Abstract
Macromolecular crowding has long been known to significantly affect protein oligomerization, and yet no direct quantitative measurements appear to have been made of its effects on the binding free energy of the elemental step of adding a single subunit. Here, we report the effects of two crowding agents on the binding free energy of two subunits in the Escherichia coli polymerase III holoenzyme. The crowding agents are found, paradoxically, to have only a modest stabilizing effect, of the order of 1 kcal/mol, on the binding of the two subunits. Systematic variations in the level of stabilization with crowder size are nevertheless observed. The data are consistent with theoretical predictions based on atomistic modeling of excluded-volume interactions with crowders. We reconcile the apparent paradox presented by our data by noting that the modest effects of crowding on elemental binding steps are cumulative, and thus lead to substantial stabilization of higher oligomers. Correspondingly, the effects of small variations in the level of crowding during the lifetime of a cell may be magnified, suggesting that crowding may play a role in increased susceptibility to protein aggregation-related diseases with aging.
Introduction
The dramatic effects of macromolecular crowding on protein oligomerization have been documented in a number of studies. Among the first to realize the importance of macromolecular crowding was Arthur Kornberg. After many attempts, his lab finally succeeded in replicating the oriC plasmid in a cell-free state, upon including a high concentration of polyethylene glycol (PEG) in the incubation mixture (1). As Kornberg (2) lucidly explains, “the PEG gel occupies most of the aqueous volume and excludes a small volume into which large molecules are crowded. This concentration is essential when several proteins are needed in the consecutive steps of a pathway.” Reflecting on his work on DNA replication, Kornberg made “Thou shalt correct for extract dilution with molecular crowding” one of his 10 commandments. More recently, it was observed that under dilute conditions, α-synuclein, a presynaptic protein implicated in Parkinson's disease, only aggregates into fibrils after a lag time of months. However, in the presence of crowding agents like PEG, dextran, or Ficoll, the lag time can be shortened to days (3,4). This observation suggests that an increased level of crowding, e.g., by a reduction in cell volume or a slowdown in protein degradation, both of which are expected to occur with aging, promotes susceptibility to Parkinson's disease. It has also been shown that when fetal hemoglobin is used as a therapy for sickle cell anemia, which is caused by polymerization of sickle hemoglobin, macromolecular crowding, arising from the high hemoglobin concentrations in red cells (totaling ∼300 g/l), has a significant impact on the effectiveness of the therapy (5). Fetal hemoglobin lessens the extent of polymerization, because it does not enter the polymer, but its crowding effect serves to shorten the lag time for sickle hemoglobin polymerization and consequently reduce the chance that red cells escape the microvasculature.
Despite these significant biological consequences, it seems that direct quantitative measurements of the effects of macromolecular crowding on the free energies of elemental (i.e., binary) binding steps have not been made. Such data are crucial for a quantitative understanding on how macromolecular crowding affects the formation of higher oligomers such as the replication complex, α-synuclein fibril, and sickle hemoglobin polymer. Here, we report results on the effects of two crowding agents, dextran and Ficoll, on the binding free energy of two proteins, namely, the ɛ- and θ -subunits (Fig. 1) of the Escherichia coli DNA polymerase III holoenzyme (Pol III).
Figure 1.
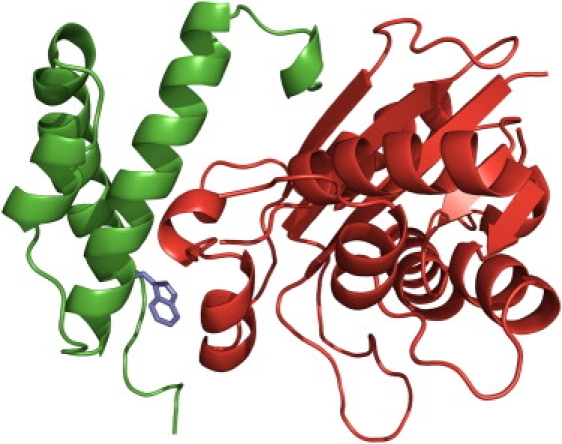
Complex formed by the θ- and ɛ-subunits, shown by the chains on the left (green) and right (red), respectively. A tryptophan residue introduced at position 27 of the θ -subunit, located within the binding interface, is shown in stick format.
We find that the crowding agents only moderately lower the binding free energy of the ɛ- and θ -subunits. The stabilization is of the order of 1 kcal/mol, corresponding to a fivefold increase in binding constant when the crowding agents are present at a concentration of 100 g/l. This modest stabilization might appear at first sight to be paradoxical in view of the above-described dramatic effects of macromolecular crowding on protein oligomerization. However, there is evidence that even such a modest level of increase in binding stability can have direct biological consequences. For example, a double mutation on a transcription factor was found to reduce its binding constant for DNA by a mere sixfold and yet resulted in distinct developmental defects (6). Zimmerman and Harrison (7) studied the 3′ → 5′ exonuclease activity of E. coli polymerase I in the absence and presence of PEG or dextran, and suggested that the <10-fold decrease in KM for nicked DNA by the crowding agents, attributed to enhanced binding between the enzyme and its DNA substrate, plays a buffering role against the inhibitory effect of an increase in intracellular ionic strength. Of more importance, we recognize that the effects of macromolecular crowding on the formation of higher oligomers are cumulative (8). That is, for energetic purposes, it may be assumed that a higher oligomer is formed by a series of binary binding steps (i.e., by adding one subunit at a time), and crowding would lower the binding free energy of each step by ∼1 kcal/mol. Then, the net stabilization of the higher oligomer can be substantial, providing a rationalization for the above dramatic effects of macromolecular crowding on protein oligomerization.
The stabilization effect of macromolecular crowding can be largely attributed to excluded-volume interactions between the binding molecules and the crowding agent (9). The total effective volume of the binding molecules is reduced upon forming a specific complex, thereby reducing the excluded-volume interactions with the crowding agent (Fig. 2). Our atomistic modeling of excluded-volume interactions for the ɛ-θ complex results in a level of stabilization that is consistent with the experimental results.
Figure 2.
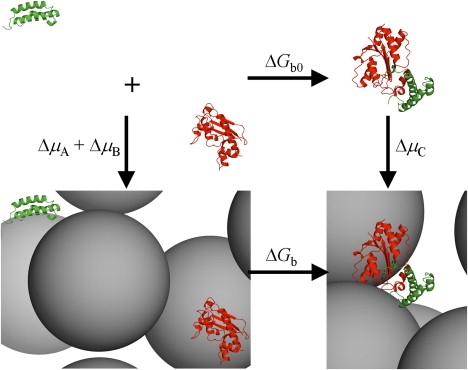
Thermodynamic cycle for calculating the effect of macromolecular crowding on binding free energy. The difference in binding free energy, ΔΔGb = ΔGb − ΔGb0, is the same as the difference in crowding-induced changes in chemical potential, ΔμC − ΔμA − ΔμB, between the bound and unbound states. The two proteins represented by red and green ribbons are ɛ186, and θ, respectively.
Materials and Methods
Expression and purification of the ɛ- and θ-subunits
Plasmids for the ɛ186 and θ -proteins (containing 186 and 76 amino acids, respectively) were gifts from Dr. Nick Dixon of Australian National University (Canberra, New South Wales, Australia). The W51Y and F27W mutations on θ were introduced using a Quick Change kit (Stratagene, La Jolla, CA). Expression and purification of the two proteins followed previously published protocols (10).
Measurement of binding constant
Binding was monitored on a Cary Eclipse fluorometer when ɛ186 was titrated into θ. Initially, 200 μl of θ at a concentration of 10 μM was placed in a cuvette with a 4-mm path length. Increasing amounts (up to 200 μl) of ɛ186 at a stock concentration of 200 μM were sequentially added to the cuvette. After each addition, we allowed 5 min for equilibration, after which a fluorescence spectrum in the wavelength range 300–400 nm was recorded (excitation wavelength at 294 nm, with bandpass filter set to 10 nm and 5 nm for emission). The proteins were dissolved either in a buffer (50 mM Tris, pH 7.6, with 100 mM NaCl and 1 mM dithiothreitol) or in a crowded medium created by adding 100 g/l of dextran of molecular mass ranging from 6 to 150 kD or Ficoll70 (purchased from Sigma-Aldrich, St. Louis, MO) to the buffer. To subtract the fluorescence of ɛ186, ɛ186 was titrated, according to the same schedule as for the first cuvette, into a second cuvette containing either buffer or crowded medium (i.e., the same content as the first cuvette but without the θ-protein). Both cuvettes were thermostated at 15°C.
The difference spectrum between the two cuvettes was taken as the total fluorescence intensity, F, of the θ-protein in the unbound and bound states. At 353 nm, the change in F by the titration of ɛ186 was maximal. This intensity, F353, as a function of the total concentration, x, of ɛ186 in the cuvettes was fitted to
| (1) |
where the two terms in parentheses are the contributions by unbound and bound θ, respectively, and Cu and Cb are their concentrations. Note that both the unbound and bound contributions have a quadratic term (which turned out to be small). The titration of ɛ186 also diluted θ in the cuvette. At a concentration of x μM for ɛ186, the dilution factor was 1 − x/200 (note that 200 μM was the stock concentration of ɛ186), and the total concentration, Ct, of θ was 10(1 − x/200) μM (the initial concentration of θ was 10 μM). If the binding constant is Ka, then the concentrations of unbound and bound θ are
| (2) |
| (3) |
The fitting, with Fu1, Fu2, Fb1, Fb2, and Ka as floating parameters, was done in gnuplot.
Homology modeling, molecular dynamics simulations, and modeling of crowding
The structure of the ɛ186-θ complex was built by homology modeling using the ɛ186-HOT complex (PDB entry 2ido) (11) as template. The sequence alignment between HOT (chain B in 2ido) and θ was

The two positions, F27 and W51, on θ that were further mutated in this study are indicated in bold in the above sequence alignment. The homology-based structure for the ɛ186-θ complex is shown in Fig. 1.
Molecular dynamics simulations of the ɛ186-θ complex in an octahedral water box (containing 13,729 water molecules) were carried out using the Amber99 force field (12). From a 16-ns trajectory at 298 K, 700 conformations were selected, at intervals of 20 ps, from the last 14 ns to do calculations on crowding. Those calculations, following an algorithm developed previously (13), yield the increase in chemical potential due to volume exclusion by crowders.
Results and Discussion
Modeled structure of the ɛ-θ complex
The ɛ-θ complex is part of the catalytic core of Pol III (a third subunit, α, contains the polymerase active site). The θ-binding site is located in the N-terminal domain (referred to as ɛ186) of the ɛ-subunit, which also contains the 3′-5′ proofreading exonuclease activity (10,14,15). The binding of θ has been found to increase the resistance of ɛ186 to thermal and chemical denaturation (10,16). Using as template the structure for the complex of ɛ186 with HOT (11), a protein sharing 55% sequence identity with θ, we built the structure for the ɛ186-θ complex (Fig. 1).
Measurement of ɛ-θ-binding constant
We used tryptophan fluorescence to monitor the ɛ186-θ-binding. ɛ186 does not contain any tryptophan residue, whereas θ contains a single tryptophan, at position 51, which is part of the hydrophobic core and somewhat away from the binding interface with ɛ186. We therefore mutated W51 into tyrosine and replaced another residue within the interface, F27, with tryptophan (Fig. 1) to use tryptophan fluorescence as a probe for binding.
In Fig. 3, we display the change in fluorescence intensity as ɛ186 is titrated into θ in buffer. By fitting the fluorescence intensity to a 1:1 binding model, we found the binding constant (Ka0) for the ɛ186 and the W51Y/F27W double mutant to be 1.4 ± 0.3 μM−1 in buffer. Correspondingly, the binding free energy ΔGb0, in units of kBT (where kB is Boltzmann's constant and T is absolute temperature), is –lnKa0 = −14 ± 0.2.
Figure 3.
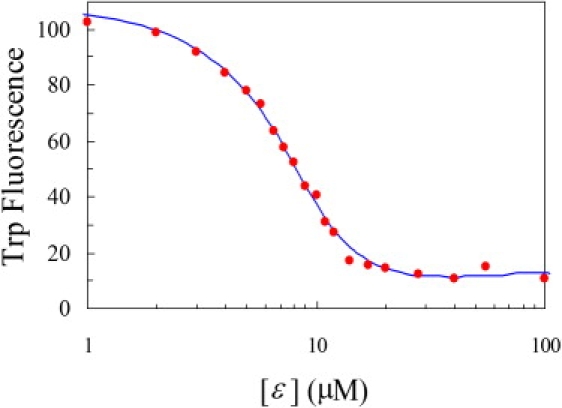
Change in fluorescence intensity when ɛ186 is titrated into θ in buffer. The symbols display raw data, and the curve shows the fit to Eq. 1.
Effects of crowding agents
Binding constants were measured in the presence of dextran of varied molecular mass or Ficoll70, each at a concentration of 100 g/l. The values were 7.5 ± 2.6, 3.2 ± 0.9, 2.0 ± 0.7, 2.9 ± 1.0, and 1.9 ± 0.5 μM−1 for dextran with molecular mass of 6, 40, 70, 100, and 150 kD, respectively, and 2.5 ± 0.7 μM−1 for Ficoll70. The increase in binding constants shows that the crowding agents all have a stabilizing effect. The corresponding decreases in binding free energy, ranging from 0.3kBT to 1.7kBT, are displayed in Fig. 4.
Figure 4.
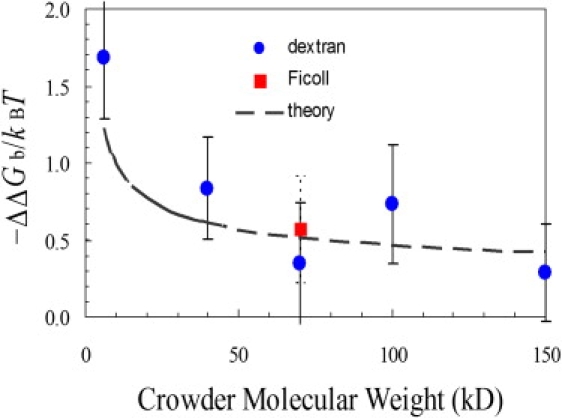
Changes in binding free energy in the presence of dextran of various molecular mass or Ficoll70, each at a concentration of 100 g/l. The symbols are experimental data, and the curve displays theoretical predictions.
The level of stabilization exerted by dextran varies systematically with the size of dextran (as signified by molecular mass). The stabilizing effect diminishes as the dextran size increases. This trend is in agreement, both qualitatively and quantitatively, with theoretical predictions given below based on atomistic modeling of macromolecular crowding. The level of stabilization due to Ficoll70 is similar to that due to dextran at the same molecular mass.
Atomistic modeling of macromolecular crowding
We have developed an efficient method for calculating the effect of macromolecular crowding on binding free energy (13). The method is based on the thermodynamic cycle illustrated in Fig. 2 and calculates ΔΔGb by obtaining the increases, due to volume exclusion by crowders, in chemical potential of the protein-protein complex and its two free subunits, which are represented in atomistic detail. In Fig. 5, we display the changes in binding free energy by the presence of crowders of three sizes, 20, 30, and 50 Å, occupying 5%, 15%, 25%, or 35% of the total volume. The crowders all have a stabilizing effect, because the protein-protein complex is more compact and hence experiences an increase in chemical potential smaller than that of the two free subunits combined.
Figure 5.
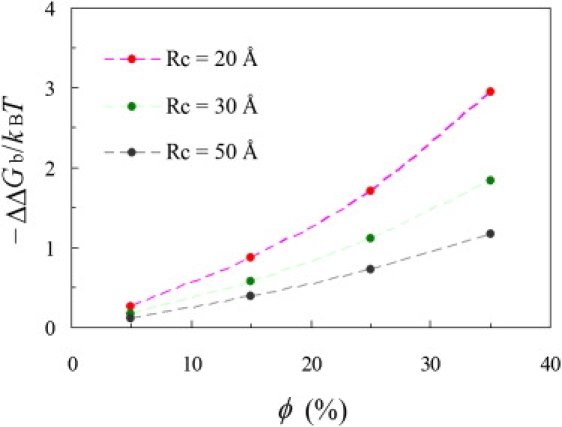
Dependence of ΔΔGb calculated for the ɛ186-θ complex on crowder size and volume occupancy. The symbols are calculations with atomistically detailed representations for the proteins, and the curves are from fitting to Eq. 4.
For a crowder of a given size, the level of stabilization increases with increasing volume occupancy. At a fixed volume occupancy, the stabilization effect diminishes with increasing crowder size. This finding can be understood as follows. It takes few copies of a large crowder to achieve the same volume occupancy achieved by many copies of a small crowder. Relative to the voids left by the many copies of a small crowder, the voids left by the few copies of the large crowder are big and hence less discriminating between the complex and the free subunits.
We found that the calculated increase in chemical potential for each species can be fitted well to an expression inspired by the scaled particle theory (17):
| (4) |
where Rc, Sc = 4πRc2, C, and ϕ = 4πRc3C/3 represent, respectively, the radius, surface area, number density, and volume occupancy of the crowder. In the scaled particle theory, a, S, and V represent the radius, surface area, and volume, respectively, of a spherical test protein. For the ɛ186-θ complex and its free subunits, we treat a, S, and V as floating parameters. The fitted values, in the order of ɛ186, θ, and their complex, are: a = 22.3, 19.6, and 24.5 Å; S = 7431.2, 4941.6, and 10656.3 Å2; and V = 28185.2, 11010.1, and 37093.1 Å3. For each species, the values of S and V are distinctly different from what is expected from a sphere with radius a. For example, modeling the ɛ186-θ complex as a sphere with a = 24.5 Å would underestimate the value of S by 29% but overestimate the value of V by 67%. The underestimation of S and overestimation of V are also true for the two subunits. The changes in binding free energy resulting from the fitting on Δμ are shown as the dashed curves in Fig. 5. The quality of the fit is demonstrated by the fact that the curves are indistinguishable from the data points.
Both our experimental data and the calculations show a decrease in stabilizing effect when the crowder size is increased. We asked the further question of whether the experimental data can be quantitatively explained by the calculations. To address this question, we modeled dextran as a spherical crowder, with the radius given by Rc = 8.26M1/3 Å, where M is the molecular mass in kD. The values of Rc given by this expression are somewhat lower than the sizes of dextran with various molecular mass measured in dilute conditions (18), to account for contraction of dextran (19,20) at the high concentration used in our experimental study. In addition, we assumed that 100 g/l of dextran corresponds to a volume occupancy of 15%. Finally, we used Eq. 4 to predict the increases in chemical potential for ɛ186, θ, and their complex, using the fitted values of a, S, and V listed above. The resulting changes in binding free energy by dextrans of different molecular mass are shown as the dashed curve in Fig. 4. It can be seen that the predictions are in reasonable agreement with the experimental data.
Biological consequences of crowding
The measured stabilizing effect due to crowding by dextran and Ficoll70 on the binding of the ɛ- and θ-subunits is modest, of the order of 1–2 kBT, or ∼1 kcal/mol. Such a level of stabilization is comparable to those exerted by these crowding agents on protein folding stability (21–24). Despite the modest amounts of stabilization, we were able to observe systematic variations with crowder size. Moreover, using our atomistic modeling, we were able to quantitatively explain these variations.
The stabilization due to crowding observed here on a binary binding step is in the range of effects of point mutations. A point mutation, when strategically located, can have significant biological consequences (6). However, the stabilization effect due to crowding is nonspecific, in that it is applicable to every binding step, regardless of the identities of the proteins undergoing binding. For the same reason, the stabilizing effects due to crowding are also cumulative when forming a higher oligomer (8): each addition of a monomer is accompanied by an increase in stability due to crowding. This cumulative nature can lead to substantial stabilization of the nucleus for protein aggregation or polymerization, which in turn results in shortening of the lag time. Dramatic shortening of lag times by macromolecular crowding has indeed been observed for α-synuclein fibrillization (3,4) and sickle hemoglobin polymerization (5).
The cumulative nature of macromolecular crowding during protein oligomerization also magnifies variations in stabilizing effect due to changes in crowder size, species, concentration, and composition. It is known that the protein degradation machinery becomes more and more sluggish and, hence, damaged proteins accumulate inside cells (25). Cell volumes may also decrease over time (26). Therefore, the level of intracellular crowding and, correspondingly, its stabilizing effect are expected to increase with time. This reasoning suggests that intracellular crowding may play a role in the increased onset of aggregation-related diseases such as Parkinson's with aging.
The ɛ-θ complex studied here is part of the catalytic core of Pol III. Although the ɛ-subunit contains the 3′-5′ proofreading exonuclease activity, the role of the θ-subunit has not been established. One suggestion is that the binding of θ stabilizes the ɛ-subunit against denaturation (10,16). Our results mean that intracellular crowding will assist such a role of θ-binding. In addition to the catalytic core, Pol III includes seven accessory subunits. Together, the 10 subunits endow the holoenzyme with extremely high efficiency and fidelity. Our study reinforces Kornberg's notion that intracellular crowding plays an important role in stabilizing such multicomponent biological machines.
The cumulative stabilizing effects of macromolecular crowding on high oligomers may impact a wide range of biological processes. For example, transcriptional regulatory proteins, such as dnaA (27) and the lac repressor LacI (28), often have multiple specific binding sites on genomic DNA. Our study suggests that macromolecular crowding may facilitate the cooperative binding of multiple copies of such regulatory proteins to genomic DNA. More generally, proteins that appear monomeric in dilute conditions may actually be oligomeric under the crowded conditions inside cells.
This study focused on the thermodynamics of protein-protein binding, but our findings also have implications for binding kinetics. As noted previously (29), macromolecular crowding presents two opposing effects on the binding rate constant. On the one hand, it slows down the translational and rotational diffusion of the binding proteins, leading to a decrease in the rate constant. On the other hand, the stabilization of protein binding observed here indicates that macromolecular crowding induces an effective attraction between the binding proteins, leading to an increase in the rate constant. Indeed, under macromolecular crowding, binding rate constants have been found to be much greater than expected from decreases in diffusion constants (30,31), demonstrating crowding-induced attraction between proteins during their binding process.
Conclusion
We have presented a direct quantitative measurement of the effects of macromolecular crowding on the binding stability of two proteins. A modest level of stabilization is observed, which is consistent with theoretical predictions based on atomistic modeling of excluded-volume interactions with crowders. The modest effects of crowding on elemental binding steps are cumulative, thus leading to substantial stabilization of higher oligomers. We suggest that an increased level of intracellular crowding with time may partly explain why susceptibility to protein-aggregation-related diseases such as Parkinson's increases with aging.
Acknowledgments
We thank Nick Dixon for providing the plasmids for the ɛ186 and θ -proteins and the protocols for their expression and purification, Daniel Spencer for first implementing the protocols in our laboratory, and Robert L. Baldwin for discussion.
This work was supported in part by National Institutes of Health grant GM058187.
References
- 1.Fuller R.S., Kaguni J.M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA. 1981;78:7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg A. Ten commandments: lessons from the enzymology of DNA replication. J. Bacteriol. 2000;182:3613–3618. doi: 10.1128/jb.182.13.3613-3618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shtilerman M.D., Ding T.T., Lansbury P.T. Molecular crowding accelerates fibrillization of α-synuclein: could an increase in the cytoplasmic protein concentration induce Parkinson's disease? Biochemistry. 2002;41:3855–3860. doi: 10.1021/bi0120906. [DOI] [PubMed] [Google Scholar]
- 4.Uversky V.N., Cooper E.M., Bower K.S., Li J., Fink A.L. Accelerated α-synuclein fibrillation in crowded milieu. FEBS Lett. 2002;515:99–103. doi: 10.1016/s0014-5793(02)02446-8. [DOI] [PubMed] [Google Scholar]
- 5.Rotter M., Aprelev A., Adachi K., Ferrone F.A. Molecular crowding limits the role of fetal hemoglobin in therapy for sickle cell disease. J. Mol. Biol. 2005;347:1015–1023. doi: 10.1016/j.jmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman S.B., Harrison B. Macromolecular crowding increases binding of DNA polymerase to DNA: an adaptive effect. Proc. Natl. Acad. Sci. USA. 1987;84:1871–1875. doi: 10.1073/pnas.84.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minton A.P. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers. 1981;20:2093–2120. [Google Scholar]
- 9.Zhou H.-X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamdan S., Bulloch E.M., Thompson P.R., Beck J.L., Yang J.Y. Hydrolysis of the 5′-p-nitrophenyl ester of TMP by the proofreading exonuclease (epsilon) subunit of Escherichia coli DNA polymerase III. Biochemistry. 2002;41:5266–5275. doi: 10.1021/bi0159480. [DOI] [PubMed] [Google Scholar]
- 11.Kirby T.W., Harvey S., DeRose E.F., Chalov S., Chikova A.K. Structure of the Escherichia coli DNA polymerase III ɛ-HOT proofreading complex. J. Biol. Chem. 2006;281:38466–38471. doi: 10.1074/jbc.M606917200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 13.Qin S., Zhou H.-X. Atomistic modeling of macromolecular crowding predicts modest increases in protein folding and binding stability. Biophys. J. 2009;97:12–19. doi: 10.1016/j.bpj.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keniry M.A., Park A.Y., Owen E.A., Hamdan S.M., Pintacuda G. Structure of the θ subunit of Escherichia coli DNA polymerase III in complex with the ɛ subunit. J. Bacteriol. 2006;188:4464–4473. doi: 10.1128/JB.01992-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pintacuda G., Park A.Y., Keniry M.A., Dixon N.E., Otting G. Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J. Am. Chem. Soc. 2006;128:3696–3702. doi: 10.1021/ja057008z. [DOI] [PubMed] [Google Scholar]
- 16.Gupta R., Hamdan S.M., Dixon N.E., Sheil M.M., Beck J.L. Application of electrospray ionization mass spectrometry to study the hydrophobic interaction between the ɛ and θ subunits of DNA polymerase III. Protein Sci. 2004;13:2878–2887. doi: 10.1110/ps.04889604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebowitz J.L., Rowlinson J.S. Thermodynamic properties of mixtures of hard spheres. J. Chem. Phys. 1964;41:133–138. [Google Scholar]
- 18.Weiss M., Elsner M., Kartberg F., Nilsson T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 2004;87:3518–3524. doi: 10.1529/biophysj.104.044263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z., Weng W., Bookchin R.M., Lew V.L., Ferrone F.A. Free energy of sickle hemoglobin polymerization: a scaled-particle treatment for use with dextran as a crowding agent. Biophys. J. 2008;94:3629–3634. doi: 10.1529/biophysj.107.117465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan J.-M., Chyan C.-L., Zhou H.-X., Chung T.-Y., Peng H. The effects of macromolecular crowding on the mechanical stability of protein molecules. Protein Sci. 2008;17:2156–2166. doi: 10.1110/ps.037325.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu Y., Bolen D.W. Efficacy of macromolecular crowding in forcing proteins to fold. Biophys. Chem. 2002;101–102:155–165. doi: 10.1016/s0301-4622(02)00148-5. [DOI] [PubMed] [Google Scholar]
- 22.Spencer D.S., Xu K., Logan T.M., Zhou H.-X. Effects of pH, salt, and macromolecular crowding on the stability of FK506-binding protein: an integrated experimental and theoretical study. J. Mol. Biol. 2005;351:219–232. doi: 10.1016/j.jmb.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Roberts A., Jackson S.E. Destabilised mutants of ubiquitin gain equal stability in crowded solutions. Biophys. Chem. 2007;128:140–149. doi: 10.1016/j.bpc.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Batra J., Xu K., Zhou H.-X. Nonaddtive effects of mixed crowding on protein stability. Proteins. 2009 doi: 10.1002/prot.22425. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuervo A.M., Bergamini E., Brunk U.T., Dröge W., French M. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 26.Bartosz G. Erythrocyte membrane changes during ageing in vivo. In: Harris J.R., editor. Blood Cell Biochemistry. Vol. 1. Plenum; New York: 1990. [Google Scholar]
- 27.Fuller R.S., Kornberg A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc. Natl. Acad. Sci. USA. 1983;80:5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villa E., Balaeff A., Schulten K. Structural dynamics of the lac repressor-DNA complex revealed by a multiscale simulation. Proc. Natl. Acad. Sci. USA. 2005;102:6783–6788. doi: 10.1073/pnas.0409387102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H.-X. Protein folding and binding in confined spaces and in crowded solutions. J. Mol. Recognit. 2004;17:368–375. doi: 10.1002/jmr.711. [DOI] [PubMed] [Google Scholar]
- 30.Drenckhahn D., Pollard T.D. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 1986;261:12754–12758. [PubMed] [Google Scholar]
- 31.Kuttner Y.Y., Kozer N., Segal E., Schreiber G., Haran G. Separating the contribution of translational and rotational diffusion to protein association. J. Am. Chem. Soc. 2005;127:15138–15144. doi: 10.1021/ja053681c. [DOI] [PubMed] [Google Scholar]


