Abstract
γ-Aminobutyric acid transaminase (GABA-T) catalyses the breakdown of GABA to succinic semialdehyde. In this report, three GABA-T isoforms were identified in the tomato (Solanum lycopersicum L.) plant. The deduced amino acid sequences of the three isoforms are highly similar over most of their coding regions with the exception of their N-terminal regions. Transient expression of the individual full-length GABA-T isoforms fused to the green fluorescent protein in tobacco suspension-cultured cells revealed their distinct subcellular localizations to the mitochondrion, plastid or cytosol, and that the specific targeting of the mitochondrion- and plastid-localized isoforms is mediated by their predicted N-terminal presequences. Removal of the N-terminal targeting presequences from the mitochondrion and plastid GABA-T isoforms yielded good recovery of the soluble recombinant proteins in Escherichia coli when they were co-expressed with the GroES/EL molecular chaperone complex. Activity assays indicated that all three recombinant isoforms possess both pyruvate- and glyoxylate-dependent GABA-T activities, although the mitochondrial enzyme has a specific activity that is significantly higher than that of its plastid and cytosolic counterparts. Finally, differential expression patterns of the three GABA-T isoforms in reproductive tissues, but not vegetative tissues, suggest unique roles for each enzyme in developmental processes. Overall, these findings, together with recent information about rice and pepper GABA-Ts, indicate that the subcellular distribution of GABA-T in the plant kingdom is highly variable.
Keywords: γ-Aminobutyrate, γ-aminobutyrate transaminase, aminotransferase, gene expression, in silico analysis, recombinant expression, substrate specificity, subcellular localization, tomato, transient expression
Introduction
γ-Aminobutyrate (GABA) is a ubiquitous, non-protein amino acid that has been implicated in stress metabolism and signalling in plants (Shelp et al., 1999, 2006; Bouché and Fromm, 2004; Fait et al., 2008). GABA is synthesized via calcium/calmodulin-dependent glutamate decarboxylase (GAD) activity (Ling et al., 1994; Snedden et al., 1995) and is catabolized to succinic semialdehyde (SSA) by the GABA transaminase (GABA-T) reaction (Van Cauwenberghe and Shelp, 1999; Van Cauwenberghe et al., 2002). SSA can be metabolized to either succinate by SSA dehydrogenase (SSADH; Busch and Fromm, 1999) or to γ-hydroxybutyrate (GHB) by SSA reductase activity (the enzyme is officially designated as GLYR for glyoxylate reductase) (Breitkreuz et al., 2003; Fait et al., 2005; Hoover et al., 2007; Simpson et al., 2008). GAD activity is localized to the cytosol, whereas GABA-T activity is traditionally thought to be localized in mitochondria with SSADH (Breitkreuz and Shelp, 1995). Two GLYR isoforms are present in plants, one in the cytosol and another in the plastid (Hoover et al., 2007; Simpson et al., 2008).
A gene encoding a mitochondrion-localized enzyme possessing both pyruvate- and glyoxylate-dependent GABA-T activities has been identified in Arabidopsis (AtGABA-T; Van Cauwenberghe et al., 2002; Clark et al., 2009). Both pyruvate- and 2-oxoglutarate-dependent GABA-T activities have been previously reported in the literature (Shelp et al., 1999); however, the pyruvate/glyoxylate-dependent enzyme appears to be the only one present in Arabidopsis, suggesting that the 2-oxoglutarate GABA-T activity is the result of multiple enzyme activities (Clark et al., 2009). AtGABA-T knockout mutants have been characterized and found to contain a reduced seed generation phenotype (Palanivelu et al., 2003). More recently, two homologous GABA-T genes have been identified in rice (Ansari et al., 2005; Wu et al., 2006), and another in pepper (GenBank accession number AAC78480). Interestingly, the pepper gene and one of the rice genes (Osl2) lack sequences encoding an N-terminal targeting presequence, making cytosolic localization likely, although neither gene product has been tested for GABA-T activity nor has its location been confirmed experimentally.
In this paper, three homologous tomato GABA-T genes were identified and the encoded proteins found to utilize both pyruvate and glyoxylate as amino acceptors. Furthermore, the three isoforms were localized to distinct subcellular compartments (i.e. mitochondrion, cytosol, or plastid), and exhibited differential expression patterns in reproductive tissues, but not vegetative tissues.
Materials and methods
Identification and RACE-PCR cloning of multiple tomato isoforms
The Arabidopsis GABA-T sequence (GenBank accession number AAK52899) was used as a template in a protein to translated nucleotide BLAST search (TBLASTN) against the tomato expression sequence tags (EST) database available on the server for the National Centre for Biotech Information (http://www.ncbi.nlm.nih.gov/). EST sequences which contained nearly 100% identity across overlapping regions were grouped together. Three distinct groups were formed and denoted SlGABA-T1, 2, and 3 (GenBank accession numbers AY240229, AY240230, and AY240231, respectively).
Total RNA was isolated from tomato (Solanum lycopersicum L. cv. MicroTom) leaves using the RNeasy kit (Qiagen, Mississauga, ON). RACE cDNA was synthesized using the SMART RACE cDNA amplification kit according to the manufacturer's instructions (Clontech, Mountain View, CA). The primers used in conjunction with the SMART RACE universal primers to amplify full-length copies of the three GABA-T isoforms from the tomato cDNA are listed in Supplementary Table S1 at JXB online. For RACE-polymerase chain reaction (PCR) amplifications, the tubes were subjected to the following conditions: 94 °C for 3 min; five cycles at 94 °C for 30 s, 70 °C for 30 s, and 72 °C for 3 min; 30 cycles at 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 3 min; and 72 °C for 10 min.
Recombinant expression, purification, and characterization of multiple tomato GABA-T isoforms
All three tomato GABA-T isoforms were cloned into the pET-15b expression vector (VWR, Mississauga, ON), with the N-terminal 46 and 60 amino acids removed from SlGABA-T1 and 3, respectively. The primers used to clone each gene are given in Supplementary Table S2 at JXB online. All three GABA-Ts were recombinantly expressed in E. coli BL-21(DE3) Rosetta (pLysS) cells (VWR, Mississauga, ON) carrying the pREP4-GroESL vector (Dale et al., 1994) and purified as described in Clark et al. (2009). Activity assays contained 50 mM N-tris(hydroxymethyl)methyl-4-aminobutanesulphonic acid (TABS; pH 9), 1.5 mM dithiothreitol, 0.625 mM EDTA, 0.1 mM pyridoxal-5-phosphate, 10% glycerol, and were conducted at 30 °C. A discontinuous assay used a final volume of 500 μl and 1 mM amino acceptor; the reaction was initiated by the addition of 1 mM amino donor or water (control), incubated in a water bath for 3 h, and then terminated by the addition of ice-cold sulphosalicylic acid to a final concentration of 60 mM (Van Cauwenberghe and Shelp, 1999). The supernatant was neutralized with 1 N NaOH and the production of specific amino acids was monitored via reverse phase HPLC as described previously (Allan and Shelp, 2006).
Analysis of the subcellular localization of multiple tomato GABA-T isoforms
In silico predictions of the subcellular localizations of various known mitochondrion and plastid proteins and the tomato GABA-T isoforms were obtained using the web-based bioinformatic programs Predotar (Small et al., 2004) (http://genoplante-info.infobiogen.fr/predotar/), TargetP (Emanuelsson et al., 2000) (http://www.cbs.dtu.dk/services/TargetP/), MitoProt (Claros and Vincens, 1996) (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html), ChloroP (Emanuelsson et al., 1999) (http://www.cbs.dtu.dk/services/ChloroP/) and PSORT (Nakai and Kanehisa, 1992) (http://www.expasy.org/tools/).
To determine the subcellular location(s) of the SlGABA-T isoforms experimentally, fusion constructs were generated with the green fluorescent protein (GFP) in pUC18/NheI-GFP, a plant expression plasmid that contains the 35S promoter and a unique NheI restriction site immediately-adjacent the 5′ end of the GFP open reading frame (ORF) (Simpson et al., 2008). Specifically, the entire ORF of each of the GABA-T isoforms was amplified via PCR using the appropriate template plasmid DNA, namely SlGABA-T1-, T2-, or T3-containing pET15 vectors (described above). The PCRs also included the appropriate forward and reverse synthetic oligonucleotide primers (see Supplementary Table S3 at JXB online) that correspond to the 5′ and 3′ ends of each GABA-T isoform ORF (minus the stop codon), as well as sequences that introduce flanking 5′ and 3′ NheI restriction sites. The resulting PCR products were then ligated in pCR2.1 TOPO (Invitrogen), followed by digestions of these vectors with NheI and cloning of the NheI fragments into NheI-digested pUC18/NheI-GFP, yielding pUC18/SlGABA-T1-GFP, pUC18/SlGABA-T2-GFP, and pUC18/SlGABA-T3-GFP. Similarly, to construct C-terminal myc-epitope-tagged versions of SlGABA-T isoforms, the NheI fragments from the pCR2.1 TOPO vectors containing individual SlGABA-T isoform sequences (see above) were cloned into NheI-digested pRTL2/NheI-myc, a plant expression vector containing a unique NheI site immediately upstream (in-frame) of the myc epitope tag sequence (Simpson et al., 2008).
Other GABA-T-GFP fusion constructs containing either the N-terminal 57 amino acids of SlGABA-T1 or the N-terminal 90 amino acids of SlGABA-T3 fused to the N terminus of GFP were generated in a similar manner. That is, sequences encoding 1–57 of SlGABA-T1 or 1–90 of SlGABA-T3, including their predicted mitochondrion and plastid targeting signals, respectively, were amplified (via PCR) using the appropriate forward and reverse primers (see Supplementary Table S3 at JXB online) and pET15/SlGABA-T1 and pET15/SlGABA-T3, respectively, as template DNA. Notably, these N-terminal sequences from SlGABA-T1 and SlGABA-T3 were slightly longer than their predicted targeting signal peptides in order to ensure the inclusion of any putative cleavage site(s) in the resulting fusion proteins and/or to maintain the N-terminal targeting signals in the proper context conferred by their endogenous upstream residues. The resulting PCR products were then ligated in pCR2.1 TOPO (Invitrogen), followed by digestions of these vectors with NheI and cloning of the NheI fragments into NheI-digested pUC18/NheI-GFP, yielding pUC18/SlGABA-T1[1–57]-GFP and pUC18/SlGABA-T3[1–90]-GFP. pUC18/SlGABA-T1[Δ2–57]-GFP and pUC18/SlGABA-T3[Δ2–90]-GFP were constructed by amplifying the SlGABA-T1 and SlGABA-T3 ORFs, but without the protein's N-terminal 2–57 or 2–90 amino acid residues. All PCRs were conducted using pET15b/SlGABA-T1 or pET15b/SlGABA-T3 as template DNA and the appropriate forward and reverse mutagenic primers that introduce 5′ and 3′ NheI sites (see Supplementary Table S3 at JXB online). The forward mutagenic primers also introduced sequences coding for an initiation (methionine) codon. The resulting PCR products were subcloned into the pCR2.1 TOPO vector, followed by digestion with NheI and ligation into NheI-digested pUC18/NheI-GFP.
pUC18/recA-CAT consisting of the N-terminal 51-amino-acid-long chloroplast transit peptide of the Arabidopsis chloroplast recA gene fused to the N terminus of bacterial chloroamphenicol acyltransferase (CAT) was constructed as follows. The recA sequence was amplified (via PCR) from the template plasmid 35S-Ct-S65T-mGFP4 (provided by MR Hanson, Cornell University; Kohler et al., 1997) using the appropriate forward and reverse primers that introduced 5′ and 3′ XmaI restriction sites. PCR products were digested with XmaI and ligated into XmaI-digested pRTL2/SmaI-CAT, a plasmid containing a modified version of CAT in which an SmaI (XmaI) site was substituted for the start codon of CAT (Flynn et al., 1998). The construction of pUC18/βATPase-CAT, consisting of the N-terminal mitochondrial targeting presequence (residues 1–60, including the cleavage site at residue 54) of the β-subunit of F1-ATPase (βATPase) fused to the N-terminus of CAT, has been described previously (Mullen et al., 1999).
All standard recombinant DNA procedures related to cloning procedures described above were performed as described by Sambrook et al. (1989). Molecular biology reagents were purchased from New England Biolabs (Mississauga, ON, Canada), Promega (Nepean, ON, Canada), Perkin-Elmer Life Sciences, Stratagene (La Jolla, CA, USA), or Invitrogen (Burlington, ON, Canada). Custom oligonucleotides were synthesized by Guelph Laboratory Services (Guelph, ON, Canada). DNA was isolated and purified using reagent kits purchased from Qiagen (Mississauga, ON, Canada). All DNA constructs were sequenced using dye-terminated cycle sequencing by Guelph Laboratory Services.
Transient-transformations of tobacco BY-2 cells and immunofluorescence microscopy were conducted as described previously (Simpson et al., 2008; Clark et al., 2009). Briefly, transient transformations of tobacco (Nicotiana tabacum L.) Bright Yellow-2 (BY-2) cells were carried out using the Biolistic Particle Delivery System (Bio-Rad) with either 10 or 5 μg of plasmid DNA for individual or co-transformations, respectively. Bombarded cells were incubated at 26 °C for ∼20 h in covered Petri dishes to allow transient expression of introduced gene(s) and protein sorting within the cells and then fixed in formaldehyde and permeabilized with pectolyase Y-23 (Kyowa Chemical Products, Osaka, Japan) and Triton X-100. Primary and fluorescent dye-conjugated secondary antibodies and sources were as follows: mouse anti-CAT IgGs (provided by S Subramani, University of California, San Diego); and, goat anti-mouse rhodamine red-X (Jackson ImmunoResarch Laboratories Inc., West Grove, PA, USA).
Microscopic visualization of labelled BY-2 cells was performed with a Zeiss Axioskop 2 MOT epifluorescence microscope (Carl Zeiss Inc, Thornwood, NY, USA) with a Zeiss 63× Plan Apochromat oil immersion objective (Carl Zeiss) and a Retiga 1300 charge-coupled device camera (Qimaging, Burnaby, BC, Canada). All images shown were deconvolved and adjusted for brightness and contrast using Northern Eclipse 5.0 software (Empix Imaging Inc., Mississauga, ON, Canada), and then composed into figures using Adobe Photoshop 8.0 (Adobe Systems, San Jose, CA). The images shown are representative of data obtained from viewing multiple (>50) transformed BY-2 tobacco cells from at least two separate biolistic bombardment experiments.
Expression analysis and in planta activity of native tomato GABA-Ts
‘MicroTom’ tomato plants were grown in controlled environment chambers (Conviron E8H, Controlled Environments Ltd., Winnipeg, Manitoba) set at 25/22 °C day/night temperatures, a photosynthetic photon flux density of 300 μmol m−2 s−1 at the top of the seedling trays (supplied by cool white fluorescent lighting, Sylvania, Mississauga, Canada), and 65% relative humidity, and supplied as necessary with tap water. Plants were grown in a Fox sandy loam (pH 6.5) under a 14 h photoperiod (i.e. long day conditions) and supplied once weekly with a modified quarter-strength nutrient solution (Shelp et al., 1992). Total RNA was isolated using the Qiagen RNeasy kit (Qiagen, Mississauga, ON). The oldest leaves were used for leaf tissue whereas the entire root was harvested for root samples. Flower parts were all harvested at the same time from open flowers within 3 d of opening and fruit tissue was harvested as each stage developed.
Real-time PCR was performed using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) with a Bio-Rad iCycler (Hercules, CA) in conjunction with the primers listed in Supplementary Table S4 at JXB online. Total RNA was treated with Turbo DNase (Ambion, Austin, TX). cDNA synthesis was conducted as described in Clark et al. (2009). Each quantitative PCR reaction used 1× SYBR Green qPCR mix (Invitrogen), 0.2 μM forward and reverse primers and 1 μl of cDNA in a 20 μl volume. All tubes were subjected to 3 min at 95 °C, followed by 40 cycles of 95 °C for 20 s, 60 °C for 20 s, and 72 °C for 20 s. SYBR Green absorbance was detected at 72 °C and all reactions were conducted in triplicate. GABA-T expression in each sample was normalized to the level of 18S ribosomal RNA (Nicot et al., 2005).
To assay crude GABA-T activity ‘Micro Tom’ tomato plants were grown in Sunshine professional growth mix (Sun Gro Horticulture Canada, Seba Beach, AB) under greenhouse conditions, supplemented with a 16 h photoperiod of 80 μmol m−2 s−1 PPFD at pot level (supplied by 400 W high pressure sodium vapour lamps, Sylvania, Mississauga, Canada), at 25/18 °C day/night temperatures. Leaf and fruit tissue was harvested after 2.5 months when breaker stage fruit began to appear on the plant. Leaf cell-free extracts were prepared by grinding tissue in 5 vols of ice-cold 50 mM TRIS-HCl buffer (pH 8.2) containing 3 mM dithiothreitol, 1.25 mM EDTA, 2.5 mM MgCl2, 10% glycerol, 6 mM 3-[3-cholamidopropyl)dimethylammonio]-1-propanesulphonate, 2% w/v polyvinylpyrrolidone, 2 μg ml−1 pridoxal-5-phosphate, 1 mM phenylmethylsulphonylfluoride and 2.5 μg ml−1 leupeptin and pepstatin A (Van Cauwenberghe and Shelp, 1999). Fruit cell-free extracts were prepared by first slicing the fruit and removing the seeds. The remaining material was homogenized in 5 vols extraction buffer (same as above) for 10 s in a Waring blender (Torrington, CT) and for 30 s at 14 000 rpm in an Ultra-Turrax T25 homogenizer (Fisher, Ottawa, ON). The homogenates from leaf and fruit tissues were incubated on ice for 20 min with gentle rocking, and centrifuged at 8000 g for 10 min at 4 °C. The supernatants were loaded onto Sephadex G25 columns (Bio-Rad Laboratories, Mississauga, CA), equilibrated with 50 mM TRIS-HCl buffer (pH 8.2) containing 3 mM dithiothreitol, 1.25 mM EDTA, 2.5 mM MgCl2, 10% glycerol, 2 μg ml−1 pyridoxal-5-phosphate, and the protein was eluted with the same buffer containing 1 mM phenylmethylsulphonylfluoride and 2.5 μg ml−1 leupeptin and pepstatin A. Total protein concentration was determined using the Bradford assay method (Bradford, 1976). GABA-T activity was measured as described above using either the TABS-based reaction mixture with 1 mM GABA and amino acceptor or a TRIS-based reaction mixture consisting of 100 mM TRIS-HCl (pH 9.0), 0.1 mM pyridoxal-5-phosphate and 10% glycerol with 2 mM GABA and 10 mM amino acceptor, as described by Akihiro et al. (2008). GABase (13.2 units ml−1; Sigma, Oakville, ON) was used as a control for both reaction assay systems.
Results and discussion
Identification, recombinant expression and substrate specificity of multiple tomato GABA-Ts
The initial BLAST search revealed 20 tomato ESTs with identity to the Arabidopsis GABA-T sequence ranging from 75–91% (see Supplementary Fig. S1 at JXB online). These sequences were used to form three distinct sequences, whose presence in the tomato genome was confirmed via RACE-PCR cloning. The isoforms, denoted SlGABA-T1, 2, and 3, contain 76–80% identity to each other and 73–82% identity to the Arabidopsis GABA-T sequence (Fig. 1). The region of greatest variability for these sequences is in the N-terminus, which is the location of the mitochondrial targeting sequence in AtGABA-T (Clark et al., 2009).
Fig. 1.
ClustalW comparison of the predicted full-length amino acid sequences for the Arabidopsis and tomato GABA-Ts. 1, 2 and 3 indicate the predicted cleavage sites for AtGABA-T, SlGABA-T1, and SlGABA-T3, respectively.
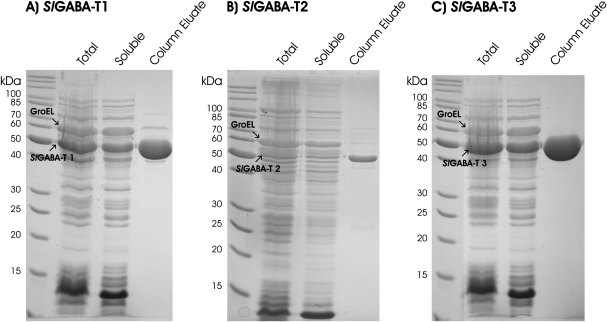
Soluble recombinant expression of the three SlGABA-T isoforms could only be achieved with co-expression of the E. coli GroES/EL chaperone complex (Dale et al., 1994), a result consistent with that found previously for AtGABA-T (Clark et al., 2009). Soluble expression of SlGABA-T1 and 3 was also facilitated via the removal of the N-terminal 46 and 60 amino acids, respectively. Purification over a nickel column provided ample supply of all three recombinant isoforms for biochemical characterization (Fig. 2). A substrate screen revealed that each isoform, like the Arabidopsis enzyme (Clark et al., 2009), has both pyruvate- and glyoxylate-dependent activities (Table 1), and utilizes GABA and Ala but not β-Ala, Orn, acetylornithine, Ser, Gly, Asn, Gln, Glu, Val, Leu, Ile, Met, Phe, His, Lys, Arg, Asp, Thr, Tyr, Trp, Pro, or Cys as amino donors (data not shown). Furthermore, the final specific activity of the purified SlGABA-T1 was in the same range as AtGABA-T (Clark et al., 2009), and two and one orders of a magnitude higher than the specific activities of SlGABA-T2 and 3, respectively (Table 1).
Fig. 2.
Purification of the recombinant proteins SlGABA-T1 (A), 2 (B), and 3 (C) using nickel resin chromatography. On each gel the prominent band corresponding to the recombinant protein and large subunit of the chaperone complex are indicated.
Table 1.
Specific activity of SlGABA-T isoforms with GABA as an amino donor and either pyruvate or glyoxylate as an amino acceptor
| Isoform | Amino acceptor | Specific activity (nmol mg−1 protein min−1) |
| SlGABA-T1 | Pyruvate | 5790±1120 |
| SlGABA-T1 | Glyoxylate | 3495±1010 |
| SlGABA-T2 | Pyruvate | 18.3±3.3 |
| SlGABA-T2 | Glyoxylate | 20.9±4.7 |
| SlGABA-T3 | Pyruvate | 206.5±64.2 |
| SlGABA-T3 | Glyoxylate | 178.5±50.4 |
Activity was determined in the presence of 1 mM pyruvate or glyoxylate with 1 mM GABA. Data represent the mean ±SE of three independent preparations.
In silico analysis of subcellular localizations of the tomato GABA-T isoforms
Prior to experimentally assessing the subcellular localization(s) of the tomato GABA-T isoforms, the accuracy of a number of web-based subcellular localization prediction programs was tested using several well-known mitochondrial or plastidial marker proteins (Table 2). Overall, Predator, TargetP, and PSORT correctly predicted the localization of most of the six marker proteins tested. MitoProt, however, predicted that all three of the plastid proteins were targeted to the mitochondrion, indicating that that program is unable to distinguish between the unique physicochemical properties within the mitochondrial and plastidial sequences. Similarly, ChloroP was able correctly to assign all three of the plastid proteins to the correct organelle, but predicted that one of mitochondrial proteins was also localized in the plastid, and that the other two mitochondrial proteins were of unknown localization. Together, these results underscore a general lack of consistency for the in silico analysis of protein localization; however, when three or more programs yield the same localization prediction it matched the experimental prediction in all cases. In terms of the predicted localization(s) of the three SlGABA-T isoforms, four of the five programs indicated that SlGABA-T1 is localized to the mitochondrion, whereas the predicted subcellular location(s) of SlGABA-T2 and SlGABA-T3 is ambiguous.
Table 2.
In silico analysis and comparison of the subcellular localization of plant GABA-Ts and various marker proteins known to target to mitochondria or plastids
| Proteina | Predotarb | TargetPc | MitoProtd | ChloroPe | PSORTf | Localizationg |
| Marker | ||||||
| ZmSOD3 | M (0.89) | M (0.80) | M (0.90) | – | PX (0.56) | M |
| AtAAT1 | M (0.65) | M (0.67) | M (0.93) | – | M (0.67) | M |
| NpβATPase | M (0.90) | P (0.79) | M (0.99) | P (0.57) | M (0.63) | M |
| AtGABA-T | M (0.87) | M (0.97) | M (0.99) | – | M (0.73) | M |
| NtRbcS | P (0.97) | P (0.69) | M (0.80) | P (0.56) | P (0.70) | P |
| AtAAT3 | – | P (0.55) | M (0.57) | P (0.51) | P (0.90) | P |
| AtrecA | P (0.95) | P (0.84) | M (0.70) | P (0.57) | C (0.45) | P |
| AtGLYR2 | – | M (0.13) | M (0.81) | – | P (0.1) | P |
| Experimental | ||||||
| SlGABA-T1 | M (0.62) | M (0.98) | M (0.95) | – | M (0.68) | M |
| SlGABA–T2 | – | – | – | – | PM (0.70) | C |
| SlGABA-T3 | – | P (0.44) | – | – | ER (0.55) | P |
Predicted targeting to the plastid (P), cytosol (C), endoplasmic reticulum (ER), mitochondrion (M), peroxisome (PX), plasma membrane (PM), or an unknown location (not determined by the program) (–). The programs used were Predotar, Target P, MitoProt, ChloroP, and PSORT (see Materials and methods for web link information on these programs). Values in parenthesis indicate the highest score obtained for the specified subcellular location with the maximum score being 1.0 in all cases.
Abbreviations and GenBank sequences used: At, Arabidopsis thaliana; Np, Nicotiana plumbaginifolia; Nt, Nicotiana tabacum; Zm, Zea mays; SOD3, superoxide dismutase isoform 3 (X12540, White and Scandalios, 1987); AAT1, mitochondrial aspartate aminotransferase isoform 1 (P46643, Schultz and Coruzzi, 1995); βATPase, β-subunit of F1-ATP synthase (P17614, Chaumont et al., 1994); GABA-T, GABA transaminase (AF351125, Clark et al., 2009); RbcS, Rubisco small subunit (M32419, Dinkins et al., 2003); AAT3, plastid aspartate aminotransferase isoform 3 (P46644, Schultz and Coruzzi, 1995); recA, RecA (M98039, Köhler et al., 1997); GLYR2, plastid glyoxylate reductase isoform 2 (AY044183, Simpson et al., 2008).
Predotar is designed to identify mitochondrial, plastid, and ER targeting sequences.
TargetP is designed to identify mitochondrial, plastid, and secretary pathway targeting sequences and may predict cleavage sites.
MitoProt is designed to test the probability of mitochondrial targeting; only sequences that contain a predictable cleavage site are listed.
ChloroP is designed to identify chloroplast transit peptides; significant values are represented by a probability value of greater than 0.5.
PSORT calculates the probability that a protein is targeted to one of 17 different subcellular locations (plant version of program).
Subcellular localization based on either previously published findings (for organelle marker proteins) or the results presented in this paper (for the SlGABA-T isoforms).
Subcellular localization of multiple tomato GABA-T isoforms in tobacco BY-2 cells
In order to discern the subcellular localization of the three SlGABA-T isoforms, tobacco (BY-2) suspension-cultured cells were used as a well-established in vivo targeting system (Banjoko and Trelease, 1995; Brandizzi et al., 2003; Miao and Jiang, 2007). Specifically, BY-2 cells were co-transformed (via biolistic bombardment) with the full-length versions of SlGABA-T1, SlGABA-T2, or SlGABA-T3 fused at their C termini to the GFP and βATPase-CAT (consisting of the N-terminal 60 amino-acid-long presequence of the β-subunit of F1-ATPase fused to the bacterial passenger protein CAT) or recA-CAT (consisting of the N-terminal 51 amino-acid-long transit peptide of the Arabidopsis chloroplast recA gene fused to CAT) serving as a well-known mitochondrion (Chaumont et al., 1994) or plastid (Kohler et al., 1997) marker fusion proteins, respectively. Co-transformed cells were then incubated for ∼20 h to allow for gene expression and protein sorting and then examined using (immuno)epifluorescence microscopy.
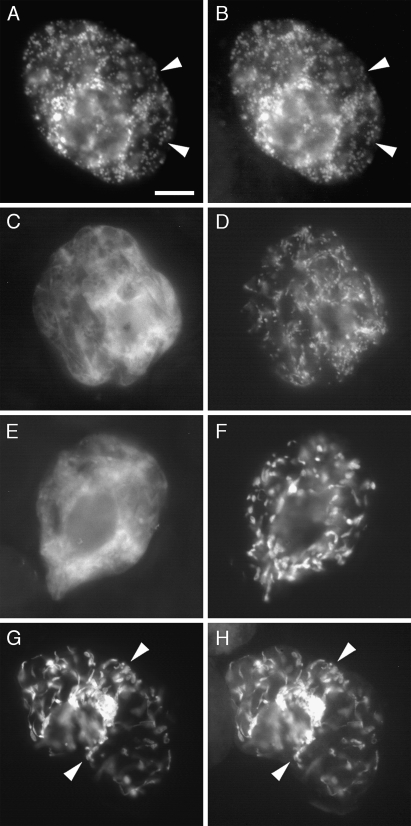
As shown in Fig. 3 (A, B), both co-expressed SlGABA-T1-GFP and βATPase-CAT yielded the same punctate fluorescence patterns in a representative transformed BY-2 cell, indicating that, consistent with the prediction results presented in Table 2, SlGABA-T1 is localized to mitochondria in plant cells. Similar results were observed when SlGABA-T1, fused at its C terminus to the myc epitope tag sequence (Fritze and Anderson, 2000) (SlGABA-T1-myc), was co-expressed with βATPase-CAT (data not shown), indicating that the mitochondrial targeting observed for SlGABA-T1-GFP was not due to the GFP moiety.
Fig. 3.
Localization of SlGABA-T1-, 2-, and 3-GFP fusion proteins in transiently co-transformed tobacco BY-2 cells. SlGABA-T1-GFP (A) co-localized with the co-expressed mitochondrial marker protein βATPase-CAT (B) in the same BY-2 cell, whereas SlGABA-T 2-GFP (C, E) failed to co-localize with co-expressed βATPase-CAT (D) or the co-expressed plastid marker protein recA-CAT (F). On the other hand, SlGABA-T3-GFP (G) co-localized with co-expressed recA-CAT (H). Arrowheads indicate obvious examples of co-calization. Bar in (A)=10 μm.
Figure 3 (C–F) shows also that SlGABA-T2-GFP, unlike SlGABA-T1-GFP, yielded a diffuse fluorescence pattern that was distinct from the fluorescence patterns attributable to either co-expressed βATPase-CAT or recA-CAT, indicating that this isoform of tomato GABA-T was not localized to mitochondria or plastids, but rather was localized throughout the cytosol. Similarly, SlGABA-T2, fused at its C terminus to a myc epitope tag (SlGABA-T2-myc) was localized throughout the cytosol in transformed BY-2 cells (data not shown). By contrast, SlGABA-T3 fused at its C terminus to either GFP (SlGABA-T3-GFP; Fig. 3G, H) or the myc epitope tag (data not shown) displayed a globular and tubular-like fluorescence pattern that was identical to that of co-expressed recA-CAT, indicating that SlGABA-T2 was localized to plastids and plastid stromules, i.e. stroma-filled tubules extending from the surface of plastids (reviewed in Natesan et al., 2005).
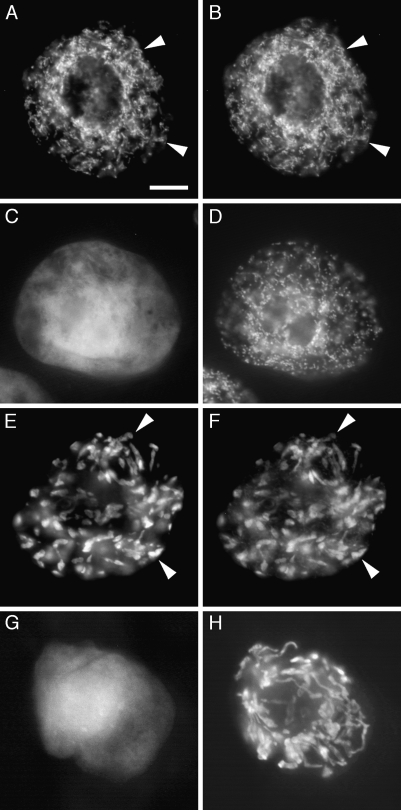
When the N-terminal 57 amino acid residues predicted to function as part of a mitochondrial targeting presequence in SlGABA-T1 (Table 2) were either removed from the SlGABA-T1-GFP (yielding SlGABA-T1[Δ1–57]-GFP) or fused alone to GFP (yielding SlGABA-T1[1–57]-GFP), the resulting modified fusion proteins were either mislocalized to the cytosol (SlGABA-T1[Δ1–57]-GFP) or localized to mitochondria (SlGABA-T1[1–57]-GFP) in a manner similar to that of the full-length SlGABA-T1-GFP (Fig. 4A–D); that is, SlGABA-T1[1–57]-GFP co-localized exclusively with the co-expressed mitochondrial marker βATPase-CAT. Together, these data reinforce the notion that SlGABA-T1 is localized exclusively to mitochondria and that the N terminus of the protein contains the molecular targeting information that is both necessary and sufficient for its proper localization to this organelle.
Fig. 4.
The N termini of SlGABA-T1 and 3 are both necessary and sufficient for targeting to mitochondria and plastids, respectively. SlGABA-T1[1–57]-GFP (A), but not SlGABA-T1[Δ2–57]-GFP (C), co-localized with the mitochondrial marker protein βATPase-CAT (B, D) in the same co-transformed BY-2 cells. Note the neighbouring SlGABA-T1[Δ2–57]-GFP and βATPase-CAT co-transformed BY-2 cell in the bottom left portion of (C) and (D). SlGABA-T3[1–90]-GFP (E), but not SlGABA-T3[Δ2–90]-GFP (G), co-localized with the co-expressed plastid marker protein recA-CAT (F, H). Arrowheads indicate obvious examples of co-localization. Bar in (A)=10 μm.
When the N-terminal 90 amino acid residues of SlGABA-T3, including its putative chloroplast transit peptide predicted by TargetP (Table 2), were fused to the N terminus of GFP, the resulting fusion protein (SlGABA-T3[1–90]-GFP) localized exclusively to plastids (and plastid stromules), as evidenced by its co-localization with co-expressed recA-CAT (Fig. 4E, F). By contrast, when SlGABA-T3-GFP was modified such that it lacked the same N-terminal region, the resulting modified fusion protein (SlGABA-T3[Δ2–90]-GFP) did not co-localize with co-expressed recA-CAT, but instead mislocalized to the cytosol (Fig. 4G, H), indicating that this region SlGABA-T3 is necessary (and sufficient) for its targeting to plastids.
Together, these data demonstrate differential localization of the three tomato GABA-T isoforms in subcellular compartments (mitochondrion, cytosol, plastid). While this is not the first suggestion of a plant GABA-T protein lacking a targeting sequence (e.g. rice (Ansari et al., 2005), and pepper (GenBank accession number AAC78480)), it is the only experimental confirmation of cytosolic localization. Moreover, this is the first evidence for a plastid-localized GABA-T in plant cells.
Developmental expression of native genes for multiple tomato GABA-Ts
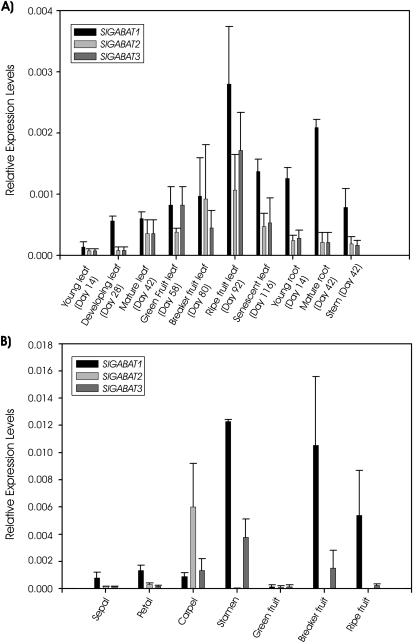
The three SlGABA-T isoforms exhibited a similar expression pattern across the vegetative plant tissues (Fig. 5A). The expression levels in leaf tissue increased up to a maximum when ripe fruit was present on the plant. With further development (i.e. senescence) GABA-T expression decreased but remained high. Throughout the vegetative tissues SlGABA-T1 expression was double those for SlGABA-T2 and 3, with the exception of green fruit and breaker fruit stages in which the expression of SlGABA-T2 and 3 matched that for SlGABA-T1. In the reproductive tissues the expression of the three isoforms was more variable (Fig. 5B). Petals and sepals had expression patterns similar to that in leaf tissue, whereas the carpel was dominated by expression of SlGABA-T2, and the stamen exhibited negligible SlGABA-T2 expression, together with the highest level of SlGABA-T1 and 3 expression found in any tissue. All three SlGABA-T transcripts were found at low levels in green fruit, whereas breaker and ripe fruit contained high levels of SlGABA-T1 transcript, moderate to low levels of SlGABA-T3 transcript, and no SlGABA-T2 transcript.
Fig. 5.
Relative abundance of SlGABA-T 1, 2, and 3 transcripts in vegetative (A) and reproductive (B) tissues. mRNA levels are expressed relative to 18S rRNA transcript levels and data represent the mean ±SE of three plants.
These results suggest that SlGABA-T1 plays the predominant role in GABA metabolism in vegetative tissue, and that the relative importance of the GABA-T isoforms varies among tissue types. It has been proposed that the reduced seed generation phenotype of the Arabidopsis GABA-T knockout mutants involves the loss of an increasing GABA gradient from the stigma to the embryo sac and, consequently, a breakdown in the growth and guidance of the pollen tube (Palanivelu et al., 2003). The expression pattern of the isoforms in reproductive tissues of tomato suggests that SlGABA-T2 is responsible for establishing the GABA gradient in the carpel, perhaps as a result of its cytosolic localization. Expression levels in tomato pollen were not tested here, but SlGABA-T1 is clearly the predominant isoform in stamens and maturing fruit tissue.
Native GABA-T activities in cell-free extracts from tomato fruits and leaves
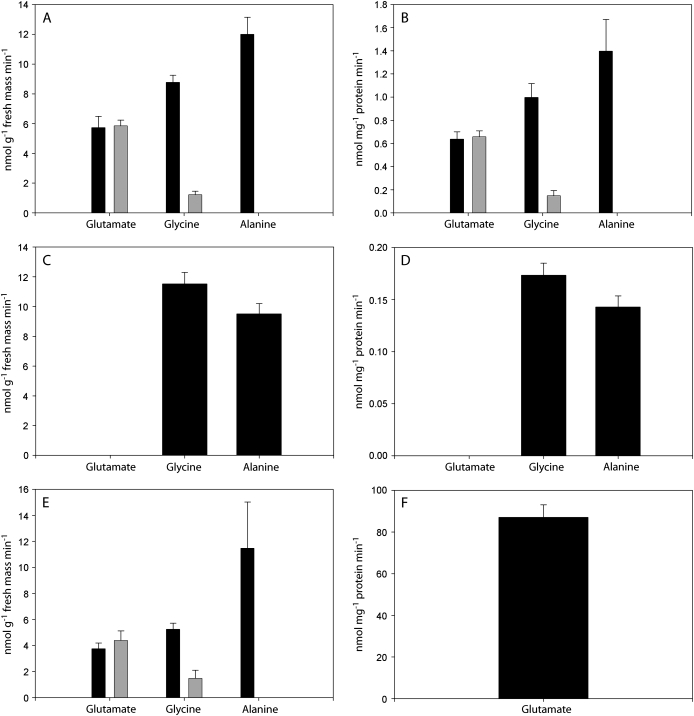
Leaves and fruits possessed both pyruvate- and glyoxylate-dependent GABA-T activities, as determined from the difference with and without GABA, in the production of Ala and Gly, respectively, by cell-free extracts (Fig. 6A–D). The specific activity was similar on a fresh mass basis in fruits and leaves (Fig. 6A, C), although on a protein basis it was higher in fruit (Fig. 6B, D) because of the overall lower protein yield from fruit tissue. While the accumulation of Glu was evident with fruit extracts in the presence of 2-oxoglutarate, it was not dependent on the presence of GABA, indicating that 2-oxoglutarate-dependent GABA-T activity is absent in tomato tissues. Similar results were observed using reaction mixtures based on TABS or TRIS- (cf. Fig. 6A, E), and 2-oxoglutarate-dependent production of Glu was detected in TABS buffer when GABase was substituted for the cell-free extract, thereby serving as a positive control for the assay (Fig. 6F).
Fig. 6.
Pyruvate-, glyoxylate-, and 2-oxoglutarate-dependent GABA-T activities in cell-free extracts from ‘Micro-Tom’ tomato fruit or leaf tissue. (A, B) Accumulation of Glu, Gly, and Ala by fruit extracts in the presence (black) or absence (grey) of GABA using a TABS-based reaction mixture. (C, D) Accumulation of Glu, Gly, and Ala by leaf extracts using a TABS-based reaction reaction mixture. (E) Accumulation of Glu, Gly, and Ala by fruit extracts in the presence (black) or absence (grey) of GABA using a TRIS-based reaction mixture. (F) Accumulation of Glu from GABase reaction in TABS assay. Data represent the mean ±SE of six plants or GABase reactions from two independent experiments and are expressed on either a fresh mass (A, C, E) or protein basis (B, D, F).
Recently, Akihiro et al. (2008) reported on the identification of a 2-oxoglutarate-dependent GABA-T activity in tomato fruit tissue, whose magnitude far exceeded that for pyruvate-dependent GABA-T activity. Previous reports indicated that 2-oxoglutarate-dependent GABA-T activity in plants is always lower than pyruvate-dependent activity (Shelp et al., 1995, 1999; Van Cauwenberghe and Shelp, 1999), and Clark et al. (2009) recently hypothesized that 2-oxoglutarate-dependent GABA-T activity in fact does not occur in plants at all. Herein, GABA-T activity was assayed in cell-free extracts from tomato fruit and leaf tissue, but no 2-oxoglutarate-dependent GABA-T activity was detected. There are several possible explanations for the discrepancy between our study and the previous study. Akihiro et al. (2008) used different assay conditions for the pyruvate- and 2-oxoglutarate-dependent activities. For 2-oxoglutarate-dependent activity, a 12 h assay was used at 37 °C, followed by the determination of Glu levels; this unusually long assay period may contribute to GABA-independent Glu accumulation, particularly with the absence of protease inhibitors throughout the purification and assay procedures. Indeed, in our own 3 h assays of fruit extracts prepared in the presence of protease inhibitors, Glu accumulated in controls that did not include GABA (Fig. 6). It is not clear that Akihiro et al. (2008) conducted such a control. Also, Akihiro et al. (2008) monitored pyruvate-dependent activity as SSA production; however, details of that assay were not included in the paper and the reference provided does not use a SSA detection method, but rather an HPLC method similar to the one used here to determine amino acid production. Overall, Akihiro et al. (2008) found lower levels of pyruvate-dependent GABA-T activity in tomato fruit than in leaves (expressed on a protein basis), whereas in the present study the activity was actually higher. Furthermore, they concluded that 2-oxoglutarate-dependent GABA-T activity is important for the breakdown of GABA that accumulates during the early stages of fruit development. Herein, there was no evidence for 2-oxogluarate-dependent GABA-T activity, but the level of pyruvate/glyoxylate-dependent activity (Fig. 6), as well as the increasing expression of the gene involved during fruit maturation (Fig. 5; Akihiro et al., 2008), indicates that pyruvate/glyoxylate-dependent GABA-T activity probably accounts for GABA catabolism.
Concluding remarks
The identification of glyoxylate-dependent GABA-T activity provides a potential link between GABA and glyoxylate metabolism in plants (Clark et al., 2009). The presence of multiple GABA-Ts in tomato, compared to the single AtGABA-T, may serve to prevent further cellular accumulation of glyoxylate, a metabolite that is highly reactive and known to inhibit Rubisco activity (Campbell and Ogren, 1990; Hausler et al., 1996). Arabidopsis is a low-light-adapted plant, typically grown at 150 μmol m−2 s−1 (Weigel and Glazebrook, 2002), whereas tomato plants can grow under high light conditions, making them more likely than Arabidopsis to be frequently exposed to high-light drought conditions, thereby resulting in higher rates of photorespiration (Wingler et al., 2000).
The identification of differentially-localized GABA-T isoforms in plants presents new possibilities for GABA metabolism. To date, the bulk of the research on GABA metabolism has been conducted with Arabidopsis, tobacco, and soybean, all of which appear to contain a single mitochondrion-localized GABA-T (Breitkreuz and Shelp, 1995; Van Cauwenberghe and Shelp, 1999; Clark et al., 2009). However, both rice (Ansari et al., 2005) and pepper (GenBank accession number AAC78480) appear to contain GABA-T genes that are predicted to localize to the cytosol (SM Clark, BJ Shelp, unpublished data), and, in this study, experimental evidence is presented for both cytosol- and plastid-localized GABA-T isoforms. It seems that the GABA pathway is more diverse in these species, involving multiple compartments. Given that GAD is located in the cytosol (Breitkreuz and Shelp, 1995) there is ample supply of GABA for GABA-T activity in the cytosol, and one can also envision that GABA is transported into the plastid, as well as the mitochondrion. The fate of GABA-derived SSA in the cytosol and plastid can perhaps be answered by revisiting the Arabidopsis model. Despite containing only a single mitochondrial GABA-T enzyme, Arabidopsis contains aldehyde reductases capable of metabolizing SSA in both the plastid and cytosol (Hoover et al., 2007; Simpson et al., 2008). If these enzymes are present in tomato and other species with additional GABA-T enzymes, then they would provide a further route for the breakdown of SSA via the synthesis of GHB. Interestingly, GABA-T localization in the cytosol was reported much earlier, but it had been reasoned that those results are due to mitochondrial breakage during extraction and fractionation (Breitkreuz and Shelp, 1995, and references therein). These more recent findings with rice, pepper, and now tomato, suggest that the cellular distribution of GABA-T in the plant kingdom is quite variable.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Synthetic oligonucleotides used for RACE-PCR amplification of SlGABA-T genes.
Supplementary Table S2. Synthetic oligonucleotides used in the construction of pET-15B expression vectors for recombinant SlGABA-T expression in E. coli.
Supplementary Table S3. Synthetic oligonucleotides used in the construction of plant expression plasmids containing SlGABA-T-GFP fusion proteins or modified versions thereof.
Supplementary Table S4. Synthetic oligonucleotides used for real-time PCR analysis of SlGABA-T gene expression.
Supplementary Fig. S1. Various expression sequence tags from tomato with homology to the Arabidopsis GABA-T and their arrangement to form three distinct groups denoted 1, 2, and 3.
Acknowledgments
The authors are grateful to PK Dhanoa (University of Guelph) for technical assistance with in vivo subcellular localization experiments. This research was supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to RTM and BJS, grants from the Ontario Ministry of Agriculture and Food and the Ontario Tomato Research Institute to BJS, and awards from the Ontario Graduate Scholarships Program to SMC and RD.
Glossary
Abbreviations
- AAT
aspartate aminotransferase
- βATPase
β subunit of F1-ATP synthase
- BY-2
Bright Yellow-2
- CAT
chloroamphenicol acyltransferase
- EST
expression sequence tag
- GABA
γ-aminobutyric acid
- GABA-T
γ-aminobutyrate transaminase
- GAD
glutamate decarboxylase
- GFP
green fluorescent protein
- GHB
γ-hydroxybutyrate
- GLYR
glyoxylate reductase
- ORF
open reading frame
- PCR
polymerase chain reaction
- RbcS
Rubisco small subunit
- SOD
superoxide dismutase
- SSA
succinic semialdehyde
- SSADH
succinic semialdehyde dehydrogenase
References
- Akihiro T, Koike S, Tani R, et al. Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant and Cell Physiology. 2008;49:1378–1389. doi: 10.1093/pcp/pcn113. [DOI] [PubMed] [Google Scholar]
- Allan WL, Shelp BJ. Fluctuations of γ-aminobutyrate, γ-hydroxybutyrate and related amino acids in Arabidopsis leaves as a function of the light–dark cycle, leaf age, and N stress. Canadian Journal of Botany. 2006;84:1339–1346. [Google Scholar]
- Ansari IM, Lee R, Chen SG. A novel senescence-associated gene encoding γ-aminobutyric acid (GABA):pyruvate transaminase is unregulated during rice leaf senescence. Physiologia Plantarum. 2005;123:1–8. [Google Scholar]
- Banjoko A, Trelease RN. Development and application of an in vivo plant peroxisome import system. Plant Physiology. 1995;107:1201–1208. doi: 10.1104/pp.107.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H. GABA in plants: just a metabolite? Trends in Plant Science. 2004;9:110–115. doi: 10.1016/j.tplants.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Irons S, Kearns A, Hawes C. BY-2 cells: culture and transformation for live-cell imaging. Current protocols in cell biology. 2003 doi: 10.1002/0471143030.cb0107s19. Chapter 1, Unit 1.7. [DOI] [PubMed] [Google Scholar]
- Breitkreuz KB, Allan WL, Van Cauenberghe OR, Jakobs C, Talibi D, Andre B, Shelp BJ. A novel γ-hydroxybutyrate dehydrogenase: identification and expression of an Arabidoposis cDNA and potential role under oxygen deficiency. Journal of Biological Chemistry. 2003;278:41552–41556. doi: 10.1074/jbc.M305717200. [DOI] [PubMed] [Google Scholar]
- Breitkreuz KE, Shelp BJ. Subcellular compartmentation of the 4-aminobutyrate shunt in protoplasts from developing soybean cotyledons. Plant Physiology. 1995;108:99–103. doi: 10.1104/pp.108.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KB, Fromm H. Plant succinic semialdehyde dehydrogenase. Cloning purification, localization in mitochondria, and regulation by adenine nucleotides. Plant Physiology. 1999;121:589–597. doi: 10.1104/pp.121.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell WJ, Ogren WL. Glyoxylate inhibition of ribulose bisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynthesis Research. 1990;23:257–268. doi: 10.1007/BF00034856. [DOI] [PubMed] [Google Scholar]
- Chaumont F, de Castro Silva Filho M, Thomas D, Leterme S, Boutry M. Truncated presequence of mitochondrial F1-ATPase β subunit from Nicotiana plumbaginifolia transports CAT and GUS proteins into mitochondria of transgenic tobacco. Plant Molecular Biology. 1994;24:631–641. doi: 10.1007/BF00023559. [DOI] [PubMed] [Google Scholar]
- Clark SM Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ. Biochemical characterization, mitochondrial localization, expression and potential functions for an Arabidopsis γ-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. Journal of Experimental Botany. 2009;60:1743–1757. doi: 10.1093/jxb/erp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Dale GE, Schonfield HJ, Langen H, Stieger M. Increased solubility of trimethoprim-resistant type S1 DHFR from Staphylococcus aureus in Escherichia coli cells overproducing the chaperonins GroEL and GroES. Protein Engineering. 1994;7:925–931. doi: 10.1093/protein/7.7.925. [DOI] [PubMed] [Google Scholar]
- Dinkins RD, Conn HM, Dirk LMA, Williams MA, Houltz RL. The Arabidopsis thaliana peptide deformylase 1 protein is localized to both mitochondria and chloroplasts. Plant Science. 2003;165:751–758. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Science. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. Highway or byway: the metabolic role of the GABA shunt in plants. Trends in Plant Science. 2008;13:14–19. doi: 10.1016/j.tplants.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Fait A, Yellin A, Fromm H. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Letters. 2005;579:415–420. doi: 10.1016/j.febslet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Flynn CR, Mullen RT, Trelease RN. Mutational analyses of a type 2 peroxisomal targeting signal that is capable of directing oligomeric protein import into tobacco BY-2 glyoxysomes. The Plant Journal. 1998;16:709–720. doi: 10.1046/j.1365-313x.1998.00344.x. [DOI] [PubMed] [Google Scholar]
- Fritze CE, Anderson TR. Epitope tagging: general method for tracking recombinant proteins. Methods in Enzymology. 2000;327:3–16. doi: 10.1016/s0076-6879(00)27263-7. [DOI] [PubMed] [Google Scholar]
- Hausler RE, Bailey KJ, Lea PJ, Leegood RC. Control of photosynthesis in barley mutants with reduced activities of glutamine synthetase and glutamate synthase. III. Aspects of glyoxylate metabolism and effects of glyoxylate on the activation state of ribulose-1,5-bisphosphate carboxylase-oxygenase. Planta. 1996;200:388–396. [Google Scholar]
- Hoover GJ, Van Cauwenberghe OR, Breitkreuz KE, Clark SM, Merrill AR, Shelp BJ. Characterization of an Arabidopsis glyoxylate reductase: general biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Canadian Journal of Botany. 2007;85:883–895. [Google Scholar]
- Köhler RH, Zipfel WR, Webb WW, Hanson MR. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. The Plant Journal. 1997;11:613–621. doi: 10.1046/j.1365-313x.1997.11030613.x. [DOI] [PubMed] [Google Scholar]
- Ling V, Snedden WA, Shelp BJ, Assmann SM. Analysis of a soluble calmodulin binding protein from fava bean roots: identification of glutamate decarboxylase as a calmodulin-activated enzyme. The Plant Cell. 1994;6:1135–1143. doi: 10.1105/tpc.6.8.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YS, Jiang L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nature Protocols. 2007;2:2348–2353. doi: 10.1038/nprot.2007.360. [DOI] [PubMed] [Google Scholar]
- Mullen RT, Lisenbee CS, Miernyk JA, Trelease RN. Peroxisomal membrane ascorbate peroxidase is sorted to a membranous network that resembles a subdomain of the endoplasmic reticulum. The Plant Cell. 1999;11:2167–2185. doi: 10.1105/tpc.11.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan SK, Sullivan JA, Gray JC. Stromules: a characteristic cell-specific feature of plastid morphology. Journal of Experimental Botany. 2005;56:787–797. doi: 10.1093/jxb/eri088. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, le Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell. 2003;114:47–59. doi: 10.1016/s0092-8674(03)00479-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbour, New York: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Schultz CJ, Coruzzi GM. The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. The Plant Journal. 1995;7:61–75. doi: 10.1046/j.1365-313x.1995.07010061.x. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, Faure D. Extracellular γ-aminobutyrate mediates communication between plants and other organisms. Plant Physiology. 2006;142:1350–1352. doi: 10.1104/pp.106.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Bown AW, McLean MD. Metabolism and functions of gamma-aminobutyric acid. Trends in Plant Science. 1999;4:446–452. doi: 10.1016/s1360-1385(99)01486-7. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Penner R, Zhu Z. Broccoli (Brassica oleracea var. italica) cultivar response to boron deficiency. Canadian Journal of Plant Science. 1992;72:883–888. [Google Scholar]
- Shelp BJ, Walton CS, Snedden WA, Tuin LG, Oresnik IJ, Layzell DB. GABA shunt in developing soybean seeds is associated with hypoxia. Physiologia Plantarum. 1995;94:219–228. [Google Scholar]
- Simpson JP, Di Leo R, Dhanoa PK, Allan WL, Makhmoudova A, Clark SM, Hoover GJ, Mullen RT, Shelp BJ. Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. Journal of Experimental Botany. 2008;59:2545–2554. doi: 10.1093/jxb/ern123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Snedden WA, Arazi T, Fromm H, Shelp BJ. Calcium/calmodulin activation of soybean glutamate decarboxylase. Plant Physiology. 1995;108:543–549. doi: 10.1104/pp.108.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenberghe OR, Makhmoudova A, McLean MD, Clark S, Shelp BJ. Plant pyruvate-dependent γ-aminobutyrate transaminase: identification of an Arabidopsis cDNA and its expression in Escherichia coli. Canadian Journal of Botany. 2002;80:933–941. [Google Scholar]
- Van Cauwenberghe OR, Shelp BJ. Biochemical characterization of partially purified gaba:pyruvate transaminase from Nicotiana tabacum. Phytochemistry. 1999;52:575–581. [Google Scholar]
- Weigel D, Glazebrook J. Arabidopsis: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- White JA, Scandalios JG. Deletion analysis of the maize mitochondrial superoxide dismutase transit peptide. Proceedings of the National Academy of Sciences, USA. 1987;86:3534–3538. doi: 10.1073/pnas.86.10.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. Photorespiration: metabolic pathways and their role in stress protection. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Zhou S, Zhang Q, Zhao W, Peng Y. Molecular cloning and differential expression of a γ-aminobutyrate transaminase gene, OsGABA-T, in rice (Oryza sativa) leaves infected with blast fungus. Journal of Plant Research. 2006;119:663–669. doi: 10.1007/s10265-006-0018-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.