Abstract
Genetic engineering of chloroplasts normally requires the stable introduction of bacterial derived antibiotic or herbicide-resistance genes as selective markers. Ecological and health concerns have been raised due to the presence of such genes within the environment or the food supply. One way to overcome this issue is the use of plant genes able to confer a metabolic or developmental advantage to the transformed cells manipulating the plant's biosynthetic pathways. We explored the feasibility of using, for plastid transformation, the selection system based on the feedback-insensitive anthranilate synthase (AS) α-subunit gene of tobacco (ASA2) as a new selective marker and the indole analogue 4-methylindole (4MI) or the tryptophan analogue 7-methyl-DL-tryptophan (7MT) as the selection agents. An expression cassette containing Prrn-ASA2 was effectively integrated into the region between accD and ycf4 of the tobacco plastome by the biolistic process. Plastid transgenic plants were obtained on medium supplemented with 300 μM 7MT or 4MI. Transplastomic plants showed normal phenotype and fertility and the resistance to the selection agents 7MT and 4MI was transmitted maternally. The plastid transformed lines also exhibited a higher level of AS enzyme activity that was less sensitive to Trp-feedback inhibition and, consequently, increased free Trp levels in leaves about 7-fold.
Keywords: Anthranilate synthase, 7-methyl-DL-tryptophan, 4-methylindole-tryptophan, non-antibiotic selection, plastid transformation, selectable marker
Introduction
The advantages of chloroplast genetic engineering are numerous and have been extensively reviewed (Bock, 2007; Lutz et al., 2007; Verma and Daniell, 2007). They include the high-level accumulation of foreign proteins (Oey et al., 2009), a lack of epigenetic effect and gene silencing, the possibility of gene stacking, and containment of the transgene due to the maternal inheritance of chloroplasts in most of the crop plants (Ruf et al., 2007).
The successful recovery of a genetically stable transplastomic plant depends on the capability to selectively amplify the one or very few plastid genome (ptDNA) copies that are transformed in the initial biolistic experiment and the key element for this process is the choice of selective markers.
There are two classes of plastid marker genes: ‘primary selective markers’ to be used for direct selection (aadA, neo, and aphA-6) and ‘secondary selective markers’ (bar or CP4) that are not suitable for direct selection if only a few ptDNA copies are transformed, but will allow selection when most of the ptDNA copies are transformed (Maliga, 2004; Lutz et al., 2007). The ‘primary selective markers’ have bacterial origins and confer resistance to an antibiotic: the aadA gene confers resistance to spectinomycin and streptomycin (Svab and Maliga, 1993; Zoubenko et al., 1994) while neo (Carrer et al., 1993) and aphA-6 (Huang et al., 2002) confer resistance to kanamycin.
The ‘secondary selective markers’ are bacterial-derived as well and confer resistance to herbicides like phosphinothricin (bar) (Lutz et al., 2001) or glyphosate (CP4) (Ye et al., 2003).
Over the past several years, consumer and environmental groups have expressed ethical and biosafety concerns about the use of antibiotic- and herbicide-resistance genes derived from micro-organisms (Miki and McHugh, 2004). Therefore, strategies have been developed either (i) to excise the marker gene, once the selection is complete, in order to obtain a marker-free transplastomic plant or (ii) to use plant-derived markers.
Four protocols have been established to date for obtaining marker-free transplastomic plants: (i) homology-based excision via direct repeated sequences (Iamtham and Day, 2000); (ii) cotransformation–segregation (Carrer and Maliga, 1995); (iii) transient cointegration of the marker gene (Klaus et al., 2004), and (iv) excision by phage site-specific recombinases (Corneille et al., 2001). In the first two protocols the final transplastomic line is identified in a genetically unstable/segregating ptDNA population making it difficult to obtain the marker-free transplastomic plants. The third protocol is an antibiotic/phenotypic selection system that utilizes plastid mutants (e.g. knockout of a photosynthetic gene that produces a chlorophyll-deficient phenotype with pale-green leaves) as target material for chloroplast transformation. The need to produce the knockout mutant prior to the actual transformation makes the system not very convenient. The last protocol uses two different site-specific recombinases (Cre and Int) for plastid marker gene excision and, with both of them, the problem is the presence of potential pseudo sites in the ptDNA recognized by the enzyme.
Recently, a spinach (Spinacia oleracea)-derived selective marker betaine aldehyde dehydrogenase (BADH) gene has been used for plastid transformation in tobacco (Daniell et al., 2001). However, these results have not been repeatable (Maliga, 2004; Whitney and Sharwood, 2008).
Anthranilate synthase (AS) catalyses the first reaction in the tryptophan (Trp) biosynthetic pathway by converting chorismate to anthranilate. Plant AS is a heterotetramer containing two large α-subunits and two small β-subunits (Romero et al., 1995) and is feedback-regulated by the end-product, Trp, which binds to the α-subunit allosteric site (Bohlmann et al., 1996; Li and Last, 1996). A naturally occurring feedback-insensitive α-subunit gene (ASA2) has been isolated from a tobacco suspension-culture cell line resistant to the toxic Trp analogue 5-methyltryptophan (5MT) (Song et al., 1998). Trp analogues are toxic since they are able to mimic the specific feedback effect of Trp on the AS, therefore inhibiting Trp biosynthesis and causing Trp deficiency for protein synthesis. It has also been shown that plant cell cultures can convert various indole analogues to Trp analogues via the action of Trp synthase (Widholm, 1981). A feedback-sensitive AS enzyme fails to discriminate completely between its normal feedback inhibitor, Trp, and an analogue; hence if the feedback-insensitive ASA2 is expressed then the cells are resistant to the analogues and can be selected for.
Previously (Barone and Widholm, 2008), the efficacy of the ASA2 gene as a selective marker was described using either 4-methylindole (4MI) or 7-methyl-DL-tryptophan (7MT) as the selection agent for tobacco nuclear transformation and now the possibility of using this non-antibiotic, native plant gene selection system for tobacco plastid transformation is investigated.
Materials and methods
Plasmid vector
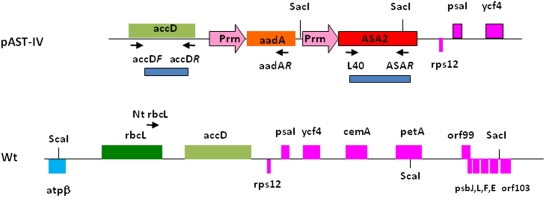
The vector pAST-IV (Fig. 1) as previously described by Zhang et al. (2001) contains a 1671 bp long modified version of the ASA2 gene (GenBank gi: 3348123) without a putative transit peptide and with an ASA2 3′-non-coding region (204 bp) as the termination sequence. The plasmid also has the aadA gene with the Chlamydomonas rbcL 3′-untranslated region. Both genes are driven by a chloroplast 16S rRNA promoter Prrn (Goldschmidt-Clermont, 1991; Eibl et al., 1999) and the site of insertion in the plastid genome is the intergenic region between accD and ycf4 at nucleotide 59029 and nucleotide 63410 (GenBank Z00044 or NC_001879), respectively (Fig. 1).
Fig. 1.
Structure of the vector pASTIV used for tobacco transformation and its corresponding region of the wild-type tobacco plastid genome based on GenBank Z00044 and NC_001879. Relevant plastid genes and their orientation are presented. Prrn: plastid 16S rRNA operon promoter, aadA: spectinomycin resistance gene with the Chlamydomonas rbcL 3′-untranslated region (500 bp), ASA2, modified version of ASA2 (1671 bp) including its termination sequence (204 bp). The locations of SacI and ScaI used for Southern hybridization are shown. Location and orientation of PCR primers are indicated by arrows. accD probe (accD F–accD R) and ASA2 probe (L40–ASAR) are shown. The figure size is not to scale.
Bombardment and regeneration of chloroplast transgenic plants
Plastid transformation was carried out as described in Svab and Maliga (1993) with some modifications. Briefly, one fully expanded, dark-green leaf of tobacco (Nicotiana tabacum) ‘Petit Havana’ grown in sterile culture was placed abaxial side up on RMOP medium (Svab et al., 1990) and the transforming DNA was introduced by the biolistic process using the biolistic device PDS1000/He and 0.6 μm gold particles (Bio-Rad, Hercules, CA). Two days after bombardment, leaf discs (6 mm in diameter each) were cut out from the leaves and transferred to RMOP medium containing either 300 μM 4MI (Acros Organics Morris Plains, NJ, USA) or 300 μM 7MT (Sigma-Aldrich, St Louis, MO, USA). Every other week the plant material was transferred onto new fresh 7MT/4MI-containing RMOP medium and further chopped into small pieces when needed in order to ensure a continuous contact between the leaf material and the selection medium. The 4MI/7MT resistant clones were identified as green shoots 6–12 weeks after bombardment. Leaves from PCR positive shoots were subjected to three additional rounds of regeneration/selection on RMOP-4MI/7MT medium in order to obtain the homoplastic condition. Resistant shoots from the third round of selection were transferred to the rooting medium with 75 μM of either 4MI or 7MT. Rooted plants were then moved to soil in a greenhouse after gently washing off the agar-solidified rooting medium. Two transformation experiments were performed.
Nucleic acids analysis
Total cellular DNA was extracted using the DNeasy plant kit (Qiagen, Valencia, CA, USA). PCR was carried out to identify the transgene insertion, with Taq DNA polymerase (New England Biolabs Inc., Ipswich, MA, USA) for 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 1.5 min. The primers used for amplification of a 4.4 kb fragment were aadAR (5′-ACCTTAGTGATCTCGCCTTTCACG-3′), located at the end of the aadA and NtrbcL (5′-TTTGCAGCAGTGGACGTTTTGGATAA-3′), located in the 3’ end of the plastid gene rbcL (Fig. 1)
For Southern hybridization analysis restriction endonuclease treatment of 5 μg DNA per sample was performed using 5 units μg−1 of ScaI or SacI (New England Biolabs Inc.) enzyme in the manufacturer's buffer, at 37 °C for 5 h. The digested DNA was separated by 0.8% agarose gel electrophoresis in 1× TAE buffer and then blotted onto a nylon membrane (Hybond-N+, GE Healthcare Limited, Little Chalfont, UK) and cross-linked to the membrane by UV. The probes were prepared by PCR using primers designed to amplify either a 815 bp fragment from the ASA2 gene (L40 5′-CTAAAAGCGGGAACTTGATTCCGC-3′ and ASAR 5′-TCTGTACACTTCAAATGGGTCAGC-3′) or a 758 bp fragment from the accD gene (accD F 5′-TATAGGGCCGTTTGTGGTGGTGAA-3′ and accD R 5′-AATGCAATGTAGGCGTTGGGTTCG-3′) (Fig. 1). The probes were purified with QIAquick PCR purification kit (Qiagen) and then radiolabelled with α-32P-labelled dCTP (3000 Ci mmol−1) through the random primer method using a Megaprime kit (GE Healthcare Limited). Prehybridization, hybridization, and subsequent washing steps were performed according to the standard protocols (Sambrook et al., 1989). Signals were detected by exposing the blots to X-ray films (Denville Scientific Inc., Metuchen NJ, USA) for 1–5 d at −70 °C.
Total RNA was extracted from young expanded leaves with RNeasy kits (Qiagen) and treated with rDNase I RNase-Free (USB Biochemicals, Cleveland, OH, USA) to eliminate any genomic DNA contamination.
For northern analysis 5 μg of total RNA were separated on a 1% agarose gel in formaldehyde/MOPS buffer, transferred to a nylon membrane and cross-linked to the membrane by UV as described above. The membrane was prehybridized for 2 h at 42 °C in 5× SSPE, 5× Denhardt's solution, 50% formamide, 0.1% SDS, and 20 mg l−1 sheared salmon sperm DNA and then hybridized with the same ASA2 probe used for the Southern analysis at 42 °C overnight. After washing, the membrane signals were detected with X-ray films as described above.
RT-PCR (reverse transcription-polymerase chain reaction) was performed on total RNA treated with rDNase I RNase-Free using the same primer set as for the PCR probe amplification (L40–ASAR). The fragments produced were cloned into a pGEM®-T Easy Vector System I (Promega Corporation, Madison, WI, USA) and sequenced. A PCR reaction was performed with the same primers without the reverse transcriptase step to demonstrate the absence of genomic DNA contamination in the samples.
Plant crosses
To obtain seeds to test for the maternal inheritance of the ASA2 gene, the anthers of closed green tobacco flowers, from either WT or the transplastomic plants were removed with tweezers prior to their dehiscence and covered with a plastic bag. At the beginning of colour formation of the petals of the emasculated flowers (1–2 d after emasculation) paternal pollen was collected from either WT or the transplastomic plants, resuspended in sterile distilled water and a 5 μl volume of pollen suspension was transferred onto the top of the stigma of each emasculated flower. Flowers were bagged again and seeds collected after 4–5 weeks. Six randomly chosen lines were used for the plant crosses three selected on 300 μM 7MT (lines 26, 27, 51) and three selected on 300 μM 4MI (A, C, F).
AS assays and Trp measurement
AS enzyme activity was measured in crude protein extracted from 2 g of leaves using the buffer of Bernasconi et al. (1994) and desalted using an Econo-Pac 10DG column (Bio-Rad). Protein concentration was determined using a protein dye-binding assay kit (Bio-Rad). AS enzyme activity was measured as the conversion rate of chorismate to anthranilate as described previously (Song et al., 1998). Either 100 mM NH4Cl or 10 mM glutamine (Gln) was added to the assay mixture to determine α-subunit activity or AS holoenzyme activity, respectively, and Trp at different concentrations to measure feedback inhibition. The total extract of a single leaf of three different plants for each line was analysed.
Free Trp was extracted with 0.1 N HCl from a young expanded leaf from the top of 35 d-old-plants grown in the growth chamber and measured as described by Cho et al. (2000).
Results
Selection and regeneration of transgenic plants
Six to twelve weeks after bombardment with pAST-IV, resistant green shoots appeared from the brown tobacco leaf pieces placed on the selection medium (Fig. 2a, b). Integration of the ASA2 gene into the chloroplast genome was first confirmed by PCR screening of transformants (Fig. 3a) using a primer set (NtrbcL–aadAR) designed to eliminate lines with nuclear integration of the transgene and escapes (Fig. 1).
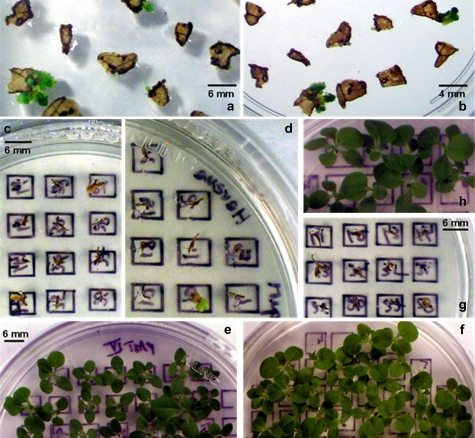
Fig. 2.
Bombarded tobacco leaf material on RMOP/7MT 300 μM (a) or RMOP/4MI 300 μM (b) with green resistant shoots. Seed germination of WT (N. tabacum, Petit Havana) on either 4MI (c) or 7MT (d) 75 μM. Seedlings from seeds of self-pollinated transplastomic plant 27 on 4MI (e) and plant C on 7MT (f) 75 μM. Germination of seeds on 7 MT 75 μM from either WT plant pollinated with pollen from transplastomic plant C (g) or from transplastomic plant C pollinated with pollen from WT (h).
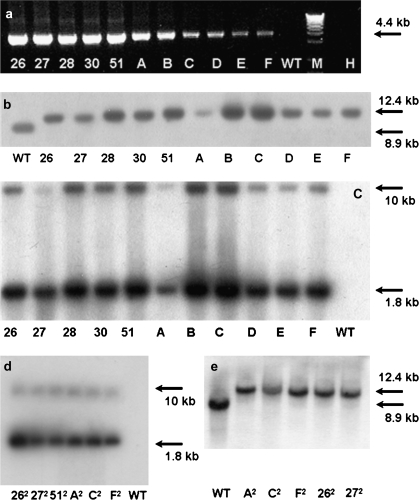
Fig. 3.
Plastid genomic analysis of site-specific integration of the ASA2 gene: (a) PCR amplification with primer aadAR (aadA-specific) and primer NtrbcL (plastid site-specific-see Fig. 1); for Southern blot DNA was digested with either ScaI and hybridized to the plastid site-specific probe accD (b, e) or SacI and hybridized to an ASA2 probe (c, d). Lines 26–51 selected on 300 μM 7MT and lines A–F selected on 300 μM 4MI. WT, untransformed control; H, blank control/water; M, DNA ladder. Lines 262–512 F1 crosses (male non-transgenic×female transgenic) germinated on 75 μM 7MT and lines A2–F2 F1 crosses (male non-transgenic×female transgenic) germinated on 75 μM 4 MI. WT, untransformed control; H: blank control/water.
In the first experiment, 24 independent resistant clones were obtained from 130 leaves bombarded with vector pAST-IV. Ten clones were recovered from medium containing 7MT and 14 from medium containing 4MI. The PCR screening showed that five (two on 7MT and three on 4MI) out of the 24 putative transformants recovered after the first round of selection were positive, as shown by the presence of the expected 4.4 kb PCR product.
In the second experiment, 32 (15 on 7MT and 17 on 4MI) independent resistant clones were recovered from 140 leaves bombarded. The PCR analysis confirmed six of them to be chloroplast transformants (three on 7MT and three on 4MI). As shown in Fig. 3a no PCR fragment was produced in the WT since primer aadAR is located in the coding region of the aadA gene. PCR positive lines were subjected to three rounds of additional regeneration/selection with either 7MT or 4MI and then the green shoots were allowed to root and grow into plantlets.
Nucleic acids analysis: integration and expression of the transgene
To prove the homoplastomic condition of the transformants after the third round of selection, Southern-blot hybridizations were carried out with either an accD gene probe or an ASA2 gene probe. As previously shown in Zhang et al. (2001) when hybridized with a 0.7 kb accD probe a ScaI DNA-digested blot results in an 8933 bp band corresponding to the native plastid accD-ycf4 region in the WT (GenBank NC_001879; Fig. 1), but generates a 12.4 kb fragment in the pASTIV plants due to the insertion of the Prrn-aadA-Prrn-ASA2 cassette in the ptDNA (Figs 1, 3b). The SacI DNA-digested gel hybridized with a 0.8 kb ASA2 gene probe shows two bands at 1.8 kb and 10 kb (Fig. 3c) for the transformant plants as a result of the two SacI sites in the Prrn-ASA2 cassette (Fig. 1) and one downstream of ycf4. No signal was detected in the WT, even though the tobacco nuclear genome contains a small AS α-subunit gene family including ASA2 (Zhang et al., 2001), probably due to the relatively low amount of total cellular DNA used. The 1.8 kb band is much stronger because the AS probe covers most of the ASA2 gene (Fig. 1). The same results were obtained with DNA extracted from F1 crosses (male non-transgenic×female transgenic) (Fig. 3d, e) confirming the insertion of the ASA2 gene in the ptDNA of the progenies and their homoplastomy.
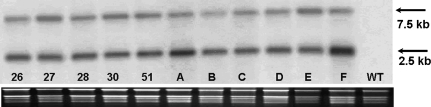
To determine the expression of the Prrn-ASA2 gene in the transformed plants northern-blot hybridization and RT-PCR were performed. The northern analysis showed two transcripts, one of 2.5 kb and one of 7.5 kb (Fig. 4). This result is in accordance with Zhang et al. (2001), indicating that the 2.5 kb transcript in the pAST-IV plants ends in the intergenic region between accD and psaI, whereas the 7.5 kb mRNA is the result of a transcribed operon that includes ASA2 and genes downstream of the ycf4 gene. ASA2 mRNA could not be detected by northern blot analysis in the leaf of WT plants (Fig. 4) most likely because as previously shown (Song et al., 1998; Zhang et al., 2001) the naturally occurring ASA2 gene is expressed at a very low level in the WT plants.
Fig. 4.
Northern blot analysis of the total RNA probed with ASA2 probe (see Fig. 1). Plants in lanes 26–51 were selected on 300 μM 7MT and plants in lanes A–F were selected on 300 μM 4MI. WT, untransformed control. The ethidium bromide-stained 26S, 18S, 16S, and 23S rRNAs are shown at the bottom to indicate RNA loading.
RT-PCR produced the expected 816 bp PCR fragments from the transgenic plants and the sequence analysis of the fragments confirmed their identity (data not shown).
Maternal inheritance
Germination of seeds from WT and self-pollinated transplastomic plants on rooting medium with either 7MT or 4MI (75 μM) resulted in small brown, WT seedlings that stopped growing after 10–14 d (Fig. 2c, d) whereas for the transformant lines the seedlings were green and normal-looking (Fig. 2e, f). Pollination of the WT plants with pollen from transplastomic plants produced progeny sensitive (Fig. 2g) to the analogues while seeds from transplastomic plants pollinated with the WT pollen were able to germinate on selective medium containing analogues (Fig. 2h) demonstrating the maternal inheritance of the ASA2 gene inserted in the ptDNA.
ASA2 enzyme activity and free Trp measurement
When the AS holoenzyme activity was measured without Trp inhibition the leaf extracts from transformed rooted plants showed activity 4-fold that of the untransformed line (line 27, 96.4±17 pmol min−1 mg−1 protein; line C, 97.2±37 pmol min−1 mg−1 protein; WT, 23.05±37 pmol min−1 mg−1 protein) suggesting that the α-subunits produced by the ASA2 gene in the plastid can functionally couple with the free β-subunits present in the chloroplast, increasing the capacity to convert chorismate into anthranilate.
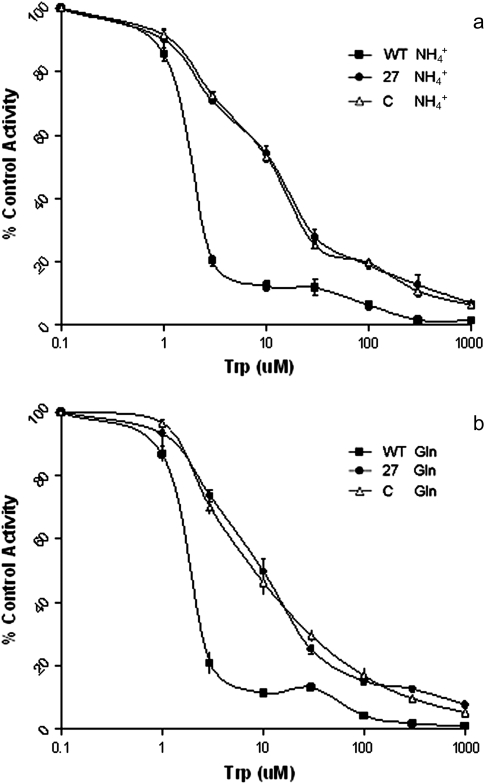
The AS activity from the transgenic plants showed less sensitivity to Trp inhibition compared to the untransformed tobacco. When the α-subunit activity was measured (Fig. 5a) at 10 μM Trp lines 27 and C still retained about 50% of the activity whereas the WT had about 15% of the activity. Similar results were found for the relative AS holoenzyme activities at 300 μM Trp where the untransformed line is almost completely inhibited while the plastid lines still maintain about 15% of the total activity (Fig. 5b). The estimated Ki values causing 50% inhibition of the AS holoenzyme activity were 2.3, 10.2, and 10.8 μM for WT, line 27, and line C, respectively.
Fig. 5.
Trp inhibition of AS enzyme activity in total leaf extracts from wild-type (WT) (filled squares) and transplastomic plants 27 selected on 7MT (filled circles) and C selected on 4MI (open triangles), with 100 mM NH4Cl (a) (α-subunit activity) or 10 mM Gln (b) (holoenzyme activity) as substrate. The values are means ±SD of three replicates.
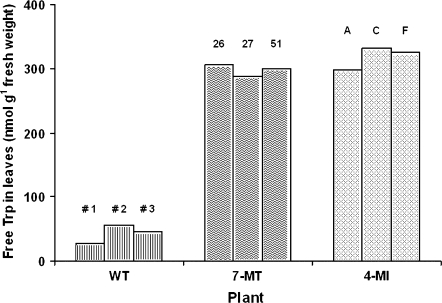
Since the AS enzyme is insensitive to feedback inhibition by Trp, the free Trp levels were measured in three WT plants, three transgenic lines selected on 7MT (27, 30, 31) and three transgenic lines selected on 4MI (B, C, D). The transplastomic lines showed a higher Trp level averaging 297±5.4 nmol g−1 fresh weight for 7MT selected lines and 318±10 nmol g-1 fresh weight for 4MI selected lines while the WT had 41±7.2 nmol g−1 fresh weight (Fig. 6).
Fig. 6.
Free Trp content in leaves of three WT plants and three transplastomic plants selected on either 7MT (26, 27, 51) or 4MI (A, C, F).
Conclusion/discussion
ASA2 has previously been used as a selectable marker gene in nuclear transformation to select the forage legume Astragalus sinicus hairy roots with the Trp analogue 5MT as the selection agent (Cho et al., 2004) and tobacco with either the Trp analogue 7MT or the indole analogue 4MI (Barone and Widholm, 2008). Zhang et al. (2001) demonstrated that tobacco transplastomic plants expressing the ASA2 gene and selected on spectinomycin were resistant to 5MT. These reports suggested that a selection system based on a feedback-insensitive AS α-subunit gene as a selectable marker could be a possible tool for chloroplast transformation as well.
Using the ASA2/7MT or 4MI selection system, it has been possible to generate transgenic tobacco plants via plastid transformation. The efficiency of transformation is comparable with the one with spectinomycin previously reported in this laboratory with the same vector pAST-IV (Zhang et al., 2001). When the leaf discs were selected for spectinomycin resistance, three individual resistant shoots were obtained from 60 leaves bombarded (Zhang et al., 2001) while under analogue selection five individual resistant lines were obtained from 130 leaves with 7MT and six individual resistant clones were obtained from 140 leaves with 4MI as reported here. The frequency of plastid transformed plants (6) among regenerated shoots (31) was 19.3% for 4MI and 20% for 7MT (5 resistant plants from 25 regenerated shoots) with the rest of the shoots being Trp analogue-resistant mutants/escapes (Widholm, 1972) or nuclear transformants. These values are comparable to the 25.3% reported for spectinomycin (20 resistant plants from 79 regenerated shoots) in Zhang et al. (2001).
The transgenic nature of the putative plastid transformants was confirmed by Southern and northern blot analysis. The gene/trait was stably and maternally inherited in the subsequent generation and the chloroplast transgenic lines grew normally, flowered, and set viable seeds showing that the over-expression of ASA2 gene did not result in any negative effects for the plants.
The plastid transformed plants have about 7-times the normal concentration of free Trp in leaves due to higher level of AS enzyme activity insensitive to Trp-feedback inhibition. Similar increases were found previously in different ASA2-expressing species like the forage legume A. sinicus (Cho et al., 2000) where a 1.3–5.5-fold increase in free Trp was observed, tobacco (Zhang et al., 2001) with free Trp 10 times higher than in the WT and soybean (Inaba et al., 2007) with a 4–5-fold increase in leaves and embryogenic tissue cultures.
The development of ASA2 as a chloroplast selective marker is important not only for the biosafety reasons reported above, but also for practical reason due to the scarcity of primary selective markers available. The use of the ASA2/7MT or 4MI selection scheme could facilitate the expansion of chloroplast transformation technology to many economically important crops like cereals that are naturally resistant to spectinomycin and for which a specific selection system still has to be established. In addition to the expression of any intended transgene(s) inserted into the plastome, increases in the essential amino acid Trp might (should) also be realized in the transformed plants as a value-added trait. For example, Zhang et al. (2001) observed a 27–29% increase in total Trp (free plus protein-bound) levels in mature, dry seeds from the transplastomic plants over-expressing ASA2.
Acknowledgments
The authors thank Wei Q. Zhong for their technical assistance and Dr. Anatoliy V. Lygin for tryptophan content measurement. This research was supported in part through The Consortium for Plant Biotechnology Research, Inc., by DOE Prime Agreement No.GO12026-175. This support does not constitute an endorsement by DOE or The Consortium for Plant Biotechnology Research, Inc., of the views expressed in this publication. This study was also supported in part by funds from the Illinois Agricultural Experiment Station, the Biotechnology Research and Development Corp. and Dow AgroSciences LLC.
Glossary
Abbreviations
- AS
anthranilate synthase
- Trp
tryptophan
- 7MT
7-methyl-DL-tryptophan
- 4MI
4-methylindole
- ptDNA
plastid genome
References
- Barone P, Widholm JM. Use of 4-methylindole or 7-methyl-DL -tryptophan in a transformant selection system based on the feedback-insensitive anthranilate synthase α-subunit of tobacco (ASA2) Plant Cell Reports. 2008;27:509–517. doi: 10.1007/s00299-007-0480-y. [DOI] [PubMed] [Google Scholar]
- Bernasconi P, Walters EW, Woodworth AR, Siehl DL, Stone TE, Subramanian MW. Functional expression of Arabidopsis thaliana anthranilate synthase subunit I in Escherichia coli. Plant Physiology. 1994;106:353–358. doi: 10.1104/pp.106.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Current Opinion in Plant Biology. 2007;18:100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Lins T, Martin W, Eilert U. Anthranilate synthase from Ruta graveolens. Duplicated ASa genes encode tryptophan-sensitive and tryptophan-insensitive isoenzymes specific to amino acid and alkaloid biosynthesis. Plant Physiology. 1996;111:507–514. doi: 10.1104/pp.111.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer H, Hockenberry TN, Svab Z, Maliga P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Molecular and General Genetics. 1993;241:49–56. doi: 10.1007/BF00280200. [DOI] [PubMed] [Google Scholar]
- Carrer H, Maliga P. Targeted insertion of foreign genes into the tobacco plastid genome without physical linkage to the selectable marker gene. Biotechnology. 1995;13:791–794. [Google Scholar]
- Cho HJ, Brotherton JE, Song H-S, Widholm JM. Increasing tryptophan synthesis in a forage legume Astragalus sinicus by expressing the tobacco feedback-insensitive anthranilate synthase (ASA2) gene. Plant Physiology. 2000;123:1069–1076. doi: 10.1104/pp.123.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, Brotherton JE, Widholm JM. Use of the tobacco feedback-insensitive anthranilate synthase gene (ASA2) as a selectable marker for legume hairy root transformation. Plant Cell Reports. 2004;23:104–113. doi: 10.1007/s00299-004-0789-8. [DOI] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Svab Z, Maliga P. Efficient elimination of selectable marker genes from the plastid genome by the CRE-lox site-specific recombination system. The Plant Journal. 2001;72:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB. Marker free transgenic plants: engineering the chloroplast genome without the use of antibiotic selection. Current Genetics. 2001;39:109–116. doi: 10.1007/s002940100185. [DOI] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, Koop H-U. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. The Plant Journal. 1999;19:333–345. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Research. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FC, Klaus SMJ, Herz S, Zuo Z, Koop HU, Golds TJ. Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Molecular Genetics and Genomics. 2002;268:19–27. doi: 10.1007/s00438-002-0738-6. [DOI] [PubMed] [Google Scholar]
- Iamtham S, Day A. Removal of antibiotic resistance genes from transgenic tobacco plastids. Nature Biotechnology. 2000;18:1172–1176. doi: 10.1038/81161. [DOI] [PubMed] [Google Scholar]
- Inaba Y, Brotherton JE, Ulanov A, Widholm JM. Expression of a feedback-insensitive anthranilate synthase gene from tobacco increases free tryptophan in soybean plants. Plant Cell Reports. 2007;26:1763–1771. doi: 10.1007/s00299-007-0381-0. [DOI] [PubMed] [Google Scholar]
- Klaus SMJ, Huang FC, Golds TJ, Koop H-U. Generation of marker-free plastid transformants using a transiently cointegrated selection gene. Nature Biotechnology. 2004;22:225–229. doi: 10.1038/nbt933. [DOI] [PubMed] [Google Scholar]
- Li J, Last RL. The Arabidopsis thaliana trp5 mutant has a feedback-resistant anthranilate synthase and elevated soluble tryptophan. Plant Physiology. 1996;110:51–59. doi: 10.1104/pp.110.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Azhagiri AK, Tungsuchat-Huang T, Maliga P. A guide to choosing vectors for transformation of the plastid genome of higher plants. Plant Physiology. 2007;145:1201–1210. doi: 10.1104/pp.107.106963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Knapp JE, Maliga P. Expression of bar in the plastid genome confers herbicide resistance. Plant Physiology. 2001;125:1585–1590. doi: 10.1104/pp.125.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Annual Review of Plant Biology. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- Miki B, McHugh S. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. Journal of Biotechnology. 2004;107:193–232. doi: 10.1016/j.jbiotec.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. The Plant Journal. 2009;57:436–445. doi: 10.1111/j.1365-313X.2008.03702.x. [DOI] [PubMed] [Google Scholar]
- Romero RM, Roberts MF, Phillipson JD. Anthranilate synthase in microorganisms and plants. Phytochemistry. 1995;39:263–276. doi: 10.1016/0031-9422(95)00010-5. [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proceedings of the National Academy of Sciences, USA. 2007;104:6879–6880. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Song H-S, Brotherton JE, Gonzales RA, Widholm JM. Tissue culture specific expression of a feedback-insensitive Nicotiana tabacum anthranilate synthase. Plant Physiology. 1998;117:533–543. doi: 10.1104/pp.117.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proceedings of the National Academy of Sciences, USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proceedings of the National Academy of Sciences, USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiology. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Sharwood RE. Construction of a tobacco master line to improve rubisco engineering in chloroplasts. Journal of Experimental Botany. 2008;59:1909–1921. doi: 10.1093/jxb/erm311. [DOI] [PubMed] [Google Scholar]
- Widholm JM. Cultured Nicotiana tabacum cells with an altered anthranilate synthetase which is less sensitive to feedback inhibition. Biochimica et Biophysica Acta. 1972;261:52–58. doi: 10.1016/0304-4165(72)90312-1. [DOI] [PubMed] [Google Scholar]
- Widholm JM. Utilization of indole analogues by carrot and tobacco cell tryptophan synthase in vivo and in vitro. Plant Physiology. 1981;67:1101–1104. doi: 10.1104/pp.67.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye GN, Colburn S, Xu CW, Hajdukiewicz PTJ, Staub JM. Persistance of unselected transgenic DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiology. 2003;133:402–410. doi: 10.1104/pp.103.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-H, Brotherton JE, Widholm JM, Portis AR. Targeting a nuclear anthranilate synthase alpha-subunit gene to the tobacco plastid genome results in enhanced tryptophan biosynthesis. Return of a gene to its pre-endosymbiotic origin. Plant Physiology. 2001;127:131–141. doi: 10.1104/pp.127.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoubenko OV, Allison LA, Svab Z, Maliga P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Research. 1994;22:3819–3824. doi: 10.1093/nar/22.19.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]