site 1) and a genitourinary medicine clinic (Ambrose King Centre in London, site 2). Participants were recruited from March to November 2007.
Participants
All men attending the two sites were invited to join the study. They were eligible for enrolment if they were aged at least 16, had not taken antibiotics in the previous month, and were able to understand the written information forms for the study. Each participant received an information sheet about the study. Participants gave written informed consent and were then interviewed confidentially about their symptoms and relevant sexual history. Participants at site 2 also filled in a written questionnaire concerning sample collection methods and preferences after they had provided a sample.
Specimen collection
Each participant provided two urine samples, the first one collected using a FirstBurst urine collection device9 that collects the first 4-5 ml of urine. Following the manufacturers’ instructions (Roche and Gen-Probe), the second urine specimen was collected with a collection cup routinely used in sexually transmitted disease clinics in the UK at least two hours after the FirstBurst specimen was collected. Although not randomised, a previous study showed that the order of collection did not significantly affect the organism load in specimens collected by urine cup and tested with the polymerase chain reaction assay.9 Each participant was shown an illustrated instruction sheet detailing collection of urine with the FirstBurst device.
The second urine sample was divided into two portions, one for polymerase chain reaction testing by an independent laboratory and the other for freezing and storage in case we needed to test discordant samples with the transcription mediated amplification assay. All urine specimens were handled and stored according to the recommendations of the relevant test manufacturers.
An independent clinical laboratory evaluated the reproducibility of the Chlamydia Rapid Test in a study using the guideline of the National Committee on Clinical Laboratory Standards.11 Two operators tested in duplicate a panel consisting of 10 randomised masked samples over a period of five days according to the procedure for the Chlamydia Rapid Test.
Testing with the Chlamydia Rapid Test
Clinic staff tested urine samples on site, with all staff having passed tester’s requirements in accordance with the National Committee on Clinical Laboratory Standards.11 A 3 ml portion of the FirstBurst urine sample was diluted with 6 ml of deionised water in a centrifuge tube and then centrifuged at 3000 g for 20 minutes at room temperature. Occasionally we performed batch centrifugation of up to 16 specimens/run. The resulting supernatant was discarded and the pellet was subjected to extraction by the sequential addition of 400 µl of reagent 1, 300 µl of reagent 2, and 100 µl of reagent 3, with mixing between additions. We then added 100 µl of the extracted sample to a tube containing lyophilised amplification and detection reagents. The resulting mixture was gently agitated until it became a clear pink solution, after which we added the test strip to the solution and allowed it to stand for 25 minutes before reading the result. The test strip contains a monoclonal antibody to chlamydial lipopolysaccharide12 at the test line and includes a separate procedural control line. The appearance of a result line on the test strip indicated the presence of chlamydia, which was further graded with a semi-quantitative in-house grading card.
Testing with polymerase chain reaction and transcription mediated amplification
The performance of the Chlamydia Rapid Test was assessed to meet the requirements for Conformité Européenne licensure, which stipulate that the comparator test should be a “state of the art” assay and should be performed with specimens approved for the test. Polymerase chain reaction testing for chlamydia, which is licensed for urine specimens, was chosen as the ideal standard test for the study. The Roche Amplicor CT/NG polymerase chain reaction assay has undergone comprehensive evaluations with male urine samples and is similar in reliability to other nucleic acid amplification methods for detection of C trachomatis in urine.13
Urine specimens were sent to a laboratory accredited by the Clinical Pathology Accreditation (UK) for C trachomatis testing with the Amplicor CT/NG polymerase chain reaction assay according to manufacturer’s instructions (Roche Diagnostic Systems, Branchburg, NJ). Samples that yielded discordant results with the Chlamydia Rapid Test and the polymerase chain reaction assay were tested with the mono-specific Aptima CT kit, a transcription mediated amplification assay (Gen-Probe, San Diego, CA) according to manufacturer’s instructions at the Sexually Transmitted Bacteria Reference Laboratory (Health Protection Agency, Colindale). In addition, 100 randomly selected specimens negative with polymerase chain reaction and 20 concordant positive samples were masked and tested by this latter assay to minimise potential bias introduced by testing discordant samples only.
Quantification of organism load
Real time quantitative polymerase chain reaction analysis was performed as described previously14 but with modifications.15 A portion (500 µl) of the FirstBurst urine sample from men with positive results with polymerase chain reaction was centrifuged at 17 860 g for 15 minutes at 25°C and the resulting pellet processed as described previously.15 A 20 µl portion of each DNA extract was used for quantitative polymerase chain reaction analysis. Estimation of organism load was determined from a reference curve constructed with a calibrated plasmid pCTL12A preparation (provided by I Clarke).
Statistical analysis
Standard statistical methods were used to analyse data with SAS V9.1 software (SAS, Cary, NC). A P value of <0.05 was considered significant.
Results
Participants and sites
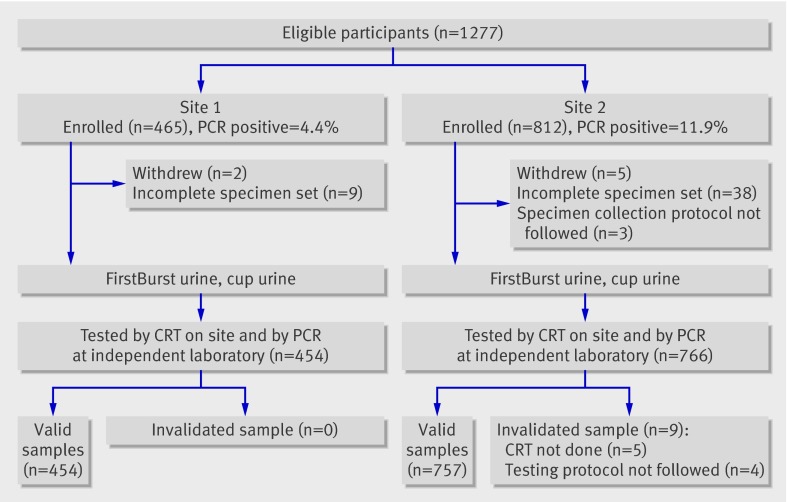
Of 1277 eligible participants at two clinical sites, 1211 contributed usable samples for this study. The sample size was based on an average prevalence of 10% and a sensitivity rate of 85%. With 120 positive results, this would mean less than 7% margin of error for our estimates.
Of the 66 excluded participants, 47 had incomplete specimen sets, seven withdrew, and the rest were excluded because of deviations from testing protocol. The mean age for the 1211 men who took part was 18.2 (range 16.0-25.0) at site 1 and 29.8 (16.5-73.4) at site 2 (P<0.001, unequal variance t test (Satterthwaite)). Figure 1 shows the recruitment and testing algorithm for the study participants at both clinical sites. Most participants at site 1 attended the sexual health centre for contraception and other reproductive health services and were asymptomatic. In contrast, 62% (467/749) of the participants at site 2 (genitourinary medicine clinic) presented with symptoms that included urethral discharge (155/741, 21%) and dysuria (169/744, 23%). In addition, 3% (20/757) of the participants at site 2 attended the clinics after being identified through contact tracing. Of these 20 patients, 30% (6/20) were positive for chlamydia with polymerase chain reaction and 25% (5/20) were positive with the rapid test.
Fig 1 Recruitment and testing algorithm for study participants. PCR=polymerase chain reaction, CRT=Chlamydia Rapid Test
Reproducibility of testing with the Chlamydia Rapid Test
There was a concordance of 100% between the expected results and the results generated from randomised and masked panels by two independent operators performing the Chlamydia Rapid Test.
Performance of the Chlamydia Rapid Test
The Chlamydia Rapid Test was performed on urine collected with the FirstBurst device, while the polymerase chain reaction was performed on urine collected with the standard cup. Positivity rates with polymerase chain reaction assay were 4% (20/454) at site 1 and 12% (90/757) at site 2. The combined unresolved sensitivity and specificity of the Chlamydia Rapid Test was 81.8% and 98.5%, respectively (table 1). After the Sexually Transmitted Bacteria Reference Laboratory tested discordant samples, the Chlamydia Rapid Test had a combined overall sensitivity and specificity of 82.6% and 98.5% (table 1). All 100 randomly selected concordant negative samples (100%, 95% confidence interval 96.38% to 100%) and 19/20 (75.13% to 99.87%) concordant positive samples were confirmed by the transcription mediated amplification assay (exact confidence interval calculated with the Monte Carlo exact method of estimation). When we took the results of the limited concordant testing into consideration, the imputed sensitivity, specificity, positive predictive value, and negative predictive value was 81.9% (range 54.0-82.6%), 98.1% (96.4-98.5%), 80.4% (63.6-84.1%), and 98.3% (94.8-98.3%), respectively. The ranges in brackets are the extreme scenarios, using both the lower and upper 95% confidence interval estimates for positive and negative concordance in our imputation.
Table 1.
Unresolved and resolved* performance with Chlamydia Rapid Test versus polymerase chain reaction. Figures are percentages (numbers) (binomial 95% exact confidence intervals)
| Site | Sensitivity (%) | Specificity (%) | Positive predictive value | Negative predictive value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Unresolved | Resolved | Unresolved | Resolved | Unresolved | Resolved | Unresolved | Resolved | ||||
| 1 (n=454) | 90.0 (18/20) (68.3 to 98.8) | 90.0 (18/20) (68.3 to 98.8) | 98.2 (426/434) (96.4 to 99.2) | 98.2 (426/434) (96.4 to 99.2) | 69.2 (18/26) (48.2 to 85.7) | 69.2 (18/26) (48.2 to 85.7) | 99.5 (426/428) (98.3 to 99.9) | 99.5 (426/428) (98.3 to 99.9) | |||
| 2 (n=757) | 80.0 (72/90) (70.2 to 87.7) | 80.9 (72/89) (71.2 to 88.5) | 98.7 (658/667) (97.4 to 99.4) | 98.7 (659/668) (97.5 to 99.4) | 88.9 (72/81) (80.0 to 94.8) | 88.9 (72/81) (80.0 to 94.8) | 97.3 (658/676) (95.8 to 98.4) | 97.5 (659/676) (96.0 to 98.5) | |||
| Total (n=1211) | 81.8 (90/110) (73.3 to 88.5) | 82.6 (90/109) (74.1 to 89.2) | 98.5 (1084/1101) (97.5 to 99.1) | 98.5 (1085/1102) (97.5 to 99.1) | 84.1 (90/107) (75.8 to 90.5) | 84.1 (90/107) (75.8 to 90.5) | 98.2 (1084/1104) (97.2 to 98.9) | 98.3 (1085/1104) (97.3 to 99.0) | |||
*All resolved values calculated from data on testing of discrepant samples.
The Chlamydia Rapid Test had overall resolved positive and negative predictive values of 84.1% and 98.3% (table 1). There were significant differences in the negative (P=0.009) and positive (P=0.028) predictive values of the Chlamydia Rapid Test between the two sites, although the 95% confidence intervals overlapped slightly for the positive predictive values. Furthermore, the likelihood ratio for a positive result from the Chlamydia Rapid Test suggests that for the low prevalence site (site 1), a positive result modifies the odds of chlamydia by 50:1, while in the higher prevalence site (site 2), the likelihood modifies it by 61:5 (table 2). In contrast, a negative result from the test modifies the original odds of chlamydia in site 1 by 0.102 to 1, and in site 2, by 0.185 to 1. Taking into account the results from limited number of concordant positive and negative samples tested, the adjusted likelihood ratios for a positive and a negative result for the combined sites were 43.1 (range 15.0-55.1) and 0.184 (0.177-0.477), respectively.
Table 2.
Positive and negative likelihood ratio for test results with Chlamydia Rapid Test
| Site | Positive | Negative | |||
|---|---|---|---|---|---|
| Unresolved | Resolved | Unresolved | Resolved | ||
| 1 (n=454) | 50.0 | 50.0 | 0.102 | 0.102 | |
| 2 (n=757) | 61.5 | 62.2 | 0.202 | 0.193 | |
| Total (n=1211) | 54.5 | 55.1 | 0.185 | 0.177 | |
Chlamydia Rapid Test in asymptomatic patients
At the time of recruitment to the study, 90% (18/20) of men positive for chlamydia with polymerase chain reaction from site 1 and 31% (28/90) from site 2 had no genitourinary symptoms. Of these asymptomatic men, 89% (16/18) and 71% (20/28), respectively, had positive results with the Chlamydia Rapid Test, giving an overall test sensitivity for asymptomatic men of 78% (36/46). With symptomatic men who were positive for chlamydia, there was a higher combined sensitivity (84%, 54/64) at both sites.
Organism load in men positive with polymerase chain reaction
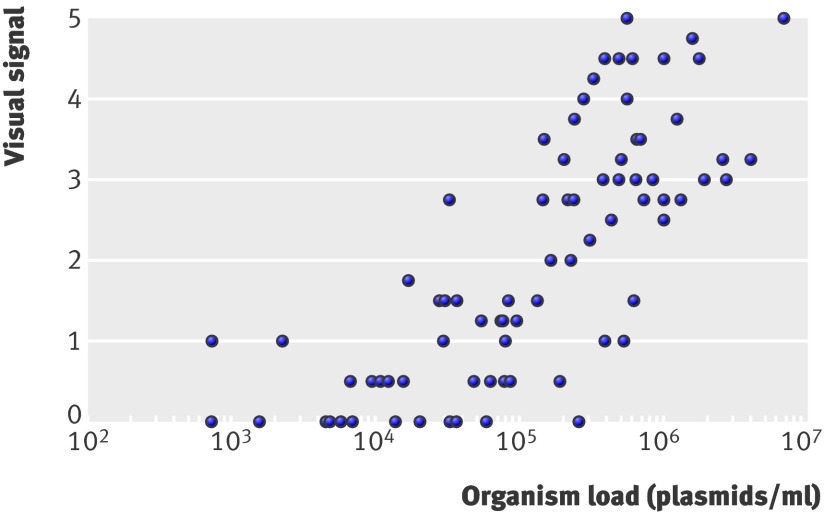
After the Chlamydia Rapid Test there was enough urine left from the FirstBurst sample to determine organism load in 80/90 men positive with polymerase chain reaction at site 2. We analysed the DNA extracted from these urine specimens with quantitative polymerase chain reaction assay with a primer set that amplifies a highly conserved sequence of the 7.5-kb cryptic plasmid of C trachomatis. The organism load for these samples ranged from 7.28×102 to 6.93×106 plasmids/ml of urine. The visual signal of the Chlamydia Rapid Test was significantly correlated (r=0.7897, P<0.001) with C trachomatis load (fig 2).
Fig 2 Correlation between C trachomatis organism load determined with quantitative polymerase chain reaction analysis and visual signal of Chlamydia Rapid Test for 80 urine specimens positive with polymerase chain reaction (r=0.7897, P<0.001)
Acceptability of FirstBurst device and waiting time for result
After specimens had been collected from the men in site 2, each participant was offered a written questionnaire. The response rate was 95% (767/812), although some of the returned questionnaires were not filled in completely, with the result that the total number of answers for each question varied slightly. Some 98% (741/759) of respondents found the instructions easy to understand, and 97% (735/755) found collection of their urine easy. With regard to preferences, 89% (619/697) of respondents preferred to give a urine sample, 7% (49/697) would have preferred to give a urethral swab, and 4% (29/697) were willing to provide either sample. Participants were not asked to give a urethral swab so for those men who had not experienced urethral swabbing their expressed preference might not be valid. As to preference for a urine collector, 76% (525/687) of respondents preferred the FirstBurst device, 18% (124/687) preferred the urine cup, and the remaining 6% (38/687) were willing to use either device. With regard to waiting time for the test result, 96% (653/683) of respondents indicated that they were willing to wait an hour or more and 4% (30/683) indicated that they would not wait more than an hour.
Discussion
Clinical performance of the new Chlamydia Rapid Test
The Chlamydia Rapid Test is suitable for diagnosis of chlamydial infection in men because of its high sensitivity and specificity. This new test provides same day results, allowing for immediate treatment of infected patients. The Chlamydia Rapid Test evaluated here for use with male urine is a modified version of that previously evaluated for use with vaginal swabs, wherein a similar performance (83.5% resolved sensitivity and 98.9% specificity) was observed.16 Its application to rapid chlamydia testing might enhance efforts to target asymptomatic men and facilitate both the treatment and contact tracing of infected individuals.
The Chlamydia Rapid Test examined was developed for the detection of C trachomatis infection in men with the use of first void urine as sample. C trachomatis infects the urethral mucosa in men, and this tissue is usually sampled either with urethral swabs or first void urine. The participants in our study preferred urine samples because of the discomfort associated with swab collection. We recently showed that the chlamydial load in first void urine does not differ significantly from that of urethral swabs.15 To facilitate the reliable collection of the first void urine samples, we used a new urine collection device (FirstBurst), which is provided with the Chlamydia Rapid Test kit. The urine sample collected with this device contains a load of C trachomatis up to six times that in specimens collected with a regular urine cup, thereby increasing the sensitivity of the test.9 The order of different collection methods did not significantly affect the organism load of the specimens of urine collected in cups tested with polymerase chain reaction.9
The evaluated Chlamydia Rapid Test incorporates a novel signal amplification system that has proved effective in enhancing the performance of this test.12 16 The level of sensitivity achieved by the Chlamydia Rapid Test in this study is unprecedented for an immunoassay based rapid test, as previously developed rapid tests for Chlamydia have shown poor sensitivity with cup collected male urine when compared with nucleic acid amplification tests.8 For example, using Ligase chain reaction as a comparison test, Clearview Chlamydia (Unipath, now Inverness) had a sensitivity and specificity of 67.7% and 95.5%, respectively, Testpack Chlamydia (Abbott) had 70.9% and 95.5%, and Surecell Chlamydia (Kodak) had 62.9% and 100%.8 The difference in performance with the previous rapid tests might in part be related to the FirstBurst sample collection.
The Chlamydia Rapid Test showed an overall unresolved sensitivity of 81.8% and a positive predictive value of 84.1%. At the young people’s sexual health centre (site 1), where the prevalence of chlamydial infection was only 4.4%, the positive predictive value of the Chlamydia Rapid Test was significantly lower (69.2 v 88.9%) than that at the genitourinary medicine clinic (site 2), where the prevalence of infection was 11.9%, although there was a slight overlap in the 95% confidence intervals for the two sites. This finding is not unexpected because of the different prevalence rates at the respective study sites. The Chlamydia Rapid Test also showed an overall high positive likelihood ratio (54.5:1), suggesting that a positive result multiplies the individual’s original likelihood of chlamydia by 54.5:1.
Efforts to control chlamydial infection among young sexually active people are unlikely to succeed without an effective testing programme for men. Given that men do not get tested for chlamydia as often as women do, as well as the inevitable delay in obtaining results with currently available testing methods, men with asymptomatic chlamydial infection continue to provide a transmission link for the disease that is missed by current testing programmes. Prevention of the spread of disease will thus require a high performance rapid test for chlamydia that allows for a test and treat regimen, in addition to the existing nucleic acid amplification tests. Emerging concerns regarding the role of chlamydia in male subfertility might encourage more asymptomatic men to come forward for screening.5 6
The performance of the Chlamydia Rapid Test is clearly related to the organism load within the specimen. Higher loads have been associated with increased chlamydia positivity in tested sexual partners, suggesting that those with higher loads are more infectious.17 This was confirmed in our study, where we observed a higher positivity rate (30% with polymerase chain reaction v 25% with the Chlamydia Rapid Test) in men who attended because of contact tracing. Men who tested positive with the Chlamydia Rapid Test are thus likely to be the most infectious, and rapid removal of these infected men from the reservoir of infection with a test and treat regimen might have an advantage over traditional screening approaches. This hypothesis warrants further investigation in appropriately designed trials.
Reliance on nucleic acid testing alone can be problematic, depending on the target used in the assay. This is evidenced by the recent demonstration of the presence of the Swedish C trachomatis variant with partial deletion of plasmid in many European countries18 as well as another distinct variant strain that could not be detected by plasmid based nucleic acid amplification tests.19 These strains would be detectable by the Chlamydia Rapid Test because the test recognises the genus specific Chlamydia lipopolysaccharide.
In developing countries, the availability of the Chlamydia Rapid Test would also allow more men to be screened and treated. Specific diagnostic tests for chlamydia are rarely available in these regions and management is often based on self reported symptoms and clinical signs, thus missing asymptomatic men altogether. Given the high prevalence of chlamydial infection among sex workers in these settings,20 male customers are likely to transmit infection to their other sexual partners and should therefore be among the primary targets for chlamydia testing.
Strengths and weaknesses of the study
This study included two centres with relatively large population sizes at sites with low and high prevalence. We used confirmatory testing on discordant samples to resolve true chlamydial positivity and additional random testing to minimise the bias from “selective” analysis of discordant samples. In addition, there was high reproducibility of testing between operators, strengthening the validity of the laboratory methods. The Chlamydia Rapid Test specimens were collected from men who had not had to hold their urine for two hours, thus providing applicability in a real world setting without the time restrictions imposed by traditional testing algorithms.
The number of men positive for chlamydia at site 1 was low because of the low prevalence nature of this site. The 95% confidence intervals for the sensitivity of the Chlamydia Rapid Test were larger at site 1 than at site 2. Also, in the determination of chlamydia load, the reference curve was based on purified plasmid preparations in buffer, therefore we could not control for inhibitory substances potentially present in some urine specimens.
Future research
Future studies should focus on determining whether the widespread use of the Chlamydia Rapid Test in screening young men for chlamydial infection in high prevalence settings can help prevent sequelae among their female sexual partners. In addition, more studies are warranted to verify whether the Chlamydia Rapid Test would reassure young men that testing can be quick, simple, and non-invasive, and this might increase uptake. The rapid test might also be used to confirm the chlamydia status in men with symptomatic non-gonococcal urethritis at the point of care, thus allowing more efficient contact tracing and focusing the health promotion resources on these individuals at the time of testing. Any potential improvement of the outcome over current approaches with nucleic acid amplified based screening should be evaluated in relation to alternative strategies. Finally, to further reduce the testing time of the rapid test, it might be worth exploring the use of urethral swab, especially in genitourinary medicine clinics that routinely collect urethral smears. This advantage, however, should be balanced with the fear in men of urethral procedures, which abounds in genitourinary medicine clinics, not only among those who have attended previously.21
Conclusions
The new Chlamydia Rapid Test for urine samples from men showed high sensitivity and specificity that can produce results within an hour. It is therefore suitable as a primary diagnostic tool, especially in settings where patients need to be tested and treated in one visit. It could also be used as a screening tool in areas with a high prevalence of infection or in settings where access to nucleic acid amplification tests is limited or where the return rate is low. Further studies are required to determine the utility and cost effectiveness of the Chlamydia Rapid Test in such roles.
What is already known on this topic
A point of care Chlamydia Rapid Test for women can achieve a high level of sensitivity and specificity with specimens from non-invasive vaginal swabs
What this study adds
The same new point of care Chlamydia Rapid Test can achieve a high level of sensitivity and specificity in men by using a collection device for male urine (FirstBurst), specifically designed to collect the first voided volume, which contains a higher organism load
The test can be an effective diagnostic tool for chlamydial infection in men, allowing immediate treatment and contact tracing
It is also potentially useful as a simple and reliable alternative to nucleic acid amplification assays for testing of male urine in chlamydial screening programmes in high prevalence settings
We thank I Clarke (University of Southampton) for providing the plasmid pCTL12A used as a standard in the quantitative analysis; S Padmore and H Nuamah for performing on-site chlamydia testing at Brook in Birmingham; V Shah, K Gujadhur, and G Wall for performing on-site chlamydia testing at Ambrose King Centre; J Wawrzyniak (University of Cambridge) for data entry and analysis; J-P Allain (University of Cambridge) and J White (Guy’s and St Thomas’ Hospital, London) for critical review of the manuscript; and the participants and clinic staff at the two study sites.
Contributors: HHL was the chief investigator and is the guarantor. E-CN wrote the first draft of the manuscript and led in the manufacture of the Chlamydia Rapid Test kits used in the study. BTG, IU-L, and PB were the principal investigators at the clinical sites. J-PM, LM-T, VL, and HHL developed the clinical plan and acted as clinical monitors. E-CN and C-EM developed the Chlamydia Rapid Test for men. J-PM and E-CN were responsible for data entry and analysis. AS was the clinician involved in the study at site 1. SA and CI tested and analysed discordant samples. All authors participated in drafting and revising the manuscript, and all approved the final version.
Funding: The study was funded by a Wellcome Trust grant to the University of Cambridge and additional support from the NIHR Cambridge Biomedical Research Centre. The funding sources had no role in study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Competing interests: J-PM, C-EM, and HHL are equity holders in Diagnostics for the Real World, which markets the rapid test technologies developed at the University of Cambridge. Both the University of Cambridge and the Wellcome Trust are also equity holders of the company.
Ethical approval: The study was approved by Moorfields and Whittington research ethics committee (05/Q0504/53) and Brook in Birmingham research ethics committee and informed consent was given by all patients.
Cite this as: BMJ 2009;339:b2655
References
- 1.Centers for Disease Control and Prevention. Sexually transmitted disease surveillance, 2006. Atlanta, GA: US Department of Health and Human Services, November 2007.
- 2.Health Protection Agency. Chlamydia statistics. www.hpa.org.uk/infections/topics_az/hiv_and_sti/Stats/STIs/chlamydia/statistics.htm.
- 3.Ku L, St Louis M, Farshy C, Aral S, Turner CF, Lindberg LD, et al. Risk behaviours, medical care, and chlamydial infection among young men in the United States. Am J Public Health 2002;92:1140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macleod J, Salisbury C, Low N, McCarthy A, Sterne JA, Holloway A, et al. Coverage and uptake of systematic postal screening for genital Chlamydia trachomatis and prevalence of infection in the United Kingdom general population: cross sectional study. BMJ 2005;330:940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Idahl A, Boman J, Kumlin U, Olofsson JI. Demonstration of Chlamydia trachomatis IgG antibodies in the male partner of the infertile couple is correlated with a reduced likelihood of achieving pregnancy. Hum Reprod 2004;19:1121-6. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez J, Ramos B, Santiso R, Agarwal A, Gonsalvez J, Gallegos G. Frequency of sperm cells with fragmented DNA in males infected with Chlamydia trachomatis and Mycoplasma sp, determined with the sperm chromatin dispersion (SCD) test. Fertil Steril 2007;88:S5. [Google Scholar]
- 7.Gift TL, Pate S, Hook EW III, Kassler WJ. The rapid test paradox: when fewer cases detected lead to more cases treated: a decision analysis of tests for Chlamydia trachomatis. Sex Transm Dis 1999;26:232-40. [DOI] [PubMed] [Google Scholar]
- 8.Chernesky M, Jang D, Krepel J, Sellors J, Mahony J. Impact of reference standard sensitivity on accuracy of rapid antigen detection assays and a leukocyte esterase dipstick for diagnosis of Chlamydia trachomatis infection in first-void urine specimens from men. J Clin Microbiol 1999;37:2777-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisniewski C, White J, Michel CE, Mahilum-Tapay L, Magbanua JP, Nadala ECB, et al. Optimal collection of first-void urine for detection of Chlamydia trachomatis infection in men. J Clin Microbiol 2008;46:1466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee HH, Smetham GTL, Stankus PJ, Hazelwood SC, Nadala ECB, Wisniewski CA. Liquid sampler and method. European patent 2003771170, November 2007.
- 11.National Committee on Clinical Laboratory Standards. User protocol for evaluation of qualitative test performance: approved guideline. No EP12-A 2002;22(14).
- 12.Michel CE, Solomon AW, Magbanua JPV, Massae P, Huang L, Mosha J, et al. Field evaluation of a rapid point-of-care assay for targeting antibiotic treatment for trachoma control: a comparative study. Lancet 2006;367:1585-90. [DOI] [PubMed] [Google Scholar]
- 13.Verkooyen AP, Noordhoek GT, Klapper PE, Reid J, Schirm J, Cleator GM, et al. Reliability of nucleic acid amplification methods for detection of Chlamydia trachomatis in urine: results of the first international collaborative quality control study among 96 laboratories. J Clin Microbiol 2003;41:3013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickett MA, Everson S, Pead PJ, Clarke IN. The plasmids of Chlamydia trachomatis and Chlamydophila (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 2005;151:893-903. [DOI] [PubMed] [Google Scholar]
- 15.Michel CE, Sonnex C, Carne CA, White JA, Magbanua JPVM, Nadala ECB, et al. Chlamydia trachomatis load at matched anatomic sites: implications for screening strategies. J Clin Microbiol 2007;45:1395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahilum-Tapay L, Laitila V, Wawrzyniak J, Alexander S, Swain A, Barber P, et al. New point of care Chlamydia Rapid Test—bridging the gap between diagnosis and treatment: performance evaluation study. BMJ 2007;335:1190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogers SM, Miller WC, Turner CF, Ellen J, Zenilman J, Rothman R, et al. Concordance of Chlamydia trachomatis infections within sexual partnerships. Sex Transm Infect 2008;84:23-8. [DOI] [PubMed] [Google Scholar]
- 18.Ripa T, Nilsson P. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill 2006;11:E061109 2. www.eurosurveillance.org/ew/2006/061109.asp#2. [DOI] [PubMed] [Google Scholar]
- 19.Magbanua JP, Goh BT, Michel CE, Aguirre-Andreasen A, Alexander S, Ushiro-Lumb I, et al. Chlamydia trachomatis variant not detected by plasmid based nucleic acid amplification tests: molecular characterisation and failure of single dose azithromycin. Sex Transm Infect 2007;83:339-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saison F, Mahilum-Tapay L, Michel CE, Buttress N, Nadala ECB, Magbanua JP, et al. Prevalence of Chlamydia trachomatis infection and performance of ChlamydiaRapid Tests among low- and high-risk Filipino women in resource-limited settings. J Clin Microbiol 2007;45:4011-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradbeer C, Soni S, Ekbote A, Martin T. You’re not going to give me the umbrella, are you? BMJ 2006;333:1287-88. [DOI] [PMC free article] [PubMed] [Google Scholar]