Abstract
The majority of proteins in eukaryotic cells are modified according to highly regulated mechanisms to fulfill specific functions and to achieve localization, stability, and transport. Protein ubiquitination is one of the major post-translational modifications occurring in eukaryotic cells. To obtain the proteomic dataset related to the ubiquitin (Ub)-dependent regulatory system in Arabidopsis, affinity purification with an anti-Ub antibody under native condition was performed. Using MS/MS analysis, 196 distinct proteins represented by 251 distinct genes were identified. The identified proteins were involved in metabolism (23.0%), stress response (21.4%), translation (16.8%), transport (6.7%), cell morphology (3.6%), and signal transduction (1.5%), in addition to proteolysis (16.8%) to which proteasome subunits (14.3%) is included. On the basis of potential ubiquitination-targeting signal motifs, in-gel mobilities, and previous reports, 78 of the identified proteins were classified as ubiquitinated proteins and the rest were speculated to be associated proteins of ubiquitinated proteins. The degradation of three proteins predicted to be ubiquitinated proteins was inhibited by a proteasome inhibitor, suggesting that the proteins were regulated by Ub/proteasome-dependent proteolysis.
Keywords: Arabidopsis seedling, MS, ubiquitin-related protein
Introduction
Ubiquitin (Ub)-mediated protein modification is a critical post-translational regulatory mechanism that occurs in all eukaryotic cells. The conserved 76 amino acid polypeptide, Ub, is covalently attached to a substrate protein as a signal molecule, and this attachment leads to various outcomes. The widely known fate of ubiquitinated proteins is degradation by 26S proteasome, one specific case being that more than four Ubs comprise a multi-Ub chain via lysine (K) 48 residues (Thrower et al., 2000). Other types of ubiquitination, such as mono-ubiquitination and non-canonical ubiquitination, are implicated in various cellular functions, including endocytosis, endosomal sorting, signal transduction, and DNA damage repair (reviewed by Haglund and Dikic, 2005). Ubiquitination of target proteins requires the sequential action of three enzymes: Ub-activating enzyme (E1), Ub-conjugating enzyme (E2 or UBC), and Ub ligase (E3) (Hershko and Ciechanover, 1998; Pickart, 2001). The completely sequenced Arabidopsis genome has enabled the prediction of plant ubiquitination enzymes. To date, the activities of two E1s (Hatfield et al., 1997) and 25 E2s (Kraft et al., 2005) have been experimentally proven, and 12 genes encoding UBC domains are predicted to be E2s on the basis of their sequences (Bachmair et al., 2001). Similar to other organisms, Arabidopsis E3s are predicted to form the largest family comprising more than 1400 genes (Mazzucotelli et al., 2006). Furthermore, the presence of additional proteins, such as an enhancer of E2, has been reported (Yanagawa et al., 2004). Clearly, numerous proteins are involved in Ub-mediated protein regulation.
To identify ubiquitinated proteins in yeasts and mammals, several proteomic approaches that utilize various purification methods and MS/MS analyses have been reported (Peng et al., 2003; Hatakeyama et al., 2005; Jeon et al., 2007). In plants, two groups recently reported the proteomics of ubiquitinated proteins from Arabidopsis cell cultures and seedlings, respectively (Maor et al., 2007; Manzano et al., 2008). Meanwhile, a Ub-related proteome that includes both ubiquitinated proteins and their associated proteins has been reported only in human cells (Matsumoto et al., 2005).
As many proteins show spatiotemporal expression during development/life cycle, it is speculated that Ub-related proteins also vary among distinct tissues at various developmental stages. Therefore, the accumulation of information of Ub-related proteins from differentiated tissues at various stages would facilitate an understanding of Ub-mediated protein regulation throughout the life cycle. In addition, to make a comparison with the reports of Maor et al. (2007) and Manzano et al. (2008) that provided useful information of ubiquitinated proteomes from non-differentiated cell cultures and Arabidopsis seedlings, respectively, the proteomic analysis of Ub-related proteins expressed in Arabidopsis seedlings was performed in this study. For the large-scale isolation of Ub-related proteins, the purification was performed under native conditions. Previous studies of Arabidopsis ubiquitinated proteomes utilized different Ub-binding domains (UBAs) and showed that each UBA has distinct specificity for ubiquitinated proteins (Maor et al., 2007; Manzano et al., 2008). In order to overcome the limited specificity for target recognition, an anti-Ub antibody was applied to isolate Ub-related proteins. This study could provide helpful information for future work related to Ub-mediated protein regulation in plants.
Materials and methods
Plant materials
Arabidopsis (ecotype Columbia) seeds were germinated and cultured with shaking in liquid Murashige and Skoog medium containing 1% sucrose and 0.5 g l−1 MES, under a 16/8 h light/dark cycle at 22 °C. MG132 (Peptide Institute, Inc., Osaka, Japan) was added to 10-d-old cultured seedlings at a final concentration of 10 μM. After 24 h treatment, the seedlings were harvested. Nicotiana benthamiana was grown in a temperature-controlled growth room maintained at 25 °C under a 16/8 h light/dark cycle. Four- to five-week-old plants were used for experiments.
Large-scale purification of Ub-related proteins
To prepare an immunoaffinity column, HiTrap NHS-activated HP (1 ml, GE Healthcare Amersham Biosciences KK, Tokyo, Japan) was coupled with 1 mg of anti-Ub antibody FK2 (Nippon Bio-Test Laboratories, Tokyo, Japan) or mouse serum (Chemicon International, Inc., California, USA) as a negative control according to the manufacturer's instructions.
To purify Ub-related proteins under native condition, Arabidopsis seedlings were ground in liquid N2 with a mortar and pestle, and the powder was further ground in buffer A [50 mM TRIS-HCl (pH 7.5), 150 mM NaCl] containing the Complete Protease Inhibitor cocktail (Roche Applied Science, GmbH, Mannheim, Germany), 5 mM 2-mercaptoethanol, and 10 μM MG132. The homogenate was centrifuged at 32 300 g for 15 min and the supernatant was centrifuged again for 5 min. The supernatant was filtered through a 0.8 μm syringe filter. The total protein extract (200–250 mg) was applied to an immunoaffinity column equilibrated with buffer A. After washing the column with 5 vols of buffer A, bound proteins were eluted with buffer B [0.1 M glycine-HCl (pH 3.0), 150 mM NaCl]. Purification of the Ub-related proteins was performed three times.
In-gel digestion of purified proteins, MS/MS analysis, and data reduction
The eluted proteins from three independent purifications were mixed and fractionated by SDS-PAGE. Protein bands were detected with Flamingo™ Fluorescent Gel Stain (Bio-Rad Laboratories, CA, USA). The protein bands were excised and other smearing regions were cut into 2-mm-long gel pieces for in-gel trypsin digestion. In-gel digestion and MS/MS analysis were performed according to the methods described by Fujiwara et al. (2006) and Nakashima et al. (2008). The gel pieces were dehydrated by washing twice with 100% acetonitrile, and dried with a vacuum concentrator. The proteins were reduced with 10 mM DTT at 56 °C for 45 min and then alkylated with 55 mM iodoacetamide at room temperature in the dark for 30 min. After washing twice with 25 mM ammonium bicarbonate, the samples were dehydrated again with 50% acetonitrile and dried. The protein samples were digested with 10 μg ml−1 proteomics-grade trypsin (Promega, Madison, WI, USA) for 12 h at 37 °C.
The digested peptides were subjected to column chromatography (PEPMAPC18, 5 μm, 75 μm internal diameter, 15 cm; Dionex, Sunnyvale, CA) using the CapLC system (Waters, Milford, MA, USA). Buffers were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). A linear gradient from 5% to 45% B for 25 min was applied, and peptides eluted from the column were introduced directly into a Q-TOF Ultima mass spectrometer (Waters) at a flow rate of 100 nl min−1. In the ESI-positive ion mode, ionization was performed at a capillary voltage of 2.2 kV with the PicoTip nanospray source (New Objective, Cambridge, MA). For survey scan, mass spectra were acquired for the two most intense ions from the precursor ion scan between m/z 400 and 1500. For collision-induced dissociation (CID), the collision energy was set automatically according to the mass and charge state of the precursor peptide. MS/MS spectra were analysed with the MASCOT server against a protein database from the National Center for Biotechnology Information. The applied MASCOT search parameters were as follows: (i) taxonomy: Arabidopsis thaliana; (ii) potential modifications: carbamidomethyl and oxidation as fixed modifications, myristoylation (N-term G, K); (iii) max missed cleavage: 1; (iv) peptide tolerance: ±0.5 Da; (v) MS/MS tolerance: ±0.2 Da; and (vi) peptide charge: 2+ and 3+. Proteins detected from peptide fragments with high reliability [MASCOT score >40 (P <0.05)] were selected as identified proteins.
The presence of putative motifs in the identified proteins was analysed using Eukaryotic Linear Motif resource (ELM; http://elm.eu.org/) for destruction-box (D-box) and KEN-box, and GENETYX-MAC software for PEST sequences.
Antibody production
To produce a polyclonal fructose biphosphate aldolase-like (FBA) antibody, the open reading frame (ORF) of Arabidopsis FBA (At3g52930) was amplified with primers ‘fructose AntiB-F’ (5′-GGAATTCCATATGTCTGCCTTCACAAGCAA-3′) and ‘fructose AntiB-R’ (5′-CGGAATTCTCAGTACTTGTAATCCTTCACG-3′), thereby introducing a NdeI site at the 5′ terminus and an EcoRI site at the 3′ terminus. The NdeI–EcoRI fragment of FBA was cloned into the expression vector pET28c (+) carrying 6×-histidine at the N terminus. The resulting plasmid was transformed into BL21 (DE3) cells. The purified histidine-tagged FBA protein was injected into a rabbit as the antigen. The antiserum obtained was used as anti-FBA antibody for immunoblot analysis.
Agroinfiltration
The ORFs of At1g12840 (de-etiolated 3; DET3) and At3g04120 (glyceraldehyde-3-phosphate dehydrogenase C subunit; GAPC) were amplified from RIKEN Arabidopsis Full-Length cDNAs (RAFL) using the following primers: ‘attB1-At1g12840-5′’ (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGACTTCGAGATAT-3′) and ‘attB2-At1g12840-3′’ (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCAGCAAGGTTGATAGT-3′), and ‘attB1-At3g04120-5′’ (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTGACAAGAAG-3′) and ‘attB2-At3g04120-3′’ (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGGCCTTTGACATGTG-3′), respectively. Transfer of the PCR products to the entry vector pDONR221 was performed by BP reaction (Gateway; Invitrogen). Each ORF fragment of At1g12840 and At3g04120 on pDONR221 was transferred to the binary vector pGWB14 (Nakagawa et al., 2007) carrying 3× HA by LR reaction (Gateway; Invitrogen).
The resulting constructs (DET3-HA and GAPC -HA) were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Agroinfiltration using N. benthamiana leaves was performed as described previously (Katou et al., 2005). Discs were collected from leaves infiltrated with Agrobacterium cells after infiltration for 2 d.
Degradation assays
For the degradation assays of DET3-HA and GAPC-HA proteins, total protein was extracted from agroinfiltrated N. benthamiana leaf discs with extraction buffer C (50 mM TRIS-HCl, 2 mM ATP, 5 mM MgCl2, 10 mM 2-mercaptoethanol, and 20% glycerol) supplemented with 30 μM MG132 or DMSO. For the FBA degradation assay, total protein from Arabidopsis seedlings was extracted with buffer C supplemented with 10 μM leupeptin (Peptide Institute) and 30 μM MG132 or DMSO. The protein extracts were incubated at room temperature for 2 h. 3× SDS sample buffer was added to stop the reaction, and the sample was used for immunoblot analysis. Anti-HA antibody was purchased from Abcam plc (Cambridge, UK). The degradation assay for each protein was replicated three times. Signal intensities of proteins detected on immunoblotted membranes were quantitated by digitizing with Image J software (http://rsbweb.nih.gov/ij/). Each quantitated value of HA or FBA signal was divided by the corresponding quantitated value of the control protein. The relative amounts of the remaining proteins (%) after incubation with MG132 or DMSO were calculated.
Results and discussion
Purification and identification of Ub-related proteins from Arabidopsis seedlings
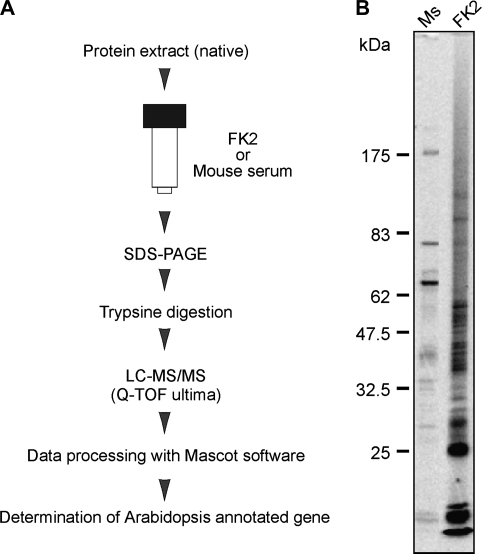
To isolate Ub-related proteins by immunoaffinity chromatography, monoclonal antibody FK2 was applied, which selectively recognizes the Ub moiety but not free Ub (Fujimuro et al., 1994). Approximately 250 mg of total protein was obtained from 50 g of Arabidopsis seedlings and applied to the immunoaffinity column under native condition (Fig. 1A). The staining pattern of eluted proteins subjected to SDS–PAGE was reproducible among the samples derived from three independent purification steps (see Supplementary Fig. S1 at JXB online). Compared to the mouse serum column, a number of discrete bands on a smeared background were detected in the purified sample eluted with the FK2 column (Fig. 1B), suggesting different mobilities in a gel caused by the heterogeneity of multi-Ub chains, as observed in a study of human cells (Matsumoto et al., 2005).
Fig. 1.
Immunoaffinity purification and identification of Ub-related proteins. (A) Flow chart of purification and identification of Ub-related proteins. (B) Immunopurified proteins with FK2 or mouse serum (Ms) from Arabidopsis seedlings were subjected to SDS-PAGE and stained with Flamingo™.
Numerous Ubs were detected by MS/MS analysis, indicating that they were probably derived from Ub-conjugated proteins. Ubs and E2 proteins were eliminated from the list of proteins isolated with the FK2 column and the proteins isolated with the mouse serum column were further subtracted. In this study, only proteins with a score of over 40 (P <0.05) were selected as candidate proteins with high reliability. Accordingly, 196 proteins, which were represented by 251 distinct genes including possible paralogs, were determined as Ub-related proteins (see Supplementary Table S1 at JXB online). Comparing the results of this study with Arabidopsis ubiquitinated proteomes reported in two studies, 33 proteins were overlapped and only one protein (β-tubulin; protein no. 137 in Supplementary Table S1 at JXB online) was common to the three studies (see Supplementary Table S1 at JXB online). The low overlapping result compared to the previous studies may be due to the difference in the differentiation state of the protein source. In addition, unstructured threshold settings for protein screening from the MS scores may account for the difference in listed proteins among the three studies. The following reasons are proposed. Immunopurification with FK2 used in this study, enabled recognition of all types of ubiquitinated proteins, whereas each UBA used in the other studies had distinct specificity to ubiquitinated proteins. Therefore, the dominant ubiquitinated proteins were preferentially trapped by FK2. In addition, our dataset included a high proportion of associated proteins of ubiquitinated proteins due to the native condition used in protein purification. In fact, proteins annotated as RING-type E3 (protein no. 109 in Supplementary Table S1 at JXB online) and putative Ub receptors, DNA-repair protein RAD23 (protein no. 119 in Supplementary Table S1 at JXB online), and UBA-like motif-containing protein (protein no. 196 in Supplementary Table S1 at JXB online), were identified in this study, suggesting that non-direct target proteins for ubiquitination were also isolated.
Characterization of Ub-related proteins from Arabidopsis seedlings
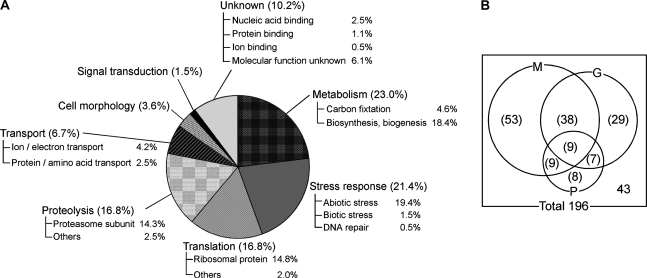
The identified proteins were categorized on the basis of the biological processes described in ‘The Arabidopsis Information Resource’ (TAIR) (Fig. 2A). The large population of proteins involved in metabolism (23.0%) as well as previous Arabidopsis ubiquitinated proteomes (Maor et al., 2007; Manzano et al., 2008) may indicate the significance of ubiquitination for protein regulation in cellular metabolism. Similar to the previous report of Manzano et al. (2008), it was found that the proteins involved in stress response (21.4%) were more abundant in seedlings than in cell cultures. This might be due to the differentiation state of the cells, because the growth condition in liquid culture seemed to be more stressful for seedlings than for cell cultures. Indeed, proteins involved in abiotic stress were dominant in this category (Fig. 2A). The majority of translational proteins identified in our study (16.8%) were ribosomes, in agreement with Maor's study (Maor et al., 2007). It has been speculated that ubiquitination might play an important role in the regulation and/or quality control of ribosomal proteins, as reported in human cells (Matsumoto et al., 2005). In addition, the recent report by Kraft et al. (2008) of a link between ubiquitination and regulated degradation of mature ribosomes in yeast also supports our results. The percentage of proteins involved in signal transduction (1.5%) was lower than that previously reported for the ubiquitinated proteome from Arabidopsis seedlings (Manzano et al., 2008). The difference may depend on the threshold for the screening of identified proteins from MS scores, as described above. Only proteins with a high reliability (95% confidence) were listed in this study, whereas Manzano's list contained proteins with low reliability (Manzano et al., 2008). The proportions of other components were similar to that found in previous Arabidopsis ubiquitinated proteomes. One significant difference compared to the previous Arabidopsis ubiquitinated proteomes was the high percentage of proteasome subunits (14.3%), which was probably dependent on the native condition for protein purification. This fact indicated that the associated proteins of ubiquitinated proteins were isolated as well, and implied that most proteins were involved in Ub/proteasome-dependent proteolysis.
Fig. 2.
Characterization of proteins identified from Arabidopsis seedlings. (A) Proportion of identified proteins categorized according to function. Each category was further subdivided according to specific function. (B) Numbers of potential ubiquitinated proteins and their associated proteins. Number outside the circles indicates the number of associated proteins of ubiquitinated proteins. M, proteins containing at least one motif; G, proteins detected in multiple gel pieces; P, proteins previously reported as ubiquitinated proteins.
Classification of identified proteins
Purification under native condition is a feasible technique to identify Ub-related proteins that play major roles in Ub-dependent regulation (Matsumoto et al., 2005). Since the protein population isolated under native condition included not only ubiquitinated proteins but also their associated proteins, they were classified as ubiquitinated proteins and their associated proteins based on the following three criteria.
First, previous Arabidopsis ubiquitinated proteomes from cell cultures and seedlings, respectively, were examined (Maor et al., 2007; Manzano et al., 2008). Of the proteins identified in our study, 33 have been reported as ubiquitinated proteins, suggesting that these were the potential direct targets of ubiquitination (Fig. 2B; see Supplementary Table S1 at JXB online).
Second, the identified proteins were classified on the basis of potential ubiquitination-targeting signal motifs (D-box, KEN-box, and PEST sequence) to predict the ubiquitinated proteins. D-box and KEN-box are short sequence elements in the substrates of the anaphase-promoting complex/cyclosome (APC/C), which is a multisubunit RING-type E3 (King et al., 1996; Pfleger and Kirschner, 2000), and indeed, RING-type E3 (protein no. 109 in Supplementary Table S1 at JXB online) was found in our study. PEST sequences that are rich in proline (P), glutamic acid (E), serine (S), and threonine (T) were found in a number of short-lived proteins controlled by proteolysis, mostly via ubiquitin-mediated degradation. Almost half of the identified proteins (109/196 proteins (55.6%)) contained at least one motif, implying that they are the potential targets of Ub/proteasome-dependent proteolysis (Fig. 2B; see Supplementary Tables S1 and S2 at JXB online).
Third, multiple detections from different gel pieces implied multi-ubiquitination of the proteins, since the heterogeneity of multi-Ub chains accounted for the different mobilities in a gel. Of the identified 196 Ub-related proteins, 83 (42.3%) were found in 2–17 gel pieces of different sizes (Fig. 2B; see Supplementary Table S1 at JXB online), suggesting that the proteins were probably tagged with heterogeneous multi-Ub chains.
Considering potential ubiquitination-targeting signal motifs, in-gel mobilities of the identified proteins, and previous reports, 153 proteins (78.0%) were predicted as the potential targets of ubiquitination, including 109 potential target proteins (55.6%) of Ub/proteasome-dependent proteolysis, whereas the remaining proteins (21.9%) were potential molecules associated with the ubiquitinated proteins.
Degradation assays of proteins predicted as ubiquitinated proteins
The results suggested that a large proportion of the identified proteins were involved in Ub/proteasome-dependent proteolysis. Therefore, the identified proteins were examined by proteasome degradation assay. According to the prediction described above, three proteins predicted as potential ubiquitinated proteins, At1g12840 (DET3, protein no. 82 in Supplementary Table S1 at JXB online), At3g04120 (GAPC, protein no. 1 in Supplementary Table S1 at JXB online), and At3g52930 (fructose biphosphate aldolase-like; FBA, protein no. 6 in Supplementary Table S1 at JXB online), were chosen for the assay.
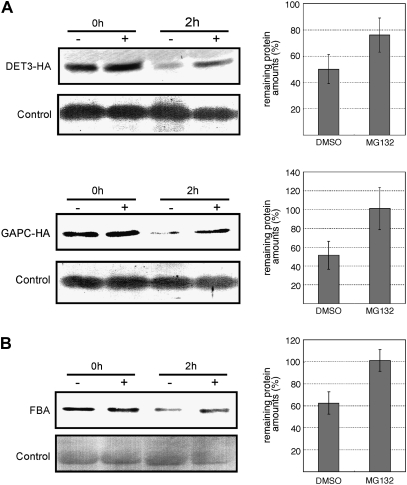
DET3 encodes subunit C of vacuolar H+-ATPase (V-ATPase). V-ATPase is one of the major proton pumps that act to acidify intracellular compartments (Sze et al., 2002) and DET3 is significantly responsible for its activity (Schumacher et al., 1999). Since DET3 contains four D-box motifs and was detected in two gel pieces (Supplementary Table S1 at JXB online), regulation of DET3 protein by Ub/proteasome-dependent proteolysis was highly expected. DET3 protein tagged with 3× HA was transiently expressed in N. benthamiana leaves by agroinfiltration. The protein extract was incubated with MG132, a 26S proteasome inhibitor. As a result, it was found that DET3 protein degradation was inhibited by MG132 treatment but not DMSO treatment (Fig. 3A), indicating that it was conjugated with canonical Ub chains and then underwent the proteasome-dependent proteolysis. Other subunits A and B of V-ATPase were respectively isolated as ubiquitinated proteins in Maor's and Manzano's studies (Maor et al., 2007; Manzano et al., 2008), whereas the anti-Ub antibody used in our study could trap all of these subunits, implying that the difference in the identified subunits of V-ATPase may be attributed to the distinct specificity of the ligands used for affinity purification. It was also suggested that each subunit is ubiquitinated by a distinct ubiquitination pathway, and at least one subunit DET3 of the V-ATPase complex was degraded by the 26S proteasome.
Fig. 3.
Degradation assays of potential ubiquitinated proteins. Protein extracts were treated with MG132 (+) or DMSO (–). Lower panels of each figure indicate loading controls. Graphs on the right represent the relative amount of remaining protein (%) after treatment with MG132 (+) or DMSO. Error bars indicate standard deviations. (A) DET3-HA and GAPC-HA proteins transiently expressed in N. benthamiana leaves were detected with anti-HA antibody. (B) Total proteins were extracted from Arabidopsis seedlings. Immunoblot analysis was performed with anti-FBA antibody.
GAPC was detected in four gel pieces, although the ubiquitination-targeting signal motif was absent (see Supplementary Table S1 at JXB online). The degradation of GAPC by the 26S proteasome has not been reported to date. Since it has also been reported as a ubiquitinated protein (Manzano et al., 2008), the degradation by the 26S proteasome of the GAPC protein, which was transiently expressed similar to DET3, was examined. As shown in Fig. 3A, the degradation of the GAPC protein was inhibited by MG132 treatment, but not DMSO treatment, indicating that it was regulated by the Ub/proteasome-dependent proteolysis.
FBA was identified in seven gel pieces, although it did not contain any potential ubiquitination-targeting signal motifs (see Supplementary Table S1 at JXB online). FBA was also identified as a ubiquitinated protein (Maor et al., 2007). Thus, it was expected that FBA would be degraded by the 26S proteasome. To examine the FBA degradation by the 26S proteasome in Arabidopsis seedlings, the protein extract was incubated with or without MG132, and FBA protein was detected with its antibody. As shown in Fig. 3B, the degradation of FBA was inhibited by MG132, demonstrating that FBA was regulated by Ub/proteasome-dependent proteolysis in Arabidopsis seedlings.
The degradation of DET3, GAPC, and FBA by the 26S proteasome was demonstrated for the first time in this study. GAPC and FBA are involved in glycolysis and are known to respond to environmental stress. Thus, the turnover of these proteins may be regulated by the Ub/proteasome-dependent proteolysis in the glycolytic pathway and/or stress response. It would be interesting to investigate the contribution of Ub-mediated regulation of these proteins in metabolism and/or stress response as a future work.
Conclusions
This study showed the Ub-related proteome of Arabidopsis seedlings. Protein purification under native conditions with an anti-Ub antibody contributed to the isolation of various Ub-related proteins that mainly comprised proteins involved in Ub/proteasome-dependent proteolysis. The protein population identified contained both the targets of ubiquitination and their associated proteins. Biochemical evidence is required to characterize exactly which protein is the direct target of ubiquitination. Nevertheless, classification of the identified proteins based on the potential ubiquitination-targeting signal motifs, in-gel mobilities, and the previous reports contributed to the prediction of ubiquitinated proteins and their associated proteins. Our results are expected to be of use to the future investigation of Ub-mediated protein regulation in plants.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Respective SDS-PAGE images of proteins immunopurified with FK2 obtained from three independent experiments (Ex1, Ex2, and Ex3). The staining pattern of the purified proteins was reproducible.
Supplementary Table S1. Ub-related proteins identified from Arabidopsis seedlings.
Supplementary Table S2. Proteins containing potential ubiquitination-targeting signal motifs for Ub/proteasome-dependent proteolysis.
Acknowledgments
We thank Dr T Nakagawa (Shimane University, Japan) for pGWB14 vector and Ms R Okanami for technical assistance. This work was supported by KAKENHI (19658041), a Grant-in-Aid for Exploratory Research from the Japan Society for the Promotion of Science (JSPS), and a Grant-in-Aid for Scientific Research from the Nara Institute of Science and Technology supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Glossary
Abbreviations
- DET3
de-etiolated 3
- FBA
fructose biphosphate aldolase-like
- GAPC
glyceraldehyde-3-phosphate dehydrogenase C subunit
- ORF
open reading frame
- Ub
ubiquitin
- V-ATPase
vacuolar H+-ATPase
References
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends in Plant Science. 2001;6:463–470. doi: 10.1016/s1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Letters. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Umemura K, Kawasaki T, Shimamoto K. Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiology. 2006;140:734–745. doi: 10.1104/pp.105.068395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO Journal. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield PM, Gosink MM, Carpenter TB, Vierstra RD. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. The Plant Journal. 1997;11:213–226. doi: 10.1046/j.1365-313x.1997.11020213.x. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Matsumoto M, Nakayama KI. Mapping of ubiquitination sites on target proteins. Methods in Enzymology. 2005;399:277–286. doi: 10.1016/S0076-6879(05)99019-8. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jeon HB, Choi ES, Yoon JH, Hwang JH, Chang JW, Lee EK, Choi HW, Park ZY, Yoo YJ. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochemical and Biophysical Research Communications. 2007;357:731–736. doi: 10.1016/j.bbrc.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Katou S, Karita E, Yamakawa H, Seo S, Mitsuhara I, Kuchitsu K, Ohashi Y. Catalytic activation of the plant MAPK phosphatase NtMKP1 by its physiological substrate salicylic acid-induced protein kinase but not by calmodulins. Journal of Biological Chemistry. 2005;280:39569–39581. doi: 10.1074/jbc.M508115200. [DOI] [PubMed] [Google Scholar]
- King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Molecular Biology of the Cell. 1996;7:1343–1357. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature Cell Biology. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiology. 2005;139:1597–1611. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Abraham Z, López-Torrejón G, Del Pozo JC. Identification of ubiquitinated proteins in Arabidopsis. Plant Molecular Biology. 2008;68:145–158. doi: 10.1007/s11103-008-9358-9. [DOI] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Molecular and Cellular Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Hatakeyama S, Oyamada K, Oda Y, Nishimura T, Nakayama KI. Large-scale analysis of the human ubiquitin-related proteome. Proteomics. 2005;5:4145–4151. doi: 10.1002/pmic.200401280. [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis AM, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo AM. The e3 ubiquitin ligase gene family in plants: regulation by degradation. Current Genomics. 2006;7:509–522. doi: 10.2174/138920206779315728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. The Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nature Biotechnology. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes and Development. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes and Development. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Schumacher K, Müller ML, Padmanaban S, Taiz L. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends in Plant Science. 2002;7:157–161. doi: 10.1016/s1360-1385(02)02240-9. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO Journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y, Sullivan JA, Komatsu S, et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes and Development. 2004;18:2172–2181. doi: 10.1101/gad.1229504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.