Abstract
Arabinogalactan proteins (AGPs) are structurally complex plasma membrane and cell wall proteoglycans that are implicated in diverse developmental processes, including plant sexual reproduction. Male gametogenesis (pollen grain development) is fundamental to plant sexual reproduction. The role of two abundant, pollen-specific AGPs, AGP6, and AGP11, have been investigated here. The pollen specificity of these proteoglycans suggested that they are integral to pollen biogenesis and their strong sequence homology indicated a potential for overlapping function. Indeed, single gene transposon insertion knockouts for both AGPs showed no discernible phenotype. However, in plants homozygous for one of the insertions and heterozygous for the other, in homozygous double mutants, and in RNAi and amiRNA transgenic plants that were down-regulated for both genes, many pollen grains failed to develop normally, leading to their collapse. The microscopic observations of these aborted pollen grains showed a condensed cytoplasm, membrane blebbing and the presence of small lytic vacuoles. Later in development, the generative cells that arise from mitotic divisions were not seen to go into the second mitosis. Anther wall development, the establishment of the endothecium thickenings, the opening of the stomium, and the deposition of the pollen coat were all normal in the knockout and knockdown lines. Our data provide strong evidence that these two proteoglycans have overlapping and important functions in gametophytic pollen grain development.
Keywords: Arabidopsis, arabinogalactan proteins, knockouts, pollen development
Introduction
Pollen ontogeny is an attractive model to study cell division and differentiation. The progression from proliferating microspores to terminally differentiated pollen is characterized by a large-scale repression of early programme genes and the activation of a unique late gene-expression programme in mature pollen. In Arabidopsis, all microsporocytes will eventually undergo meiosis, resulting in tetrads of haploid microspores, each of which will, in turn, develop into a pollen grain. This pattern of development is quite clear, but little is known about the underlying molecular mechanisms. Only now, are the roles of specific genes involved in male gametogenesis beginning to be understood (Yang et al., 1999; Honys et al., 2006; Quan et al., 2008; Toller et al., 2008).
Arabinogalactan proteins (AGPs) are a class of structurally complex proteoglycans, present at the surface of cells throughout the plant kingdom. The protein backbones of AGPs are rich in proline/hydroxyproline, serine, alanine, and threonine and are modified by the addition of type II arabinogalactan polysaccharides and arabinose oligosaccharides (Showalter, 2001). AGPs can be divided into several classes: classical AGPs, lysine-rich AGPs, AG peptides, fasciclin-like AGPs (FLAs), and other chimeric AGPs. AGP6 and AGP11 are classical AGPs as they have the characteristic 85–151 amino acids and consist of an N-terminal signal peptide, a Pro/Hyp-rich AGP central domain and a predicted C-terminal glycosylphosphatidylinositol (GPI) lipid anchor addition sequence (Schultz et al., 1998; Showalter, 2001). By contrast, lysine-rich AGPs have a small lysine-rich region within the classical AGP domain; AG peptides are typically only 10–15 amino acids in length; and FLAs contain both AGP and fasciclin-like domains (Schultz et al., 2002; Yang and Showalter, 2007). GPI anchored AGPs are believed to be tethered to the external surface of the plasma membrane and are then released to the extracellular matrix as a consequence of the cleavage action of specific phospholipases. GPI anchoring and release may confer localized or polarized targeting and alter the functional properties of proteins (Lalanne et al., 2004).
AGPs have been implicated in diverse developmental processes, including somatic embryogenesis (van Hengel and Roberts, 2002), cell proliferation (Serpe and Nothnagel, 1994), cell expansion (Willats and Knox, 1996; Vissenberg et al., 2001), programmed cell death (Motose et al., 2004), wound responses (Guan and Nothnagel, 2004), root morphology (Seifert et al., 2002), pollen tube growth (Cheung et al., 1995; Wu et al., 1995; Levitin et al., 2008), and plant hormonal signalling pathways (Suzuki et al., 2002). Evidence implicating AGPs in sexual reproduction has been obtained by our group, for several plant species (Coimbra and Salema, 1997; Coimbra and Duarte, 2003; Coimbra et al., 2005). Recently, the selective labelling obtained with AGP glycan monoclonal antibodies during Arabidopsis pollen and pistil development, suggested that some AGPs glycan epitopes are markers for gametophytic cell differentiation (Coimbra et al., 2007).
Despite the presence of tissue-specific carbohydrate epitopes on AGPs, these investigations do not allow the characterization of a single type of AGP, but a set of AGPs with similar epitopes. Antibodies bind to carbohydrate epitopes that are present on AGPs with different protein backbones, and at the same time, individual protein backbones may be differentially glycosylated.
The expression profiles of AGPs in Arabidopsis were obtained from several microarray hybridization data sets (Zimmermann et al., 2004). Microarray experiments are extremely useful in identifying targets for further analysis, but such experiments should be viewed as starting points and must be confirmed by independent means. It was possible to validate the results showing that AGP6 and AGP11 are pollen grain and pollen tube specific (Pereira et al., 2006).
AGP6 and AGP11 are closely related, encoding proteins that share 68% amino acid sequence identity, suggesting that they are paralogous genes and that their functions may overlap. More recently, Levitin et al. (2008) showed that these two AGPs are required for pollen tube growth and stamen function, by using RNAi lines with reduced AGP6 and AGP11 expression.
In the present work, null mutants and transgenic plants were used as an approach to identify phenotypic traits attributable to either or both AGP6 and AGP11, during pollen grain development.
This work was also complemented by gene silencing studies. Two Arabidopsis transgenic lines, obtained by RNAi technology, and silenced for both AGP6 and AGP11 were used. The constructions were made under the control of the 35S promoter and also under the control of the native promoter for the AGP6 gene. Our results indicate that AGP6 and AGP11 are integral to pollen grain development, but also that some functional redundancy exists.
Materials and methods
Plant materials and growth conditions
Ds-tagged Arabidopsis lines Ds54-4754-1 and Ds11-4025-1 were obtained from the RIKEN GSC Arabidopsis Ds transposon tag line collection (Kuromori et al., 2004), and designated as agp6 and agp11, respectively. PCR amplification products encompassing regions of the AGP6 or AGP11 coding sequences and the transposon sequence were gel extracted and sequenced. The Ds transposon in agp6 and in agp11 are inserted just after a guanine base in position 130 bp and an adenine base in position 192 bp from the initiation codon, respectively. RNAi and amiRNA transformation experiments were performed in an Arabidopsis Col-0 background. Plants were germinated and grown in Murashige and Skoog (1962) medium with 0.7% agar. Plantlets were then transferred to soil and kept in a growth chamber at 21 °C under long days (16/8 h light/dark) and 60% relative humidity. Pollen tube germination and growth were according to Coimbra et al. (2007).
Light microscopy
Anthers were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in phosphate buffer (0.025 M, pH 7, with one micro drop of Tween 80), placed under vacuum for 1 h and then at 4 °C overnight. After dehydration in a graded ethanol series, the material was embedded in LR White. Thick sections (0.5 μm) were obtained with a Leica Reichert Supernova microtome, placed on glass slides, and stained with a solution of 1% Methylene blue or with Calcofluor white (fluorescent brightener; Sigma). Sections for fluorescence microscopy were mounted with Vectashield (Vector laboratories, Petersborough, UK), and sections for bright field microscopy were mounted with Eukitt quick-harder (Fluka). Images were captured with a Leica DFC420 camera, and processed with Leica Application Suite software.
Electron microscopy
For Transmission Electron Microscopy (TEM), anthers were fixed as described above, and post-fixed in OsO4 (1%) in the same buffer for 2 h, dehydrated in a graded ethanol series and embedded in EPON resin. Ultra-thin sections (40–50 nm), obtained with a Leica Reichart Supernova microtome, were mounted on 400 mesh copper grids (G2400C Agar Scientific Ltd.), stained in uranyl acetate followed by lead citrate, and examined in a Zeiss 902 electron microscope. Images were recorded with an Axiocam camera and treated with Axiovision 4 software.
For Scanning Electron Microscopy (SEM), anthers were fixed as described for TEM, dehydrated in a graded ethanol series and then mounted on stubs. After coating with gold, the samples were examined in a Jeol JSM-35C scanning electron microscope.
RNAi Arabidopsis
For preparation of stable AGP6 RNAi lines of Arabidopsis, the AGP6 RNAi vector with 450 bp of AGP6 coding sequence, in the sense and antisense orientations, were amplified and inserted into pK7GWIWG2(I) vector (Karimi et al., 2002). AGP6 shows 74% nucleotide sequence similarity with AGP11. The vector was under the control of the 35S promoter and the nopaline synthase terminator and was used to transform Arabidopsis plants via Agrobacterium tumefaciens (LBA4404) transformation (Clough and Bent, 1998).
amiRNA Arabidopsis
For preparation of stable amiRNA, AGP6+AGP11-specific sequences were identified with the WMD Web MicroRNA Designer (www.weigelworld.org). The amiRNA was constructed according to Schwab et al. (2006) using the following sequences: I miR-s gaTTGGGGAGGAGACTGTGGGTGtctctcttttgtattcc; II miR-a gaCACCCACAGTCTCCTCCCCAAtcaaagagaatcaatga; III miR*s gaCAACCACAGTCTCGTCCCCATtcacaggtcgtgatatg; IV miR*a gaATGGGGACGAGACTGTGGTTGtctacatatatattcct; pRS300-A 5′-CTGCAAGGCGATTAAGTTCGGTAAC; and pRS300-B 5′-GCGGATAACAATTTCACACAGGAAACAG. The resulting miR precursor was initially transferred into the pENTR-D-TOPO vector (Invitrogen, KanR). The ProAGP6 was PCR amplified and transferred into the pENTR-5′-TOPO (TA) vector. Both sequences were then transferred via a single LR recombination reaction into the pK7m24GW3 (SepR in bact & KanR in plant) vector (Karimi et al., 2002). The ProAGP6:AGP6 + AGP11 amiRNA sequence was transferred into Arabidopsis plants via Agrobacterium (C58) transformation (C58) (Clough and Bent, 1998).
Real-time PCR
Anther mRNA extracts were reverse transcribed using Promega Reverse Transcription System and poly(dT)12–18 to prime the reactions. cDNA was amplified using the iQ™ SYBR® Green Supermix on the iQ™5 Real-Time PCR Detection System (Bio-Rad).
Real-time RT-PCR reactions were run in duplicates. After 3 min at 95 °C followed by a 10 s denaturation step at 95 °C, samples were run for 40 cycles of 10 s at 95 °C and 30 s at 60 °C. After each run, a dissociation curve was acquired to check for amplification specificity by heating the samples from 60 °C to 95 °C. Serial dilutions of pure genomic DNA from Arabidopsis ecotype Nossen were used to set up a calibration curve, which was used to quantify plant DNA in each sample.
At the end of the PCR cycles, the data were analysed with the iQ5 2.0, Standard Edition Optical System Software v2.0.148.060623 (Bio-Rad).
Accession numbers
The Arabidopsis Genome Initiative locus identifiers are as follows: AGP6 (At5g14380), AGP11 (At3g01700), PRF5 (At2g19770), and UBC9 (At4g27960).
Results
The expression pattern of AGP6
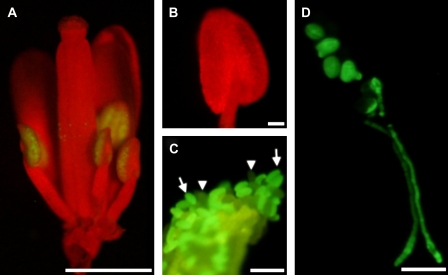
In earlier RT-PCR experiments it was shown that AGP6 is expressed in pollen grains and in germinating pollen tubes (Pereira et al., 2006). Data obtained from the Affymetrix AG and ATH1 GeneChip arrays (www.genevestigator.ethz.ch) also indicate that AGP6 is pollen specific. To determine with greater accuracy the temporal expression profile of AGP6, Arabidopsis plants were transformed with a ProAGP6:GFP gene construct. GFP fluorescence was absent in all vegetative plant parts, but became clearly visible just after the appearance of the locules in anthers (Fig. 1A, B). This stage corresponds to stage 9 as described by Smyth et al. (1990) and comprises the rapid lengthening of all organs, including stamens. GFP fluorescence was restricted to pollen and pollen tubes and could be clearly distinguished, particularly in heterozygous plants, from the green yellow autofluorescence typical of the exine and of the endothecium lignin thickenings of the anther wall (Fig. 1C). GFP fluorescence persisted through to the mature pollen grains and was observed in growing pollen tubes (Fig. 1D).
Fig. 1.
Transgenic Arabidopsis plants expressing a ProAGP6:GFP construct. (A) Low-power binocular fluorescence microscope of a flower in a stage where the anthers have their locules already formed and are extending. GFP fluorescence is exclusively visible in pollen grains. (B) Anther at the immediately preceding stage of development, with reference to the stage in (A). No fluorescence could be observed. Typically in this stage the locules are formed and microspores are released from tetrads. (C) Mature dehiscent anther, showing GFP-labelled (arrows) and unlabelled pollen grains (arrow heads). The anther wall is also visible as green-yellow autofluorescence. (D) Fluorescence microscopy of germinating mature pollen. GFP fluorescence is visible in all pollen grains and along all pollen tube extension. Bars: (A) 1 mm; (B, C) 500 μm; (D) 50 μm. (This figure is available in colour at JXB online.)
Mutations in both AGP6 and AGP11 affect pollen development
AGP6 expression early in male gametophyte biogenesis suggested that the absence of the proteoglycan may disrupt the process. A reverse genetics approach was used to characterize the biological function of both AGP6 and AGP11 in Arabidopsis. Ds tagged lines for AGP6 and AGP11 were verified as having a transposon inserted in the coding sequences. The mutants showed no detectable accumulation of AGP6 or AGP11 transcripts in RT-PCR (Fig. 2A), indicating that they are null mutations. However, neither agp6 nor agp11 mutants showed obvious defects in morphology as assessed by optical and electron microscopy.
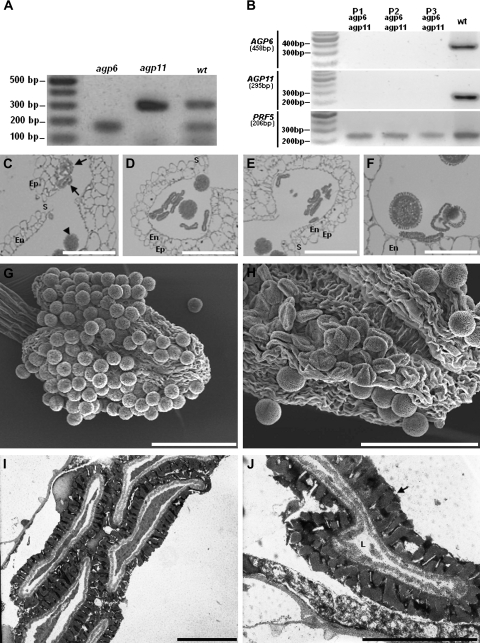
Fig. 2.
Ds transposon tag lines for AGP6 and AGP11. (A) Duplex RT-PCR amplification products of AGP6 and AGP11 mRNA transcripts in pollen of Arabidopsis wild-type (wt), and of Arabidopsis tag lines containing the Ds transposon in AGP6 (agp6) and in AGP11 (agp11) genes. Only in wild-type plants are the two expected amplification products visible. (B) RT-PCR amplification products of AGP6 and AGP11 mRNA transcripts in pollen of wild-type Arabidopsis (wt), and of three agp6 agp11 double mutant plants (P1, P2, P3). Figures under AGP and reference gene names refer to expected sizes of PCR amplification products. (C) Light micrograph of an anther of F1 plants, heterozygous for AGP6 and AGP11. Pollen grains show a collapsing phenotype (arrows) that contrasts with the ones that are roughly spherical pollen grains, and show no phenotype (arrow head). (D) Light micrograph of an anther of an agp6 F2 plant (and heterozygous for AGP11). At the time of dehiscence, more than 50% of the pollen grains show a collapsed phenotype. (E) Light micrograph of an anther of an agp11 F2 plant (and heterozygous for AGP6), exhibiting the same pollen morphology shown in (D). (F) Light micrograph of an anther of an agp6 agp11 double mutant F3 plant. Collapsing of pollen grains is evident, while some pollen grains still show a normal morphology. (G) Scanning electron micrograph of a wild-type dehiscent anther, showing normal roundish pollen grains. (H) Scanning electron micrograph of an agp6 agp11 double mutant dehiscent anther. The collapsed pollen grains are evident. (I) Transmission electron micrograph of an agp6 agp11 double mutant. The collapse of the pollen grain is evident. (J) High magnification transmission electron micrograph of pollen grains from of an agp6 agp11 double mutant. The pollen grain in the image shows a reduced cell lumen and a well-developed exine wall (arrow). En, endothecium; Ep, epidermis; L, cell lumen; S, stomium. Bars: (C, D, E, F) 20 μm; (G, H) 60 μm; (I, J) 5 μm.
To determine whether the biological roles of AGP6 and AGP11 are redundant, the two homozygous lines were crossed to obtain agp6 agp11 double mutants, using agp6 as the male to fertilize emasculated agp11 plants. F1 plants were, as expected, all heterozygous for both insertions. Optical microscopy of anthers showed that some of the F1 pollen grains had a collapsed, aberrantly shaped phenotype (Fig. 2C). F2 plants homozygous for one of the insertions and heterozygous for the other were subsequently used for pollen grain developmental studies. In these plants more than 50% of the pollen grains inside each locule were devoid of content and had a collapsed appearance (Fig. 2D, E). This dramatic effect became apparent just before the first pollen asymmetric cell division, corresponding with the timing of the onset of GFP fluorescence in the ProAGP6:GFP transgenic plants (Fig. 1). The F3 generation obtained by self-pollination of agp6–/– agp11+/– F2 plants, produced 20.35% of plants which were homozygous for both agp6 and agp11 (n=113, χ2=4.69, P <0.05).
Comparable results were obtained by self-pollination of agp6+/– agp11–/–. The observed genotypic distortions of the offspring was mainly due to a higher than expected percentage of heterozygotes. Tests were performed by PCR for the specific transposons. The absence of expression of both genes in double null mutants was confirmed by RT-PCR studies for some plants (Fig. 2B). The observation of pollen development in these plants showed the same aberrant development of pollen, with more than 50% of pollen grains collapsing during development (Fig. 2F). It is worth noting that, in all lines, anther wall tissues were found to be well preserved and the anthers dehisced similarly to wild-type plants (Fig. 2C–F), indicating the pollen-specific nature of the phenotype. The opening of the anthers was further evaluated by scanning electron microscopy observations (Fig. 2G, H). The collapsed pollen grains phenotype was also characterized by transmission electron microscopy (Fig. 2I, J), which clearly showed the degeneration of pollen contents.
Down-regulation of AGP6 and AGP11 in transgenic plants
To complement this work, RNAi and amiRNA approaches were used to knock down the expression of the two genes in transgenic plants. 35S promoter-driven RNAi and ProAGP6-driven amiRNA constructs were generated, due to the controversy related to the efficiency of the 35S promoter in pollen. The sequences used had homology to both AGP6 and AGP11, so as to obtain transgenic plants down-regulated for both genes.
The down-regulation of both AGP6 and AGP11 transcript levels were confirmed in the anthers of the RNAi plants by conventional RT-PCR and in the anthers of the amiRNA plants by real-time RT-PCR (Fig. 3A, B).
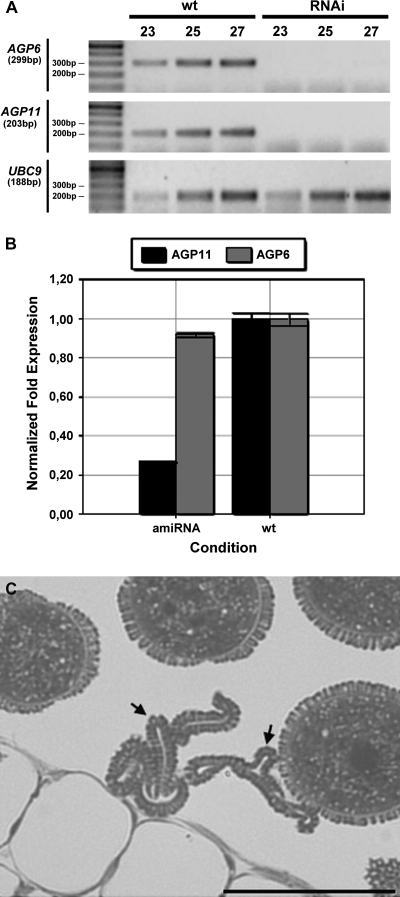
Fig. 3.
Down-regulation of AGP6 and AGP11 in RNAi and amiRNA plants. (A) RT-PCR amplification products of AGP6 and AGP11 mRNA transcripts in mature anthers of wild-type (wt) and RNAi Arabidopsis. Figures in the top row refer to PCR cycle numbers. UBC9 was used as the reference gene. (B) Real-time RT-PCR amplification products of AGP6 and AGP11 mRNA transcripts in anthers of wild-type (wt) and amiRNA Arabidopsis. In the panel each bar represents an average of two independent reactions and technical replicates. The anthers were at stage 10 of pollen development according to Smyth et al. (1990). AGP6 and AGP11 transcript levels were normalized to UBC9 levels. (C) Light micrograph of an anther from a plant exhibiting an amiRNA construction. Some pollen grains show a collapsing phenotype (arrow) while others look phenotypically normal. Bar: 20 μm.
Optical microscopy of pollen grains in the anthers of the amiRNA transgenic lines showed aborted pollen grains. These pollen grains were found to be collapsed and with no contents (Fig. 3C). The anther wall was well preserved, exactly as for the knockout mutants (Fig. 2F).
To evaluate further the timing when degeneration starts in these transgenic lines, pollen development in wild-type plants (Fig. 4A) was compared with pollen development in plants with AGP6 and AGP11 down-regulated by RNAi. The phenotype of these transgenic lines was analysed further by electron microscopy observations of anthers. Both types of plants show a normal pattern of development up to the young microspore stage. No differences were observed inside the anther locules, during the whole process of meiotic division up to the tetrad stage of pollen development (Fig. 4B). The collapse of pollen grains starts at the stage of the young microspore and is accompanied by extensive shrinkage of the cytoplasm (Fig. 4C), blebbing of the plasma membrane, and a striking separation of the plasma membrane from the intine wall (Fig. 4D) compared to the wild type (Fig. 4F). In some of the observed pollen grains, asymmetric cell division occurred, although the second pollen mitosis was never observed and lytic vacuoles appeared in the entire pollen cytoplasm (Fig. 4E). At later stages of development, the collapse of pollen was even more remarkable (Fig. 4G, H) due to the degeneration of all the pollen grain contents. Although with an empty body, pollen grains are prepared to be shed as in the wild type, with a well-developed layer of pollen coat deposited by the tapetum cells (Fig. 4H).
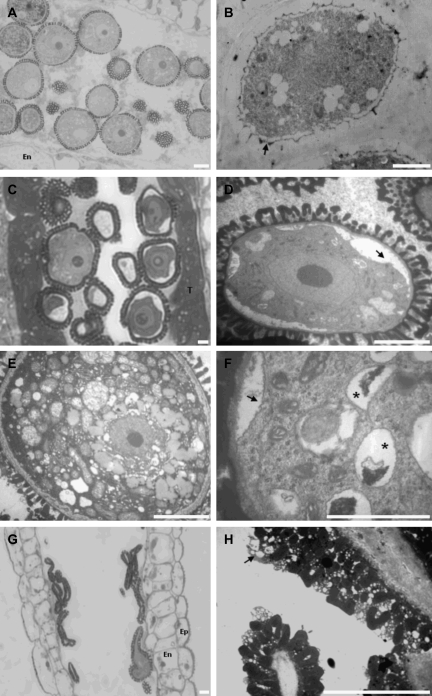
Fig. 4.
Microscopy images of plants down-regulated for AGP6 and AGP11 by RNA interference (RNAi). (A) Light micrograph of an anther from a wild-type plant showing normal pollen morphology at the stage of mature microspores. (B) Electron micrograph of an anther from an RNAi plant at the tetrad stage of development. The young microspore, surrounded by the callose wall, shows the initiation of the building of the exine wall (arrow). (C) Light micrograph of an anther from an RNAi plant at the beginning of the microspore stage of development. The anther shows a well-developed tapetum. As expected for this stage of development the endothecium is not differentiated yet. (D) Electron micrograph of a pollen grain from the anther in (C); the retraction of the plasma membrane is evident (arrow). The exine wall shows its final architecture. (E) Electron micrograph of a wild-type pollen grain. (F) Higher magnification of one of the pollen grains in (C), showing the presence of lytic vacuoles (*) as well as the cytoplasm retraction (arrow). (G) Light micrograph of an anther from an RNAi plant at the end of pollen development. It is evident the collapsing of all the pollen grains observed inside the pollen sac. (H) Electron micrograph detail from the same anther in (G). The anther releases empty pollen grains completely devoid of contents, with a well-developed pollen coat (arrow). Ep, epidermis; En, endothecium; T, tapetum. Bars: 5 μm.
Discussion
AGP6 and AGP11 encode arabinogalactan proteins that belong to the classical AGP subfamily. Both are strongly and specifically expressed in pollen (Pereira et al., 2006). By careful inspection of the images collected of ProAGP6:GFP plants, no evidence was found of GFP fluorescence outside pollen grains or pollen tubes. The green yellow light emitted by the anther walls at later stages of development are due to the autofluorescence typical of the lignin thickenings of the endothecium. These data contribute to the increasingly comprehensive view of the behaviour of individual genes in each gene family and of the gametophytic reprogramming of gene expression in Arabidopsis.
A reverse genetics approach was undertaken in an attempt to characterize further the biological significance of the two genes, AGP6 and AGP11, during pollen development. In the single T-DNA insertional null mutants of either AGP6 or AGP11, no macroscopic or microscopic phenotypic alterations were observed, as compared to wild-type Arabidopsis, corroborating a suggestion hypothesized earlier that these two genes have redundant functions (Pereira et al., 2006). These conclusions are conflicting to those reached by Levitin et al. (2008) who used lines of Arabidopsis carrying single point mutations in AGP6. It is not clear how single point mutations in AGP genes could affect function so dramatically. Even if a proline hydroxylation site or an O-Hyp glycosylation site was affected, the polypeptide chains of AGPs are thought to be abundantly and repetitively glycosylated by the specific population of glycosyltransferases. One sugar chain less may be predicted to have minor consequences.
The production of double or multiple gene null mutants is of crucial importance to the study of gene function in Arabidopsis.
In plants homozygous for one of the insertions and heterozygous for the other and in double homozygous mutant lines, many of the pollen grains failed to develop normally and collapsed, indicating that the genes are important gametophytically for pollen development.
Mutants are gametophytic if they disrupt genes that act after meiosis in the haploid phase of pollen development, and thus only the pollen grains carrying the mutant allele are expected to be affected. Whereas this may explain the presence of both normal and abnormal pollen in the F1 heterozygous lines, in the homozygous agp6 agp11 mutants there was some pollen viability, since a low percentage of the pollen grains were able to develop and germinate into functional pollen tubes, as assessed by the presence of seeds in self-pollinated plants. This indicates either that AGP6 and AGP11 are non-essential for stabilizing pollen grain development, or that an alternative pathway, potentially involving the ectopic or up-regulation of the expression of other AGPs, is able partially to compensate for the loss of the two proteins. Indeed, the Arabidopsis AGP family contains at least 47 members and several AGPs have been shown to be expressed in pollen grains (Schultz et al., 2002; Lalanne et al., 2004). In many multigene families, redundancy frequently thwarts efforts to obtain a phenotype and thus infer a function for the protein. Edelman and Gally (2001) suggested that gene degeneracy should be considered in addition to redundancy, signifying that important development processes may be ‘covered’ by several proteins, any of which might accomplish the task in the absence of another. For instance, although the MADS box gene, AGAMOUS (AG), is essential for stamen and carpel development, it has been shown that, in some mutant backgrounds, even in the absence of AG, sepals can be converted into carpelloid organs (Pinyopich et al., 2003). This indicates an AG-independent pathway for carpel development. This may help to explain why some pollen grains developed in the homozygous agp6 agp11 background.
Levitin et al. (2008) recently showed that AGP6 and AGP11 are involved in pollen tube growth. The authors suggest that the reduction of AGP6 and of AGP11 expression by RNAi caused inhibition of pollen tube growth and hampered pollen release. By contrast, it has been shown here that the reduction in fertility is due to the abortion of pollen grains during development. Knockout and knockdown mutants have been studied in detail and the only morphological trait affected by the absence or down-regulation of AGP6 and AGP11 is pollen abortion.
In recent years, a number of AGP mutants have been generated that are helping to identify the function of the products of various gene family members and to provide a framework to explore the underlying mechanisms of AGP function. A study of the overexpression of tomato LeAGP1 linked this protein to cytokinin signalling (Sun et al., 2004). AGP30 was shown to be required for root regeneration and seed germination and to enhance the plants response to abcisic acid (van Hengel and Roberts, 2003; van Hengel et al., 2004). AGP18 is essential for female gametogenesis as functional megaspores in RNAi plants failed to enlarge and divide, resulting in ovule abortion and reduced seed set (Acosta-Garcia and Vielle-Calzada, 2004). AGP17 might influence Agrobacterium binding, either by providing a binding site on the root surface or by reducing free salicylic acid levels through a signal transduction pathway involving the GPI anchor (Gaspar et al., 2004).
As all classical AGPs possess GPI anchors, it has been suggested that AGPs are involved in interactions with other membrane proteins, in cell–cell recognition events and in signal transduction pathways (Seifert and Roberts, 2007).
Lalanne et al. (2004) have shown that preventing GPI anchor addition results in pollen tube growth defects. Heterozygous GPI biosynthetic knockout mutations in Arabidopsis had no effects on sporophytic development and megagametogenesis, but showed male gametophyte-specific effects that almost completely blocked transmission through pollen. Similar experiments (Alfieiri et al., 2003) were used to show that GPI-anchored proteins were important for sperm-egg adhesion in mammals.
On the whole, the evidence presented here indicates that AGP6 and AGP11 are functionally redundant genes with important roles in Arabidopsis pollen grain development.
Acknowledgments
We thank Raquel Figueiredo for helping with the amiRNA constructs. This work was supported by funding from FCT (Fundação para a Ciência e Tecnologia, Portugal) within the project PTDC/AGR-GPL/67971/2006.
References
- Acosta-Garcia G, Vielle-Calzada JP. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. The Plant Cell. 2004;16:2614–2628. doi: 10.1105/tpc.104.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieiri JA, Martin AD, Takeda J, Kondoh G, Myles DG, Primakoff P. Infertility in female mice with an oocyte-specific knockout of GPI-anchored proteins. Journal of Cell Science. 2003;116:2149–2155. doi: 10.1242/jcs.00430. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wang H, Wu HM. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell. 1995;82:383–393. doi: 10.1016/0092-8674(95)90427-1. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa M, Pereira LG. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. Journal of Experimental Botany. 2007;58:4027–4035. doi: 10.1093/jxb/erm259. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Monteiro L, Pereira LG, Sottomayor M. Arabinogalactan proteins as molecular markers for generative cell differention and development in Arabidopsis thaliana. Acta Biologica Cracoviense. 2005;47(Supplement 1):36. [Google Scholar]
- Coimbra S, Duarte C. Arabinogalactan proteins may facilitate the movement of pollen tubes from the stigma to the ovules in Actinidia deliciosa and Amaranthus hypocondriacus. Euphytica. 2003;133:171–178. [Google Scholar]
- Coimbra S, Salema R. Immunolocalization of arabinogalactan proteins in Amaranthus hypocondriacus L. ovules. Protoplasma. 1997;199:75–82. [Google Scholar]
- Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proceedings of the National Academy of Sciences, USA. 2001;98:13763–13768. doi: 10.1073/pnas.231499798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar YM, Nam J, Schultz CJ, Lee LY, Gilson PR, Gelvin SB, Bacic A. Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of Agrobacterium transformation. Plant Physiology. 2004;135:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Nothnagel EA. Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiology. 2004;135:1–21. doi: 10.1104/pp.104.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Oh S-A, Renak D, Donders M, Solcova B, Johnson JA, Boudova R, Twell D. Identification of microspore-active promoters that allow targeted manipulation of gene expression at early stages of microgametogenesis in Arabidopsis. BMC Plant Biology. 2006;6:31. doi: 10.1186/1471-2229-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Sciences. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, Akiyama K, Kamiya A, Ito T, Shinozaki K. A collection of 11800 single-copy Ds transposon insertion lines in Arabidopsis. The Plant Journal. 2004;32:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GH, Lilley KS, Dupree P, Grossniklaus U, Tweel D. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. The Plant Cell. 2004;16:229–240. doi: 10.1105/tpc.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Moriyah Z. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. The Plant Journal. 2008;56:351–363. doi: 10.1111/j.1365-313X.2008.03607.x. [DOI] [PubMed] [Google Scholar]
- Motose H, Suriyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Pereira LG, Coimbra S, Oliveira H, Monteiro L, Sottomayor M. Expression of arabinogalactan protein genes in pollen tubes of Arabidopsis thaliana. Planta. 2006;223:374–380. doi: 10.1007/s00425-005-0137-4. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Quan L, Xiao R, Li W, Oh SA, Kong H, Ambrose JC, Malcos JL, Cyr R, Twell D, Ma H. Functional divergence of the duplicated AtKIN14a and AtKIN14b genes: critical roles in Arabidopsis meiosis and gametophyte development. The Plant Journal. 2008;53:1013–1026. doi: 10.1111/j.1365-313X.2007.03391.x. [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Gilson P, Oxley D, Youl J, Bacic A. GPI anchors on arabinogalactan-proteins: Implications for signaling in plants. Trends in Plant Science. 1998;3:426–431. [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiology. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Barber C, Wells B, Dolan L, Roberts K. Galactose biosynthesis in Arabidopsis: genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Current Biology. 2002;12:1840–1845. doi: 10.1016/s0960-9822(02)01260-5. [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annual Review of Plant Biology. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- Serpe MD, Nothnagel EA. Effects of Yariv phenylglycosides on Rosa cell suspensions: evidence for the involvement of arabinogalactan-proteins in cell proliferation. Planta. 1994;193:542–550. [Google Scholar]
- Showalter AM. Arabinogalactan proteins: structure, expression and function. Cell and Molecular Life Sciences. 2001;58:1399–1417. doi: 10.1007/PL00000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Kieliszewski MJ, Showalter AM. Overexpression of tomato LeAGP1 arabinogalactan protein promotes lateral branching and hampers reproductive development. The Plant Journal. 2004;40:870–881. doi: 10.1111/j.1365-313X.2004.02274.x. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kitagawa M, Knox JP, Yamaguchi I. A role for arabinogalactan proteins in gibberellin-induced α-amylase production in barley aleurone cells. The Plant Journal. 2002;29:733–741. doi: 10.1046/j.1365-313x.2002.01259.x. [DOI] [PubMed] [Google Scholar]
- Toller A, Brownfield L, Neu C, Twell D, Schulze-Lefert P. Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. The Plant Journal. 2008;54:911–923. doi: 10.1111/j.1365-313X.2008.03462.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Barber C, Roberts K. The expression patterns of arabinogalactan protein AtAGP30 and GLABRA2 reveal a role for abscisic acid in the early stages of root epidermal patterning. The Plant Journal. 2004;39:70–83. doi: 10.1111/j.1365-313X.2004.02104.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. The Plant Journal. 2002;32:105–113. doi: 10.1046/j.1365-313x.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. AtAGP30, an arabinogalactan protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. The Plant Journal. 2003;36:256–270. doi: 10.1046/j.1365-313x.2003.01874.x. [DOI] [PubMed] [Google Scholar]
- Vissenberg K, Feijó JA, Weisenseel MH, Verbelen JP. Ion fluxes, auxin and the induction of elongation growth in Nicotiana tabacum cells. Journal of Experimental Botany. 2001;52:2161–2167. doi: 10.1093/jexbot/52.364.2161. [DOI] [PubMed] [Google Scholar]
- Willats WGT, Knox JP. A role for arabinogalactan proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. The Plant Journal. 1996;9:919–925. doi: 10.1046/j.1365-313x.1996.9060919.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang H, Cheung AY. A pollen tube growth simulator glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;82:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Yang J, Showalter AM. Expression and localization of AtAGP18, a lysine-rich arabinogalactan-protein in Arabidopsis. Planta. 2007;226:169–179. doi: 10.1007/s00425-007-0478-2. [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes and Development. 1999;15:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W. Geneinvestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]