Abstract
Using atomic force microscopy, we examined the contribution of cardiac myosin binding protein-C (cMyBP-C) to thick-filament length and flexural rigidity. Native thick filaments were isolated from the hearts of transgenic mice bearing a truncation mutation of cMyBP-C (t/t) that results in no detectable cMyBP-C and from age-matched wild-type controls (+/+). Atomic force microscopy images of these filaments were evaluated with an automated analysis algorithm that identified filament position and shape. The t/t thick-filament length (1.48 ± 0.02 μm) was significantly (P < 0.01) shorter than +/+ (1.56 ± 0.02 μm). This 5%-shorter thick-filament length in the t/t was reflected in 4% significantly shorter sarcomere lengths of relaxed isolated cardiomyocytes of the t/t (1.97 ± 0.01 μm) compared to +/+ (2.05 ± 0.01 μm). To determine if cMyBP-C contributes to the mechanical properties of thick filaments, we used statistical polymer chain mechanics to calculate a per-filament-specific persistence length, an index of flexural rigidity directly proportional to Young's modulus. Thick-filament-specific persistence length in the t/t (373 ± 62 μm) was significantly lower than in +/+ (639 ± 101 μm). Accordingly, Young's modulus of t/t thick filaments was ∼60% of +/+. These results provide what we consider a new understanding for the critical role of cMyBP-C in defining normal cardiac output by sustaining force and muscle stiffness.
Introduction
The cardiac isoform of myosin binding protein-C (cMyBP-C) is incorporated into the thick filament at every third myosin crown in the cross-bridge bearing C-zone of the half sarcomere (1,2). Evidence suggests that cMyBP-C regulates the radial disposition and kinetics of cross-bridges (3,4), and sarcomeric stiffness and viscosity (5,6). The C-terminus of MyBP-C interacts with the myosin rod (7–9), titin (10), and other MyBP-C (11,12). These binding properties of the C-terminus appear to underlie a functional role for cMyBP-C in stabilizing and stiffening the thick filament (13–15). Understanding the influence of cMyBP-C on thick-filament properties may be an important factor in elucidating how mutations in cMyBP-C underlie some cases of hypertrophic cardiomyopathy (16).
Compared to wild-type controls (+/+), the absence of cMyBP-C in transgenic truncated protein (t/t) or knockout (−/−) mice has been shown to reduce sarcomere stiffness under rigor conditions (6), to decrease order of the myosin heads (1,17), and to diminish and abbreviate systolic elastance in vivo (5,18). These data imply that cMyBP-C provides some rigidity to the thick filament, thereby facilitating force transmission between the acto-myosin cross-bridges and the M-line (5,6). However, the degree to which thick-filament stiffness depends upon the presence of cMyBP-C is not yet known.
In this study, we examined native cardiac thick filaments isolated from t/t and +/+ populations. To obtain a quantitative measurement of thick-filament length and flexural rigidity, we acquired atomic force microscopy (AFM) images of hydrated filaments and analyzed their curvature using statistical polymer chain mechanics (19,20). We found that thick-filament length was reduced by 5%, and flexural rigidity was reduced by almost half when cMyBP-C is absent.
Materials and Methods
Mouse model
All procedures were reviewed and approved by the Institutional Animal Care and Use Committees of the Harvard Medical School and the University of Vermont Medical School and complied with the Guide for the Use and Care of Laboratory Animals, published by the National Institutes of Health. The t/t and +/+ mice used in this study were of the 129SvEv strain, ages 10–30 weeks (21).
Solutions
Chemicals and reagents were obtained from Sigma Corp. (St. Louis, MO) unless otherwise noted. Concentrations are expressed in mmol/L−1 unless otherwise noted. Protease Inhibitor Cocktail Set III (Calbiochem, LaJolla, CA): 100.0 AEBSF Hydrochloride, 0.08 Aprotinin, 5.0 Bestatin, 1.5 E-64, 2.0 Leupeptin, 1.0 Pepstatin A. Calpain solution (Calbiochem): 1.26 mg/mL−1 calpain-1 from porcine erythrocytes, 20.0 imidazole-HCl, 5.0 β-mercaptoethanol, 1.0 EDTA, 1.0 EGTA, 30% glycerol, pH 6.8. Relaxing solution: pCa 8.0, ionic strength 50 (for t/t preps) or 100 (for +/+ preps), pH 7.1, 15.0 EGTA, 5.0 ATP, 2.0 Mg2+, 5.0 Pi, 10.0 2,3-butanedione monoxime (BDM), 2% (v/v). Protease Inhibitor Cocktail Set III: 0.05 blebbistatin (Toronto Research Chemicals, Toronto, Ontario, Canada). Skinning solution: 1% (v/v) Triton X-100 added to relaxing solution. Rigor solution: same as relaxing solution but with pCa 4.5 and without ATP. Imaging solution: same as rigor solution but at ionic strength 150 (for both +/+ and t/t preps) and the Protease Inhibitor Cocktail Set III was replaced with 1 Complete Mini Pill (Roche, Palo Alto, CA) per 10 mL solution.
Thick filament isolation protocol
Cardiac myocytes were isolated from the left ventricle (LV) using retrograde perfusion of collagenase as described previously (22) with the addition of 30 BDM and 0.002 blebbistatin to inhibit myosin ATPase. Next, myocytes were skinned for 1 h at room temperature in skinning solution. Skinned myocytes were then washed twice by centrifugation in relaxing solution followed by one wash in rigor solution. The resulting pellet was then resuspended in calpain solution plus 10 μM free Ca2+ and incubated for 1 h at room temperature (23). Enzymatic treatment was stopped by the addition of relaxing solution containing 25 μM calpain inhibitor-1 (Calbiochem). Filaments were released from sarcomeres by shearing the suspension. +/+ preparations were sheared three times with a 20 gauge needle attached to a 1 mL syringe, and t/t preparations were sheared 15 times with a 100 μL pipettor. The filament suspension was subsequently centrifuged at 3500 rpm for 4 min. The supernatant was adjusted to 100 ionic strength for the t/t preparations. This suspension was then examined by AFM.
Sarcomere length
Myocytes (isolated as described above) were examined for mean sarcomere length by calculating the Fourier Transform of video images attained by light microscopy as described elsewhere (22).
AFM
A 100 μL aliquot of filament sample was placed onto freshly cleaved mica (Ted Pella, Redding, CA) and allowed to incubate for 5 min. The filament sample was immediately washed once with 100 μL imaging solution and rehydrated in 100 μL imaging solution. All images were obtained in imaging solution under ambient conditions with a Dimension 3000 Bioscope using the Nanoscope IIIa controller (Veeco Instruments, Santa Barbara, CA) operating in Tapping Mode. Images were acquired at 512 × 512 pixels, and scan sizes ranged between 3.6 and 6.0 μm providing pixel resolutions between 7 and 12 nm. All images were acquired at scan rates between 1 and 2 Hz. Triangular cantilevers (Budget Sensors, Sofia, Bulgaria, BS-SiNi) 100 μm in length with oxide sharpened tips were used for imaging.
Image analysis
Most images revealed individual and overlapping filaments, but only isolated, nonoverlapping thick filaments were selected for analysis. AFM height images were first flattened and smoothed twice in WSxM Scanning Probe Microscopy Software (Nanotech Electronica, Madrid, Spain) before export as an ASCII matrix file to preserve true image intensity values in the third dimension. Subsequently, images were analyzed with an automated program scripted in MATLAB embedded in a LabVIEW graphical user interface. The filament position was found using a method similar to that of Brangwynne et al. (24). The filament image was digitally rotated to be horizontal, and vertical image intensity profiles were generated roughly perpendicular to the filament contour at each pixel along the length of the filament (Fig. 1). The highest point of the intensity profile was identified, and the intensity of those points about the peak resulting from the thickness of the filament were represented by a Gaussian distribution. A point is only identified if there is at least one point on each side of the peak and the points are above the baseline noise. If that minimum requirement is not satisfied, a point is not identified. All other points were flattened to zero to eliminate noise and improve signal/noise (shown as squares in Fig. 1 A). The peak of the Gaussian fit (resolution amplified 100×) identified the vertical, or y-coordinate, location of the filament for each profile view at each horizontal, or x coordinate, location.
Figure 1.
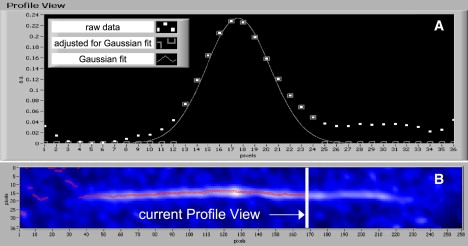
Image processing and analysis. The x,y coordinates of filaments were identified with software that scanned the profile of filaments roughly perpendicular to their longitudinal axis. The profile in A corresponds to intensity along the current Profile View line in B. Peaks of the Gaussian fit, which identify filament location, are shown as circles in panel B. The image was cropped to isolate the filament from spurious contaminating intensities (i.e., circles left of the filament).
From these x,y coordinates identifying filament location, filament length was determined by fitting a fourth-order polynomial to the coordinates. This polynomial order also has been found suitable for tracking biopolymer filament shapes such as microtubules and actin filaments (24).
To retain information about the variability of persistence lengths within a population of similar filaments, the analysis we implemented was carried out on each individual filament. This allowed us to compare persistence length measurements between populations, permitting us to use standard statistical methods to infer whether there are significant differences between groups. Because this approach differs from the more conventional approach of calculating persistence length by pooling data from multiple filaments at a given contour length (25), we will refer to the calculated value as specific persistence length. This permitted us to identify and discard filaments with shapes that were not within the resolution limits of our analysis protocol. Hence, the x,y coordinates along the contour of every filament was fit to a second-order polynomial along contour lengths (s) increasing in 100 nm increments. From each fitted contour, the end-to-end length (R) and angle between the unit tangent vectors at the beginning and end of the fitted contour (θ) were calculated as described previously (25,26). For filaments confined to two-dimensional substrates, we used the following equation to estimate persistence length (19):
| (1) |
where 〈R2 (s)〉 is the mean squared end-to-end length, s is the contour length at which R and θ were measured, and λ is the specific persistence length. For each filament, specific persistence length was estimated from fits of the data to Eq. 1 using a nonlinear Marquardt-Levenberg algorithm.
To determine the bend angle distribution as a function of axial position, we plotted the bend angle along the entire length of each filament, where distance from the center of the filament refers to the center of a 300 nm contour length from which the bend angle was measured. Next, the filament centers were aligned, and the filament bend angles in relation to the center of the filament were averaged. However, because bend angles were not symmetric along each half of every filament, the average bend angle could be influenced by filament direction whereby flipping some filaments could bias the data. To eliminate this directional bias, bend angles were “folded” at the middle of each filament hence doubling the number of bend angles for each distance along every filament's length. At this point bend angles were averaged over successive 100 nm intervals of distance from the center of the filament. A 300 nm contour length was chosen for this analysis because it approximates the length of the C-zone wherein cMyBPC is located.
The effective Young's Modulus (E) for each filament was calculated using the following equation (27):
| (2) |
where kB is the Boltzmann constant, T is the absolute temperature, and I is the area moment of inertia of the filament. For a circular cross section, I = π (filament diameter)4/64 (28). We assumed a filament diameter of 30 nm, a nominal estimate based on electron microscopic studies examining mammalian cardiac thick filaments (17).
Statistical analysis
Data are presented as mean ± SE. The t-test was used to assess statistical significance of differences in thick-filament lengths and sarcomere lengths. Between group comparisons of persistence length were performed using the Mann-Whitney U-test, which operates on nonnormal distributions. In addition, the distributions of persistence lengths were fit to the lognormal probability distribution, and estimates for the means and modes of these distribution were compared between groups with 95% confidence intervals.
Results
Thick filaments
Mouse cardiac thick filaments in physiologic solution imaged with AFM appeared intact and displayed properties of native thick filaments including tapered ends and a visible central bare zone in 29 out of 58 +/+ and 3 out of 69 t/t filaments imaged in this study (Fig. 2). Mean thick-filament lengths reported (Table 1) are similar to values reported in the literature from electron microscopic measurements of cardiac thick filaments from mouse (17) and from other mammalian species (29,30). The helical periodicity is not evident in these AFM images, however Zoghbi et al. (1) followed a similar calpain-based method to isolate intact mouse cardiac thick filaments that by three-dimensional reconstruction show near helical array of motor domains. Athough shear forces produced by a 20 gauge needle were appropriate to liberate thick filaments from +/+ myofibrils, we found that this treatment resulted in fragmentation of filaments from the t/t preparation and instead adopted a more gentle shearing with a 100 μL pipettor (see Materials and Methods). Decreasing the ionic strength from 100 mM to 50 mM also improved the yield of liberated thick filaments from the t/t preparation.
Figure 2.
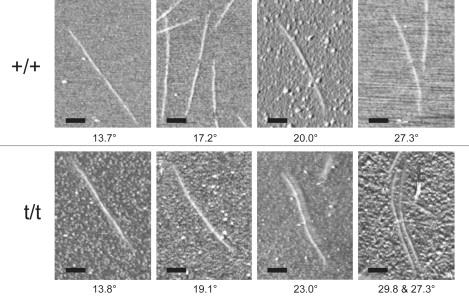
AFM images of mouse cardiac thick filaments. Top panels, +/+ thick filaments. The bare zone is visible in 50% of the filaments. Bottom panels, t/t thick filament. The bare zone is visible in only 4% of the filaments. Differences in bare zone visibility may reflect the extent to which M line material has been removed by the combination of calpain and the shearing forces used in the preparation of thick filaments. The panels display an assortment of different filament conformations and indicate the maximum bend angle found along the length of each filament, calculated from contour length = 600 nm. Scale bars = 250 nm.
Table 1.
Thick-filament length, sarcomere length, specific persistence length, and Young's modulus for isolated thick filaments imaged by AFM
| +/+ | t/t | |
|---|---|---|
| Thick-filament length (μm) | 1.56 ± 0.02 (n = 58, 3) | 1.48 ± 0.02∗ (n = 69, 3) |
| Sarcomere length (μm) | 2.05 ± 0.01 (n = 135, 3) | 1.97 ± 0.01∗ (n = 135, 3) |
| Specific persistence length (μm) | 639 ± 101 (n = 58, 3) | 373 ± 62† (n = 69, 3) |
| Young's modulus (MPa) | 65.3 ± 10.3 | 38.1 ± 6.3† |
n = number of samples, number of mice.
P < 0.01 by t-test.
P < 0.05 by Mann-Whitney U-test.
In general, +/+ and t/t filaments appear indistinguishable in AFM images (Fig. 2). This is consistent with the results of Kensler and Harris (17), who showed that cardiac filaments from cMyBP-C knockout mice (cMyBP-C−/−) have a similar appearance to those from wild-type mice (cMyBP-C+/+). However, we detect a small but significant difference in filament length (Table 1). Filaments from t/t are, on average, 5% shorter than those from +/+, a difference that is also manifested at the level of the sarcomere (Table 1).
Statistical polymer chain analysis can be used to assess the energetic equilibrium of filament conformations on a two-dimensional support by examining the ratio of the fourth moment to the squared second moment of filament fluctuations (θ) at varying contour length (s) (20). We have performed this assessment on the filament samples and have found the ratio to be near the value of three in the range 200 < s < 1500 nm (Fig. 3), a value corresponding to a Gaussian distribution function (deviations from 3 indicate the filaments are not fully energetically equilibrated in two dimensions, thus Eq. 1 provides an apparent persistence length (20)). Values at s < 200 nm and s >1500 nm significantly deviate from the expected value of 3, the former due to finite pixel size and the latter due to fewer data points (20).
Figure 3.
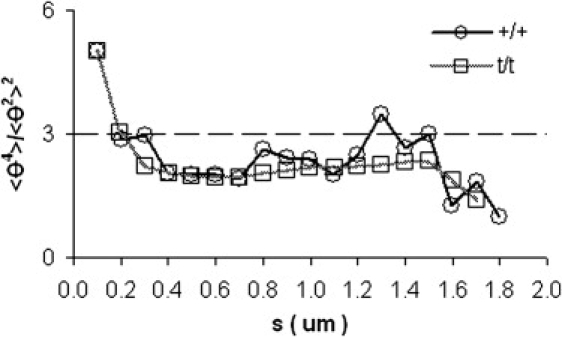
Ratio of the fourth moment and square of the second moment of the angle distribution plotted as a function of contour length. A value of three indicates a Gaussian distribution of angles. Deviations from this value at very short (<200 nm) and very long (>1500 nm) contours length are expected (see text and Rivetti et al. (20)).
Fig. 4 examines the frequency of bend angles and also their distribution along the length of the thick filament. Fig. 4 A shows the Von Mises distribution (circular analog of the Gaussian distribution) fit to filament bend angles (θ) from t/t and +/+ thick filaments. Differences in filament flexibility are revealed by comparing their distributions of bend angles at all contour lengths. Larger bend angles are more likely to occur within a t/t filament, whereas smaller bend angles occur more frequently in +/+ filaments. The standard deviation for these distributions were 0.182 radians for t/t compared to 0.158 radians for +/+, suggesting filaments containing cMyBP-C are more rigidly stable and not able to assume bend angles as varied as those measured from filaments lacking cMyBP-C. Fig. 4 B shows how the bend angles change along the length of the thick filament. For both filament types, bend angles are largest at the bare zone and taper off near the ends of the filament, indicating that most of the filaments' flexibility originates at the bare zone. Filaments from +/+ are more flexible at the bare zone compared to filaments from t/t, perhaps a reflection of the preponderance of visible bare zones among +/+ filaments. At the C-zone, where cMyBP-C is putatively located, the bend angles of t/t filaments were significantly larger than those of +/+ filaments, suggesting that cMyBPC imparts rigidity within this area of the thick filament.
Figure 4.
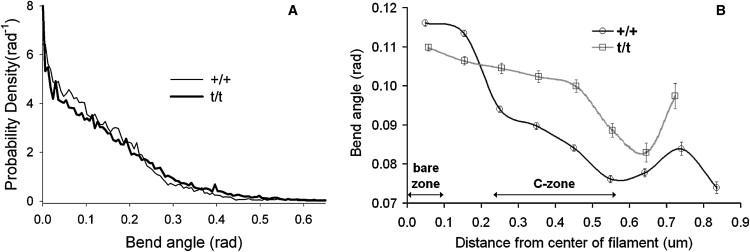
(A) Von Mises distribution fit to filament bend angles from +/+ and t/t thick filaments. Note that larger bend angles (>∼0.23 radians) are more likely observed for t/t thick filaments, whereas there is a greater likelihood for smaller bend angles (<∼0.19 radians) to be observed for +/+ thick filaments. (B) The average bend angle along half of a thick filament for contour lengths = 300 nm, which approximates the length of the C-zone wherein cMyBP-C is found. At each distance from the center of the filament, bend angles are significantly different (P < 0.001 for all distances, except between 0.6 and 0.7 μm whereby P < 0.05) between the +/+ and t/t. Near the C-zone, t/t bend angles are larger than the +/+. On the other hand, +/+ bend angles are larger near the bare zone. As the distance from the center of the filament increases past 0.6 μm, fewer bend angles contribute to the mean, explaining the more irregular bend angles found at the end of the filaments. Error bars are ± SE.
Specific persistence lengths
To determine the limits of our analysis procedure used for calculating specific persistence length, we simulated straight, infinitely stiff filaments with a pixel resolution of 12 nm (the largest pixel size for the images that were analyzed). These simulated filaments were 1.57 μm in length (roughly similar to +/+ filament length) and oriented at various angles (0°, 20°, 45°, 60°, 90°) in the x,y-plane. From these simulated analyses we found that straight filaments led to estimated values of specific persistence length ranging between 3500 μm and 4200 μm. Based on this exercise, we concluded our methodology can not differentiate from infinity estimated values for specific persistence lengths >3500 μm. We therefore excluded specific persistence lengths of actual thick filaments >3500 μm from our statistical analysis. This exclusion criterion resulted in one filament (out of 70) from the t/t group and two filaments (out of 60) from the +/+ group being discarded from the analysis.
Fig. 5 displays fits of Eq. 1 to the data for all filaments used in this analysis. Note that the range for acceptable specific persistence lengths is wide (+/+: 17–3294 μm, t/t: 6–2533 μm), yet data from both sets are tight, indicating that the fits of Eq. 1 to the data is a highly robust method of calculating specific persistence length. Differences in filament stiffness are more easily recognized at larger contour lengths, whereby anomalies resulting from the pixel resolution and scanning parameters (such as integral and proportional gains) had a smaller influence on the contour length and end-to-end length measurements. In contrast, differences in filament stiffness are not as easily discernable at short contour lengths where these anomalies are more influential.
Figure 5.
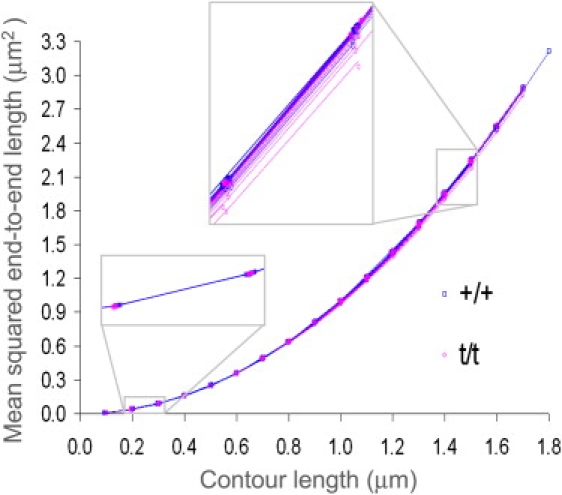
Specific persistence length was calculated from the relationship between end-to-end length and contour length using Eq. 1. The symbols represent the mean squared end-to-end values of the filaments and the solid lines represent the fit to Eq. 1. All filaments, +/+ and t/t, were fit to Eq. 1. Insets are 4× magnified views at contour lengths between 0.2 and 0.3 μm and 1.4–1.5 μm, respectively.
The mean specific persistence length for the t/t was found to be <60% of the +/+ (Table 1). Among +/+ filaments, there was no significant difference in specific persistence length between filaments with bare zones (597 ± 128 μm) and those without (681 ± 158 μm). We found, however, that the distribution of specific persistence length values were highly skewed, and the mean specific persistence length did not occur at the mode specific persistence length. We therefore characterized the distributions of apparent specific persistence lengths by fitting the cumulative distribution of specific persistence lengths to a lognormal cumulative distribution function as shown in Fig. 6 A. The probability density function (PDF) of the lognormal distribution is given as follows:
| (3) |
Figure 6.
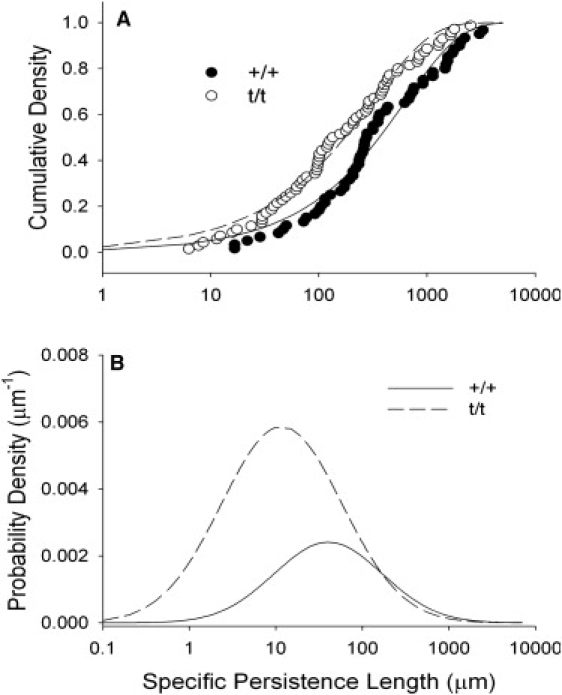
Distribution of specific persistence length values (abscissa-log scale). (A) The lognormal cumulative density function was fit to +/+ and t/t specific persistence length values. (B) The probability density functions for +/+ and t/t specific persistence length values.
The estimated parameters for the lognormal distribution, a and b, provided a basis for the calculation of means and modes of the distributions for +/+ and t/t (Table 2). The lognormal probability density functions are plotted in Fig. 6 B. Using this methodology, we estimated that the mean t/t specific persistence length was ∼64% of the +/+ (Table 2), which is similar to the result we found based on the mean statistic presented in Table 1. The mode for the t/t specific persistence lengths was <30% of the +/+ (Table 2).
Table 2.
Distribution parameters of thick filament-specific-persistence lengths estimated by nonlinear least squares fit of log-normal distribution
| +/+ (n = 58, 3) | t/t (n = 69, 3) | |
|---|---|---|
| Mean of specific persistence length (μm) | 621 (595, 643) | 396 (384, 410) |
| Mode of specific persistence length (μm) | 40.3 (31.7, 50.7) | 11.7 (10.3, 13.4) |
| Mean Young's modulus (MPa) | 63.5 (60.8, 65.7) | 40.4 (39.2, 41.9) |
| Mode Young's modulus (MPa) | 4.1 (3.2, 5.2) | 1.2 (1.1, 1.4) |
n = number of samples, number of mice. Numbers in parentheses indicate estimated value (95% confidence intervals).
Young's modulus
Calculated values for the mean and mode of Young's modulus are provided in Tables 1 and 2. Because Young's modulus is linearly related to the persistence length (Eq. 2), the statistical analysis for comparing Young's moduli is the same as that for specific persistence lengths. The Young's modulus for the t/t thick filaments, depending on our method of calculating specific persistence length given in Tables 1 and 2, was ∼60% that of +/+. This assumes that +/+ and t/t filaments have similar diameters, as previously reported (17).
Discussion
The results presented in this study reveal that cMyBP-C is required for normal length and flexural rigidity of isolated native filaments. AFM images of native thick filaments in physiologic solution showed that filaments lacking cMyBP-C are ∼5% shorter than wild-type filaments. Sarcomere length, measured in the presence of blebbistatin and BDM, is similarly reduced (∼4%) in t/t raising the possibility that cMyBP-C contributes to establishing thick-filament length that in turn is a primary determinant of sarcomere length. The relationship between sarcomere length and isolated thick-filament length suggest that the thick-filament isolation protocol used in this study preserved the native properties of the filaments. Our results agree with a previous study showing that sarcomere length in isolated myocytes from cMyBP-C knockout (−/−) mice is ∼6–7% shorter than in wild-type myocytes (31). Pohlmann et al. (31) reported that residual cycling cross-bridges in diastole are partly to account for the shorter sarcomere length, but even after treatment with BDM, diastolic sarcomere length is shorter in myocytes from the knockout as compared to the wild-type. A second possibility is that filaments lacking cMyBP-C are less stable and more susceptible to depolymerization, as has been observed by others (32). Our observation that needle shearing results in greater fragmentation of t/t filaments than of +/+ filaments is consistent with the results of Kulikovskaya et al. (32), who showed that filaments with reduced content of cMyBP-C are less resistant to shear forces.
Several studies have concluded that cMyBP-C plays an important role in proper thick-filament formation and/or stability (1,11,13,33,34). Our results are consistent with those conclusions and, in addition, provide direct evidence that cMyBP-C contributes to native thick-filament stiffness. The values reported here for specific persistence length are within the expected range in comparison to those reported for other biopolymers, i.e., several hundredfold higher than that of actin filaments (35,36) and intermediate filaments (25) and lower than that of microtubules (36,37). Schmid and Epstein (38) had previously reported persistence length values for thick filaments that are substantially lower than the values reported here. The differences are likely due to several factors including the source of the thick filaments (nematode whole body and rabbit psoas in their study versus mouse cardiac in this study) and differences in the methodology (measuring from electron microscopy versus AFM images or substrate used) and analytical approach. Although it remains difficult to establish with certainty the absolute persistence length, the results presented here show unambiguous differences in specific persistence length between +/+ and t/t filaments.
The absence of cMyBP-C reduces thick filament stiffness and the corresponding Young's Modulus between 36% and 71%, and significantly increases the bending propensity of the filament throughout the C-zone. The significant reduction in filament stiffness reveals the importance of cMyBP-C in providing structural integrity to the thick filament, and its role as an integral component for force transmission between cross-bridges and the M-line. Although the contribution of cMyBP-C to filament stiffness arises from its interaction with myosin, titin, or itself is not yet known, it is significant when one considers the molar ratio of cMyBP-C to myosin is ∼1:7. In accord with results from this study, Palmer et al. (2004) found that the cross-bridge-dependent stiffness (i.e., rigor minus relaxed) of skinned mouse myocardium from the t/t was nearly half that of the +/+ (6).
cMyBP-C has been shown to interact with two regions of the myosin rod, the light meromyosin and the S2 hinge (7,39,40). Given that the hinge is the most flexible region of the coiled-coil, binding of cMyBP-C to this region may underlie its ability to stiffen the thick filament. The binding between the N-terminal region of cMyBP-C and the myosin hinge is regulated by protein kinase A phosphorylation of cMyBP-C, which also has been shown to influence the disposition of the cross-bridges and the structure of the thick filament (41). Although we did not examine the state of cMyBP-C phosphorylation in the +/+ preparations, others have shown that an unphosphorylated cMyBP-C is associated with unstable thick filaments with disordered cross-bridges that extend away from the filament backbone (32). In contrast, a phosphorylated cMyBP-C is associated with filaments that show well-ordered cross-bridges lying uniformly along the filament backbone (32). Because at least some of the cMyBP-C are phosphorylated in the intact hearts, it is possible that the disposition of the cross-bridges with respect to the backbone may influence the flexural rigidity of the filaments.
Prior studies examining the mechanical effects of cMyBP-C on whole hearts, trabeculae, and skinned fibers have demonstrated a significant role for cMyBP-C in maintaining LV elastance during systole to support the necessary LV pressure required for systolic ejection (5,6,18). The mechanism by which cMyBP-C maintains LV systolic elastance is not yet clear, although the C-terminus of the cMyBP-C, which is incorporated into the thick filament, appears to be necessary for this role (18). Based on our results, the reduced stiffness of the thick filament lacking cMyBP-C would elevate the series compliance of the sarcomere, reduce the mechanical power transmitted from the acto-myosin cross-bridges, and thereby contribute to a reduced LV elastance during systole.
In addition to cMyBP-C phosphorylation, a second factor that has been shown to influence thick-filament structure is the myosin heavy chain (MHC) isoform composition. In particular, phosphorylation of cMyBP-C has a greater impact on the structure of filaments composed of α-MHC as opposed to filaments composed of β-MHC (30). It has been noted that MHC α/β-isoform ratio changes from 90:10 in +/+ to 50:50 in t/t (21). However, it is unlikely this isoform transition influences the specific persistence length, because molecular mechanics studies have shown that the two isoforms of myosin have similar stiffness (42–44). In addition, expression of titin, myomesin, and M-protein are similar in t/t and +/+ (5). We conclude that the reduction in t/t flexural rigidity is independent of the MHC isoform ratio or of other secondary changes in thick-filament composition that may arise from the absence of cMyBP-C.
Acknowledgments
We are grateful to Anthony Quinn and the members of the University of Vermont Microscopy Imaging Center for their expert assistance with atomic force microscopy imaging. We also recognize Alan Howard for assistance with the statistical analysis, Ting Yi for measuring sarcomere lengths, and Maria Zoghbi and John Contompasis for helpful comments.
This study was funded by National Institutes of Health grant HL59408 (D.W.M. and B.M.P.) and Nationa Science Foundation grants MCB 0315865 and IOB 0718417 (J.O.V.).
References
- 1.Zoghbi M.E., Woodhead J.L., Moss R.L., Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett P., Craig R., Starr R., Offer G. The ultrastructural location of C-protein, X-protein and H-protein in rabbit muscle. J. Muscle Res. Cell Motil. 1986;7:550–567. doi: 10.1007/BF01753571. [DOI] [PubMed] [Google Scholar]
- 3.Colson B.A., Bekyarova T., Fitzsimons D.P., Irving T.C., Moss R.L. Radial displacement of myosin cross-bridges in mouse myocardium due to ablation of myosin binding protein-C. J. Mol. Biol. 2007;367:36–41. doi: 10.1016/j.jmb.2006.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stelzer J.E., Fitzsimons D.P., Moss R.L. Ablation of myosin-binding protein-C accelerates force development in mouse myocardium. Biophys. J. 2006;90:4119–4127. doi: 10.1529/biophysj.105.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer B.M., Georgakopoulos D., Janssen P.M., Wang Y., Alpert N.R. Role of cardiac myosin binding protein C in sustaining left ventricular systolic stiffening. Circ. Res. 2004;94:1249–1255. doi: 10.1161/01.RES.0000126898.95550.31. [DOI] [PubMed] [Google Scholar]
- 6.Palmer B.M., McConnell B.K., Li G.H., Seidman C.E., Seidman J.G. Reduced cross-bridge dependent stiffness of skinned myocardium from mice lacking cardiac myosin binding protein-C. Mol. Cell. Biochem. 2004;263:73–80. doi: 10.1023/B:MCBI.0000041849.60591.45. [DOI] [PubMed] [Google Scholar]
- 7.Moos C., Offer G., Starr R., Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J. Mol. Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 8.Okagaki T., Weber F.E., Fischman D.A., Vaughan K.T., Mikawa T. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starr R., Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem. J. 1978;171:813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freiburg A., Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 11.Flashman E., Redwood C., Moolman-Smook J., Watkins H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 2004;94:1279–1289. doi: 10.1161/01.RES.0000127175.21818.C2. [DOI] [PubMed] [Google Scholar]
- 12.Moolman-Smook J., Flashman E., de Lange W., Li Z., Corfield V. Identification of novel interactions between domains of Myosin binding protein-C that are modulated by hypertrophic cardiomyopathy missense mutations. Circ. Res. 2002;91:704–711. doi: 10.1161/01.res.0000036750.81083.83. [DOI] [PubMed] [Google Scholar]
- 13.Winegrad S. Cardiac myosin binding protein C. Circ. Res. 1999;84:1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- 14.Oakley C.E., Chamoun J., Brown L.J., Hambly B.D. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int. J. Biochem. Cell Biol. 2007;39:2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 15.McClellan G., Kulikovskaya I., Flavigny J., Carrier L., Winegrad S. Effect of cardiac myosin-binding protein C on stability of the thick filament. J. Mol. Cell. Cardiol. 2004;37:823–835. doi: 10.1016/j.yjmcc.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 16.Richard P., Charron P., Carrier L., Ledeuil C., Cheav T. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 17.Kensler R.W., Harris S.P. The structure of isolated cardiac Myosin thick filaments from cardiac Myosin binding protein-C knockout mice. Biophys. J. 2008;94:1707–1718. doi: 10.1529/biophysj.107.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagayama T., Takimoto E., Sadayappan S., Mudd J.O., Seidman J.G. Control of in vivo left ventricular (correction) contraction/relaxation kinetics by myosin binding protein C: protein kinase A phosphorylation dependent and independent regulation. Circulation. 2007;116:2399–2408. doi: 10.1161/CIRCULATIONAHA.107.706523. [DOI] [PubMed] [Google Scholar]
- 19.Flory, P. J. 1969. Statistical mechanics of chain molecules. New York.
- 20.Rivetti C., Guthold M., Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996;264:919–932. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 21.McConnell B.K., Jones K.A., Fatkin D., Arroyo L.H., Lee R.T. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 1999;104:1235–1244. doi: 10.1172/JCI7377. (Erratum in J. Clin. Invest. 1999 104:1771) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer B.M., Chen Z., Lachapelle R.R., Hendley E.D., LeWinter M.M. Cardiomyocyte function associated with hyperactivity and/or hypertension in genetic models of LV hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H463–H473. doi: 10.1152/ajpheart.00310.2005. [DOI] [PubMed] [Google Scholar]
- 23.Lakey A., Labeit S., Gautel M., Ferguson C., Barlow D.P. Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J. 1993;12:2863–2871. doi: 10.1002/j.1460-2075.1993.tb05948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brangwynne C.P., Koenderink G.H., Barry E., Dogic Z., MacKintosh F.C. Bending dynamics of fluctuating biopolymers probed by automated high-resolution filament tracking. Biophys. J. 2007;93:346–359. doi: 10.1529/biophysj.106.096966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mucke N., Kreplak L., Kirmse R., Wedig T., Herrmann H. Assessing the flexibility of intermediate filaments by atomic force microscopy. J. Mol. Biol. 2004;335:1241–1250. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 26.Round A.N., Berry M., McMaster T.J., Stoll S., Gowers D. Heterogeneity and persistence length in human ocular mucins. Biophys. J. 2002;83:1661–1670. doi: 10.1016/S0006-3495(02)73934-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landau L.D., Lifshitz E.M. Pergamon Press; Oxford, NY: 1986. Theory of elasticity. [Google Scholar]
- 28.Gere J., Timoshenko S. PWS-Kent Publishing; Boston: 1990. Mechanics of Materials. [Google Scholar]
- 29.Kensler R.W. Mammalian cardiac muscle thick filaments: their periodicity and interactions with actin. Biophys. J. 2002;82:1497–1508. doi: 10.1016/S0006-3495(02)75503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisberg A., Winegrad S. Relation between crossbridge structure and actomyosin ATPase activity in rat heart. Circ. Res. 1998;83:60–72. doi: 10.1161/01.res.83.1.60. [DOI] [PubMed] [Google Scholar]
- 31.Pohlmann L., Kroger I., Vignier N., Schlossarek S., Kramer E. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ. Res. 2007;101:928–938. doi: 10.1161/CIRCRESAHA.107.158774. [DOI] [PubMed] [Google Scholar]
- 32.Kulikovskaya I., McClellan G.B., Levine R., Winegrad S. Multiple forms of cardiac myosin-binding protein C exist and can regulate thick filament stability. J. Gen. Physiol. 2007;129:419–428. doi: 10.1085/jgp.200609714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koretz J.F. Effects of C-protein on synthetic myosin filament structure. Biophys. J. 1979;27:433–446. doi: 10.1016/S0006-3495(79)85227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebillon P., Bonne G., Flavigny J., Venin S., Rouche A. COOH-terminal truncated human cardiac MyBP-C alters myosin filament organization. C. R. Acad. Sci. III. 2001;324:251–260. doi: 10.1016/s0764-4469(00)01292-0. [DOI] [PubMed] [Google Scholar]
- 35.Isambert H., Venier P., Maggs A.C., Fattoum A., Kassab R. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 36.Gittes F., Mickey B., Nettleton J., Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pampaloni F., Lattanzi G., Jonas A., Surrey T., Frey E. Thermal fluctuations of grafted microtubules provide evidence of a length-dependent persistence length. Proc. Natl. Acad. Sci. USA. 2006;103:10248–10253. doi: 10.1073/pnas.0603931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmid M.F., Epstein H.F. Muscle thick filaments are rigid coupled tubules, not flexible ropes. Cell Motil. Cytoskeleton. 1998;41:195–201. doi: 10.1002/(SICI)1097-0169(1998)41:3<195::AID-CM1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 39.Gruen M., Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 40.Kunst G., Kress K.R., Gruen M., Uttenweiler D., Gautel M. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ. Res. 2000;86:51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- 41.Levine R., Weisberg A., Kulikovskaya I., McClellan G., Winegrad S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys. J. 2001;81:1070–1082. doi: 10.1016/S0006-3495(01)75764-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyska M.J., Warshaw D.M. The myosin power stroke. Cell Motil. Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 43.Sugiura S., Yamashita H. Functional characterization of cardiac myosin isoforms. Jpn. J. Physiol. 1998;48:173–179. doi: 10.2170/jjphysiol.48.173. [DOI] [PubMed] [Google Scholar]
- 44.Malmqvist U.P., Aronshtam A., Lowey S. Cardiac myosin isoforms from different species have unique enzymatic and mechanical properties. Biochemistry. 2004;43:15058–15065. doi: 10.1021/bi0495329. [DOI] [PubMed] [Google Scholar]


