Abstract
Microtubule-nucleation activity and structural integrity of the centrosome are critical for various cellular functions. The γ-tubulin ring complexes (γTuRCs) localizing to the pericentriolar matrix (PCM) of the centrosome are major sites of microtubule nucleation. The PCM is thought to be created by two cognate large coiled-coil proteins, pericentrin/kendrin and CG-NAP/AKAP450, and its stabilization by Kizuna is essential for bipolar spindle formation. However, the mechanisms by which these proteins are recruited and organized into a proper structure with microtubule-organizing activity are poorly understood. Here we identify a centrosomal protein Cep72 as a Kizuna-interacting protein. Interestingly, Cep72 is essential for the localization of CG-NAP and Kizuna. Cep72 is also involved in γTuRC recruitment to the centrosome and CG-NAP confers the microtubule-nucleation activity on the γTuRCs. During mitosis, Cep72-mediated microtubule organization is important for converging spindle microtubules to the centrosomes, which is needed for chromosome alignment and tension generation between kinetochores. Our findings show that Cep72 is the key protein essential for maintaining microtubule-organizing activity and structural integrity of the centrosome.
Keywords: centrosome, Cep72, kizuna, microtubule, mitotic spindle
Introduction
The centrosome is the major microtubule-organizing center (MTOC) of the animal cell. During interphase, the centrosome regulates cell shape, motility and polarity, and the intracellular transport and positioning of organelles by organizing microtubules. During mitosis, the centrosome is located at the spindle pole and is involved in the formation of the bipolar spindle. Defects in the centrosome number and function can lead to an improper mitotic spindle assembly and, therefore, contribute to genomic instability and cancer progression (Nigg, 2002; Sluder and Nordberg, 2004).
The centrosome consists of a pair of nine triplets of microtubules called the centrioles and a surrounding electron-dense matrix, the pericentriolar material (PCM) (Bornens, 2002; Bettencourt-Dias and Glover, 2007). One of the important roles for the PCM is to provide binding sites for γ-tubulin ring complexes (γTuRCs) that act as microtubule nucleation templates (Moritz et al, 1995; Zheng et al, 1995). In mammalian cells, several proteins, including pericentrin/kendrin, CG-NAP/AKAP450, ninein, Nlp (ninein-like protein), and GCP-WD/NEDD1 have been implicated in the attachment of γTuRCs to the centrosome (Dictenberg et al, 1998; Mogensen et al, 2000; Takahashi et al, 2002; Casenghi et al, 2003; Haren et al, 2006; Luders et al, 2006). Pericentrin and CG-NAP, structurally related huge coiled-coil proteins, are thought to associate with each other at the centrosome to form the structural framework of the PCM and to provide binding sites for the γTuRCs (Dictenberg et al, 1998; Takahashi et al, 2002). There are reports showing that pericentrin shows a mitosis-specific property of attaching γ-tubulin at spindle poles (Dictenberg et al, 1998; Zimmerman et al, 2004). A recent report showed that pericentrin is recruited to the centrosome in a microtubule–dynein–PCM-1-dependent manner (Dammermann and Merdes, 2002). In contrast to pericentrin, the mechanism by which CG-NAP, another major PCM component, is loaded to the centrosome has remained unknown. Functional differences between these two structurally related proteins also remain to be addressed.
The centrosome duplicates once per cell cycle, which ensures the establishment of a bipolar spindle (Nigg, 2007). At the onset of mitosis, centrosomes undergo a critical reorganization termed centrosome maturation, in which the PCM expands through rapid recruitment of additional components and nucleates sufficient numbers of microtubules for spindle organization (Palazzo et al, 2000; Blagden and Glover, 2003). Furthermore, to establish stable spindle poles, centrosomes need to structurally fortify themselves during mitosis. We previously showed that the expanded PCM is stabilized by Kizuna (Kiz) (Oshimori et al, 2006). In Kiz-depleted cells, the components of the PCM, such as pericentrin and CG-NAP, which are physically ripped apart from the centrioles by the forces exerted through microtubules attached to the chromosomes, resulted in multipolar spindle formation. The Polo-like kinase 1 (Plk1)-mediated phosphorylation of Kiz, which enhances the Kiz–pericentrin association, is essential for Kiz's function. However, not much is known about the mechanisms by which Kiz is recruited to and localized at the centrosome. Here, we identify the previously uncharacterized centrosome protein called Cep72 (centrosomal protein 72 kDa) as a Kiz-interacting protein. Cep72 targets Kiz as well as γTuRC and CG-NAP to the centrosomes to ensure centrosomal microtubule organization throughout the cell cycle. During mitosis, the Cep72-mediated centrosomal MTOC activity helps connect spindle microtubules to the centrosome so that forces generated by chromosome movement along microtubules converge on the PCM. The Cep72-recruited Kiz ensures structural integrity of the PCM to endure the microtubule-mediated forces. Our data further suggest that Cep72-mediated spindle-pole formation is essential to align chromosomes at the metaphase plate and to generate tensions between kinetochores.
Results
Cep72 binds to and co-localizes with Kiz at and around the centrosomes
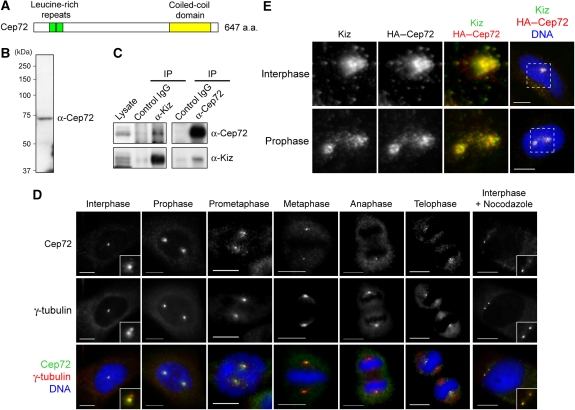
Kiz is important for the maintenance of spindle-pole integrity during mitosis (Oshimori et al, 2006). To clarify the molecular mechanisms underlying Kiz-mediated regulation, we searched for proteins directly associated with Kiz. By a yeast two-hybrid screen using Kiz as the bait, we identified the previously uncharacterized protein Cep72, which had been previously reported as a component of the centrosome in a proteomic study (Andersen et al, 2003). The Cep72 cDNA encodes a protein of 647 amino acids with two leucine-rich repeats in its N-terminal region and a putative coiled-coil domain in its C-terminal region (Figure 1A). Database searches identified orthologues of Cep72 in vertebrates and sea urchins, but not in other invertebrates or yeasts (data not shown).
Figure 1.
Cep72 associates and co-localizes with Kiz at and around the centrosome. (A) Schematic diagram of the Cep72 protein. Cep72 has two leucine-rich repeats (green, amino acids (a.a.) 55–76 and 77–98) and a potential coiled-coil domain (yellow, a.a. 476–620). (B) Western blotting of HeLa cell lysates for Cep72. (C) HeLa cell lysates were immunoprecipitated with control IgG or with anti-Cep72 or anti-Kiz antibodies. The co-precipitated proteins were analyzed by immunoblotting using the indicated antibodies. (D) Immunostaining of HeLa cells for Cep72, γ-tubulin, and DNA. (E) To examine the co-localization of Cep72 and Kiz, HeLa cells were transfected with an expression vector for HA–Cep72, fixed, and immunostained for HA (Cep72), Kiz (endogenous), and DNA. Magnified images are of the area within the boxes in the right panels. Scale bar is 10 μm.
To characterize the Cep72 protein, we generated Cep72-specific polyclonal antibodies (Figure 1B). Reciprocal immunoprecipitation experiments showed that endogenous Cep72 was associated with Kiz (Figure 1C). The immunofluorescence analysis of Cep72 in HeLa cells showed that Cep72 localized to the centrosome and centrosome-surrounding particles throughout the cell cycle (Figure 1D). These particles disappeared after microtubules were depolymerized using nocodazole, suggesting that Cep72-associating particles localized in a microtubule-dependent manner (Figure 1D, interphase+nocodazole). The exogenously expressed Myc-tagged Cep72 showed similar localization patterns, confirming the specificity of the anti-Cep72 antibodies (Supplementary Figure S1). The pattern of Cep72 localization closely resembled that of Kiz (Oshimori et al, 2006). Indeed, we observed strong co-localization of exogenously expressed HA-tagged Cep72 and endogenous Kiz at and around the centrosomes (Figure 1E).
Cep72 is required for the recruitment of Kiz to the centrosomes
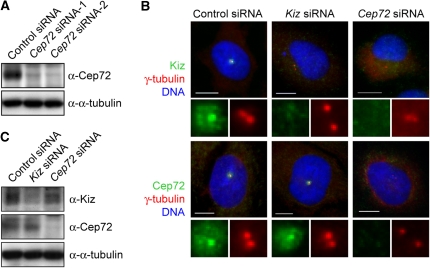
To examine whether Kiz localization is regulated by Cep72, or vice versa, we carried out RNA interference (RNAi) experiments. The transfection of two independent Cep72 siRNAs efficiently reduced the expression of Cep72 protein as shown by western blotting (Figure 2A). In more than 90% of siRNA-treated cells, Cep72 was undetectable by immunofluorescence microscopy. The centrosomal signals of Kiz disappeared or were significantly decreased in Cep72-depleted cells (Figure 2B), although western blotting of cell lysates showed almost no change in the Kiz protein levels (Figure 2C). In contrast, Kiz depletion did not affect the localization of Cep72 (Figure 2B). These results suggest that the centrosomal localization of Kiz is dependent on Cep72. The gel filtration analysis revealed two major populations of Kiz-containing complex (Supplementary Figure 2). In contrast, Cep72, as well as γ-tubulin were found in broader area, suggesting that Cep72 forms multiple complexes with multiple partners. A Kiz mutant with an altered Plk1 phosphorylation site (KizT379A) associated with Cep72 in a similar manner as wild-type Kiz, indicating that Kiz phosphorylation by Plk1 is not involved in the Kiz–Cep72 interaction (Supplementary Figure S3).
Figure 2.
Cep72 is required for the centrosomal localization of Kiz. (A) At 60 h after transfection with control or two independent Cep72 siRNAs, HeLa cells were lysed and analyzed by immunoblotting with the indicated antibodies. (B) Control, Kiz, or Cep72 siRNA-transfected HeLa cells were fixed and immunostained for γ-tubulin, DNA, and Kiz (upper panels) or Cep72 (lower panels). Bottom panels of each show enlarged images of the centrosomal area. (C) Control, Kiz, or Cep72 siRNA-transfected cells were lysed and analyzed by immunoblotting using the indicated antibodies. Scale bar is 10 μm.
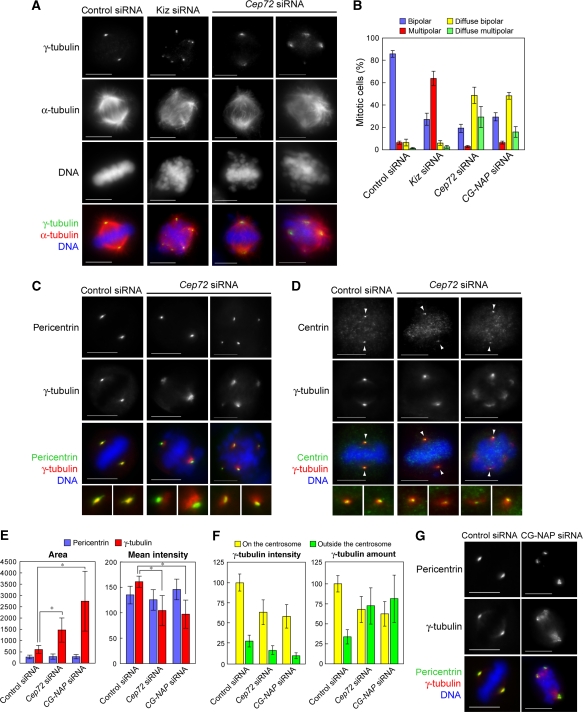
Cep72 is involved in the γTuRC localization and microtubule organizing activity of the centrosome
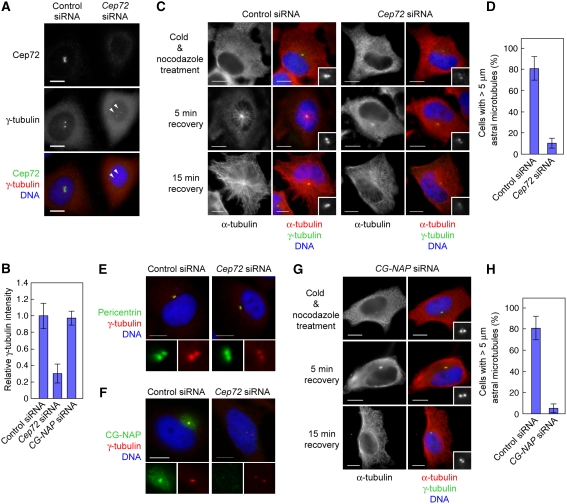
We also found that in the majority of interphase Cep72-depleted cells (78.3%, n=138), the intensity of γ-tubulin signals at the centrosomes waned markedly (see Figure 2B). To precisely compare the γ-tubulin signals, we seeded control and Cep72-depleted cells on the same cover glasses and measured γ-tubulin intensities (Figure 3A). The relative intensity of γ-tubulin staining in Cep72-depleted cells was reduced to about 30% compared to control cells (Figure 3B). The level of GCP2, another γTuRC component, was also decreased by Cep72 depletion (Supplementary Figure S4A).
Figure 3.
Cep72 is involved in the centrosomal localization of γTuRC and CG-NAP, and microtubule nucleation from centrosomes. (A) Control or Cep72 siRNA-transfected HeLa cells were plated on the same cover slips, fixed, and immunostained for Cep72, γ-tubulin, and DNA. The arrowheads indicate centrosomes in a Cep72-depleted cell. (B) Comparison of the γ-tubulin intensity of the centrosomes in control, Cep72-depleted, and CG-NAP-depleted cells. To evaluate the γ-tubulin intensity, centrosomal areas were encircled by the edge of γ-tubulin staining and the mean intensity in each area was measured and the background levels subtracted. The data indicate the relative intensity in the centrosomal area and represent the mean±s.d. (n=28). (C) Microtubule re-nucleation assays of control or Cep72 siRNA-transfected interphase cells. After recovering from cold and nocodazole treatment for 0, 5, or 15 min, the cells were fixed and stained for γ-tubulin, α-tubulin, and DNA. Insets show enlargements of the centrosomal area stained for γ-tubulin. (D) The proportion of the length (more than 5 μm) of re-growing microtubules from control or Cep72-depleted centrosomes. Data represent mean±s.d. of three experiments (n>140 for each experiment). (E, F) Control or Cep72-depleted cells were immunostained for γ-tubulin and DNA together with pericentrin (E) or CG-NAP (F). The lower panels show magnified images of the centrosomes. (G) Microtubule re-nucleation assays of CG-NAP siRNA-transfected interphase cells. Compare to the control in (C). (H) The proportion of the length (more than 5 μm) of re-growing microtubules from control or CG-NAP-depleted centrosomes. Data represent mean±s.d. of three experiments (n>100 for each experiment). Scale bar is 10 μm.
As γTuRC is essential for microtubule nucleation, we examined the microtubule-nucleating activity of the interphase centrosomes in the absence of Cep72. We carried out re-growth assays of centrosomal microtubules that had been depolymerized once by cold treatment in the presence of nocodazole. At 5 min after recovery, microtubules were nucleated from centrosomes and over 80% of control cells had microtubules longer than 5 μm. In contrast, we found that only 10.3±2.6% of Cep72-depleted cells had microtubules longer than the threshold (Figure 3C and D). At 15 min after recovery, control cells had numerous radial microtubule arrays extending from the centrosomes to the cell cortex. In contrast, radial arrays of microtubules, nucleated from the centrosomes, were rarely seen in Cep72-depleted cell, whereas some microtubules nucleated from other microtubule-organizing sites were observed throughout cytoplasm (Figure 3C, 15 min recovery). Moreover, we could not detect significant difference in overall microtubule networks in interphase Cep72-depleted cells, although it seems that the intensity of microtubules around centrosomes is less than control cells (Supplementary Figure S4B). These results indicate that the centrosomes in Cep72-depleted cells are largely devoid of microtubule-organizing activity. The Kiz depletion did not affect the localization of γ-tubulin (Figure 2B) and the MTOC activity of centrosomes (Oshimori et al, 2006). Thus, in addition to recruiting Kiz to the centrosomes, Cep72 seems to have important roles in regulating γTuRC localization and the MTOC activity of the interphase centrosomes.
Cep72 is necessary for the localization of CG-NAP
The γTuRCs are thought to be anchored to the huge coiled-coil proteins composing the PCM, such as pericentrin and CG-NAP (Dictenberg et al, 1998; Takahashi et al, 2002). Therefore, we examined whether Cep72 contributes to the centrosomal localization of these proteins. Pericentrin signals were not obviously affected by Cep72 depletion (Figure 3E). In contrast, localization of CG-NAP to the centrosomes as well as to the Golgi apparatus (Supplementary Figure S5A) was significantly impaired in Cep72-depleted cells (Figure 3F). Western blotting of cell lysates showed little change in the overall CG-NAP protein level on Cep72 depletion (Supplementary Figure S5B), indicating that Cep72 depletion caused mis-localization of CG-NAP. We also compared the localization of Cep72 and the golgin97, a Golgi marker protein, and found that although the majority of the Cep72 signals are found within the area encompassed by Golgi membranes, some centrosome-surrounding Cep72 particles were found to be very close to the Golgi membranes (Supplementary Figure S5C). Note that CG-NAP depletion did not alter the centrosomal localization of Cep72 (Supplementary Figure S5D). These results indicated that Cep72 is necessary for CG-NAP localization to the centrosomes and the Golgi apparatus during interphase.
We next examined whether CG-NAP mis-localization caused by Cep72 depletion was relevant to the reduction in centrosomal γTuRCs and the microtubule-organizing activity. Unexpectedly, the majority of interphase cells treated with CG-NAP siRNA (92.6%, n=136) had normal γ-tubulin and GCP2 signals at the centrosomes (Figure 3G and Supplementary Figure S6) and the relative intensity of γ-tubulin staining in CG-NAP-depleted cells was hardly affected as compared with control cells (Figure 3B). Nevertheless, in the re-growth assay, microtubules longer than the threshold were observed in only 5.3±2.4% of CG-NAP-depleted cells at 5 min after recovery from microtubule depolymerization (Figure 3G and H), reflecting a marked decrease in centrosomal microtubule-nucleating activity. These results indicate that although γTuRCs are able to localize to the centrosomes without CG-NAP, the resulting centrosomes fail to nucleate microtubules and organize them into radial arrays. Taken together, Cep72 is involved in targeting γTuRCs and CG-NAP, to ensure the microtubule-organizing activity of interphase centrosomes.
Cep72-mediated CG-NAP localization is essential for aster formation at the onset of mitosis
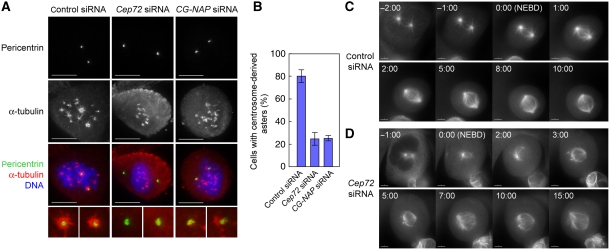
Next, we sought to clarify the roles of Cep72 in spindle formation. As Cep72 depletion led to poor centrosomal MTOC activity during interphase, we hypothesized that Cep72 depletion also affects the microtubule organization during mitosis, leading to the failure in proper spindle formation. To test this, we first carried out microtubule-re-growth assays in mitotic cells (Figure 4A). At 5 min after recovery from the depolymerizing condition, microtubules were generated prominently from the two centrosomes and from many sites on chromosome mass in control cells. In Cep72-depleted cells, however, centrosome-dependent microtubule nucleation was severely depressed, whereas microtubules emanated from chromosomes at the same level as in control cells (Figure 4A and B). We next carried out time-lapse observations of fluorescent α-tubulin-expressing cells in the presence or absence of Cep72. In control cells, microtubule asters were invariably present at the centrosomes during prophase. Soon after nuclear envelope breakdown (NEBD), centrosomal asters drove the rapid formation of a bipolar spindle (Figure 4C and Supplementary Movie S1). In contrast, Cep72-depleted cells did not generate obvious microtubule asters before NEBD. Although the asters gradually became apparent and chromosome-derived microtubules were assembled into spindle after NEBD, spindle poles were very unstable and unfocused (Figure 4D and Supplementary Movie S2).
Figure 4.
Cep72 is required for microtubule aster formation at the onset of mitosis. (A) Microtubule re-growth assays of control, Cep72, or CG-NAP siRNA-transfected mitotic cells. After recovering from cold and nocodazole treatment for 5 min, the cells were fixed and stained for pericentrin, α-tubulin, and DNA. (B) The proportion of control, Cep72-, or CG-NAP-depleted cells with the centrosome-derived microtubule asters at 5 min after re-growth. Data represent the mean±s.d. of three experiments (n>70 for each experiment). (C, D) Time-lapse imaging of mitotic U2OS cells expressing Venus α-tubulin transfected by control (C) or Cep72 siRNA (D). The frame showing NEBD is labelled as 0 min (time in min:s).
Similar to interphase, we found that CG-NAP signals disappeared or were remarkably diminished from the spindle poles in Cep72-depleted cells (Supplementary Figure S7). Furthermore, CG-NAP depletion also resulted in a specific decrease in centrosome-dependent microtubule nucleation as measured by the microtubule-re-growth assay (Figure 4A and B). These observations suggest that Cep72 or CG-NAP depletion impaired centrosome-specific microtubule nucleation and/or microtubule organization, which would lead to the generation of unstable and unfocused spindle poles.
Cep72-depletion induces abnormal spindles with diffuse and multiple poles during mitosis
As in the case of the interphase cells, Kiz is almost lost from centrosomes in Cep72-depleted mitotic cells (Supplementary Figure S8). However, it is striking that in Cep72-depleted cells γ-tubulin and GCP2 were diffusely distributed around each spindle pole irrespective of the spindle-pole number, whereas these signals remained focused in the individual fragmented poles in Kiz-depleted cells (Figure 5A, B and Supplementary Figure S9A, Oshimori et al, 2006). Furthermore, the spindle microtubules were not concentrated at spindle poles in Cep72-depleted cells (Figure 5A). It should be noted that the proportion of multipolar spindles in Cep72-depleted cells (32.2±8.6%, Figure 5A and B) was only about half of that in Kiz-depleted cells (66.7±5.4%). Nevertheless, multipolar spindles in Cep72-depleted cells shared common features with those in Kiz-depleted cells (Oshimori et al, 2006): pericentrin localized to all of the spindle poles (Figure 5C), whereas centrin, a marker for centrioles (Paoletti et al, 1996), overlapped with only two of the poles, regardless of pole number (Figure 5D). The data suggest that Cep72 depletion causes fragmentation and dissociation of the PCM from the centrioles by inducing mis-localization of Kiz, though the degree of PCM fragmentation is low.
Figure 5.
Cep72 depletion causes spindle-pole fragmentation and diffuse γTuRC distribution around each spindle pole. (A) Control, Kiz, or Cep72 siRNA-transfected cells were fixed and immunostained for α-tubulin, γ-tubulin, and DNA. (B) The proportion of spindle phenotypes in control, Kiz, Cep72, or CG-NAP siRNA-transfected cells are categorized into four types: bipolar with focused (blue) or diffuse (yellow) γ-tubulin signals, and multi-polar with focused (red) or diffuse (green) γ-tubulin signals. Data represent the mean±s.d. of four experiments (n>100 for each experiment). (C, D) Control or Cep72-depleted HeLa cells were fixed 10–12 h after release from double thymidine block and immunostained for γ-tubulin and DNA, together with pericentrin (a marker for the PCM) (C) or centrin (a centriole marker) (D). The lower panels show magnified images of two spindle poles from each upper panel. Arrowheads indicate the centrioles. (E) Quantification of the area and intensity of pericentrin or γ-tubulin labelling at the poles in bipolar spindles in control, Cep72, and CG-NAP siRNA-treated cells. The stained areas were encircled by the edge of pericentrin or γ-tubulin staining and mean intensities of pericentrin and γ-tubulin in each area were measured. Data show the average intensity in the centrosomal area and represent the mean±s.d. (n>32). *P<0.001. (F) Quantification of the intensity of γ-tubulin signals inside and outside of the pericentrin-staining area. Total amount of γ-tubulin were results from multiplying the values of area and intensity. Data show the average intensities and amounts and the mean±s.d. (n=20). (G) Control or CG-NAP-depleted cells were immunostained for pericentrin, γ-tubulin, and DNA. Scale bar is 10 μm.
Cep72-dependent aster formation ensures the transport of chromosome-derived spindle microtubules
Next we addressed the underlying mechanisms of the diffuse, unfocused spindle-pole formation caused by Cep72 depletion. To clarify the significance of Cep72 function in spindle formation, we compared the mitotic centrosomes and spindle poles among control, Kiz-depleted, Cep72-depleted, and CG-NAP-depleted cells. As shown in Figure 5E, both the size and intensity of the pericentrin signals at the poles of a bipolar spindle in Cep72-depleted or CG-NAP-depleted cells were indistinguishable from those in control cells, suggesting that the recruitment of pericentrin during centrosome maturation occurred normally without Cep72 or CG-NAP. However, in contrast with the tight overlap of pericentrin and γ-tubulin staining in control and Kiz-depleted cells (Oshimori et al, 2006), the area of γ-tubulin staining was broader than that of pericentrin in Cep72-depleted and CG-NAP-depleted cells (Figure 5C and G and Supplementary Figure S9B). The area and the mean intensity of γ-tubulin staining at each spindle pole in Cep72-depleted cells were about 2.5-fold broader and 35% lower, and in CG-NAP-depleted cells were about 4.5-fold broader and 40% lower, respectively, than those of control cells (Figure 5E). To further evaluate the amount of γ-tubulin assembled at the spindle poles, we measured γ-tubulin intensities focused on the centrosomes (i.e. area overlapping with pericentrin staining) and outside of the centrosomes (i.e. diffuse signals) separately (Figure 5F). In Cep72-depleted and CG-NAP-depleted cells γ-tubulin intensities of both populations were less than 70% of those in control cells. However, total amount of γ-tubulin at the spindle pole was not significantly different among those cells. These results suggested that Cep72 and CG-NAP depletion impaired γ-tubulin loading on the centrosomes.
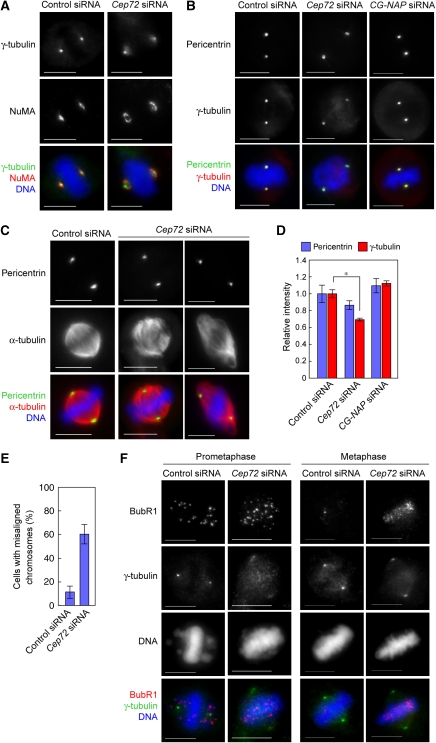
During prometaphase, microtubules nucleating near chromosomes are captured by astral microtubules nucleating from centrosomes (Khodjakov et al, 2003; Maiato et al, 2004; O'Connell and Khodjakov, 2007). Following the capture, they are incorporated into spindle poles by NuMA–dynein-dependent mechanisms that expedite proper spindle formation (Khodjakov et al, 2003; Tulu et al, 2003; Wadsworth and Khodjakov, 2004). As centrosomes in both Cep72-depleted and CG-NAP-depleted cells had poor aster formation (Figure 4A and B), results described above raised the possibility that non-centrosomal microtubules and their minus-end-associated γTuRCs could not be effectively transported or anchored to the centrosome in the absence of astral microtubules, resulting in a diffuse distribution of γTuRCs.
To test this possibility, we examined the localization of NuMA, which has a function in focusing microtubule minus ends and tethering centrosomes to the body of the spindle (Compton and Cleveland, 1994; Merdes et al, 1996, 2000). In control cells, NuMA showed focused localization in the vicinity of the compact γ-tubulin signal. In contrast, NuMA was detected as an annular-shaped signal at the periphery of the diffuse γ-tubulin signal in Cep72-depleted cells (Figure 6A), suggesting that the minus ends of microtubules were not anchored to the PCM. Consistent with this idea, all of the diffuse γ-tubulin signals outside the pericentrin area (Figure 5C and G) disappeared on microtubule depolymerization in both Cep72- and CG-NAP-depleted cells (Figure 6B). After 30 min of recovery from microtubule depolymerization, centrosomes detected by pericentrin antibodies were not necessarily positioned at the spindle poles in the absence of Cep72, although the bipolar spindle was able to re-form (Figure 6C). Thus, one of the primary roles of the Cep72-mediated recruitment pathway is to confer microtubule-nucleation activity on the centrosomes so that the non-centrosomal microtubules can converge on the common spindle poles.
Figure 6.
Cep72-mediated attachment of spindle microtubules and mitotic centrosomes is required for proper chromosome alignment and segregation. (A) Control or Cep72-depleted cells were fixed and stained for NuMA, γ-tubulin, and DNA. (B) Control, Cep72, or CG-NAP siRNA-transfected cells were incubated in cold media containing 1 μM nocodazole for 30 min to depolymerize microtubules. Cells were fixed and immunostained for pericentrin, γ-tubulin, and DNA. (C) Microtubules were depolymerized as in (B). Mitotic cells were transferred to warm media to allow microtubules to re-grow. After recovering for 30 min, the cells were fixed and immunostained for pericentrin, α-tubulin, and DNA. Pericentrin staining indicates the position of the centrosomes. (D) Quantification of the relative intensity of γ-tubulin labelling at the centrosome in microtubule-depolymerized mitotic cells. Centrosomal areas were encircled by the edge of pericentrin staining and the mean intensities of pericentrin and γ-tubulin in each area were measured. Data show the average intensity in the centrosomal area and represent the mean±s.d. (n=20). *P<0.01. (E) The proportion of cells with misaligned chromosomes in control or Cep72-depleted cells with a bipolar spindle. Data represent the mean±s.d. of four experiments (n>20 for each experiment). (F) Control or Cep72 siRNA transfected HeLa cells were immunostained with BubR1, γ-tubulin, and DNA. Scale bar is 10 μm.
Microtubule-depolymerizing experiments also showed that PCM-associated γ-tubulin signals were weaker in Cep72-depleted cells than in control cells, whereas pericentrin signals did not significantly change (Figure 6B and D). Despite its importance in microtubule nucleation, CG-NAP depletion hardly affected γ-tubulin intensity both during interphase and mitosis (Figure 6D and Supplementary Figures S6A and B). These results indicate that Cep72 is involved in the recruitment of basal levels of γTuRCs throughout the cell cycle in a CG-NAP-independent mechanism, though efficient microtubule nucleation from γTuRCs largely depends on CG-NAP.
Cep72-mediated spindle-pole formation is essential for chromosomal alignment and tension generation between sister chromatids
In Cep72-depleted cells, spindle microtubules were not well focused on the spindle poles, which seemingly reduced the microtubule-generated forces that otherwise converges on the centrosome. Accordingly significant number of Cep72-depleted cells could produce bipolar but unfocused spindle without Kiz in the PCM. However, even when Cep72-depleted cells retained apparent spindle bipolarity, these cells tended to have misaligned chromosomes (60.3±8.3%) (Figures 5A and 6E). To monitor the microtubule-mediated tension, we used an antibody against BubR1, a marker protein for the tension checkpoint (Figure 6F; Skoufias et al, 2001). In prometaphase, both control and Cep72-depleted cells have many BubR1 signals at kinetochores. In control cells, BubR1 mainly localized to kinetochores of chromosomes that had not aligned on the metaphase plate. In contrast, BubR1 stayed on chromosomes that aligned at an apparent metaphase plate in Cep72-depleted cells. During metaphase, both cells had aligned chromosomes on the metaphase plate. However, in Cep72-depleted cells, kinetochore BubR1 signals remained strong, whereas they disappeared from almost all chromosomes in control cells. The data suggest that Cep72-depleted spindles could not generate sufficient tension between sister kinetochores, leaving the spindle checkpoint active. Actually, Cep72-depleted cells showed severe cell-proliferation defects, and in many cases, cells were arrested in elongated prometaphase and dying (Supplementary Figure S10A and B). Thus, we concluded that Cep72–Kiz-mediated formation of rigid and focused spindle poles guarantees the proper alignment of the chromosomes at metaphase.
Discussion
Accumulating data show the functional importance of many of the centrosomal proteins, yet it remains largely unclear how centrosomal proteins are recruited and organized to maintain structural integrity and to show MTOC activity. Here, we show that Cep72 has key roles in regulating the centrosomal localization of γ-tubulin complex components, CG-NAP, and Kiz. We also show that Cep72 provides centrosomal microtubule-nucleation activity and has critical roles in forming a focused bipolar spindle, which is needed for proper tension generation between sister chromatids.
Cep72 depletion causes severe impairment in the centrosomal MTOC activity during both interphase and mitosis. During interphase, the amount of γTuRC associated with centrioles is markedly decreased in Cep72-depleted cells, which may contribute to the reduction of the centrosomal MTOC activity. At the onset of mitosis, the amount of centrosomally localized γTuRC is decreased but significant amount remains associated with PCM in Cep72-depleted cells (Figure 5F). Apparently, γTuRCs are recruited to the centrosome even in the absence of Cep72, possibly by a pericentrin-dependent mechanism (Zimmerman et al, 2004). Nevertheless, mitotic centrosomes in Cep72-depleted cells show little MTOC activity. This is because Cep72 is essential for the centrosomal localization of CG-NAP, and CG-NAP is crucial for microtubule aster formation from the centrosomes throughout the cell cycle. Thus, Cep72 guarantees centrosomal MTOC activity by recruiting γTuRCs, and independently but simultaneously their activator CG-NAP to the centrosomes. CG-NAP associates with kinases (PKA, PKCɛ, PKN, and CK1), phosphatases (PP1 and PP2A) (Takahashi et al, 2002), and the mitotic regulator RanGTPase (Keryer et al, 2003). Thus, by anchoring these regulatory proteins, CG-NAP may activate γTuRCs and/or stabilize nucleated microtubules. Further studies are needed to clarify the mechanism by which CG-NAP activates centrosomal MTOC.
Our data show that CG-NAP provides centrosomal MTOC activity and ensures effective transport of non-centrosomal microtubules and their minus-end-associated γTuRCs to the mitotic centrosome. These transported γTuRCs are thought to be anchored to the PCM (Khodjakov et al, 2003; Maiato et al, 2004; O'Connell and Khodjakov, 2007). However, after microtubules depolymerization, amount of γTuRCs associated with PCM of the mitotic centrosomes are unaffected by the presence or absence of CG-NAP (Figure 6D). These observations raise the possibility that CG-NAP does not act as a direct γTuRC anchoring protein and that the transported γTuRCs are not stably built in the PCM structure, but are reversibly bound to the mitotic PCM in a microtubule-dependent manner.
In contrast to Kiz and CG-NAP localization, which is completely disrupted by Cep72 depletion, localization of γTuRC is partially altered by Cep72 depletion. This result is not surprising, as numerous centrosome proteins, including pericentrin, centrosomin/Cep215/CDK5RAP2 (Fong et al, 2008), and Cep192 (Gomez-Ferreria et al, 2007; Zhu et al, 2008) have been implicated in the targeting or anchoring of γTuRC. Although most of these proteins co-localize with γ-tubulin at the PCM, Cep72 staining only partially overlaps with that of γ-tubulin (Figure 1D). Instead, Cep72 localizes to a specific site near centrioles and centrosome-satellite-like particles (Figure 1D), suggesting that Cep72 may control γTuRC localization differently from other γTuRC regulators.
CG-NAP and pericentrin are the principal components of the PCM and associate with each other to form the structural framework (Takahashi et al, 2002). As suggested by previous study on AKAP350, a variant of CG-NAP/AKAP450 (Larocca et al, 2006), loss of CG-NAP does not affect the localization of pericentrin. Our data further show that CG-NAP and pericentrin are recruited or anchored to the centrosome by distinct pathways: a Cep72-dependent and a PCM-1-dependent pathway, respectively (Dammermann and Merdes, 2002). Moreover, at the onset of mitosis, CG-NAP and pericentrin have essential but distinct functions in organizing spindle poles: pericentrin is essential for centrosome maturation (Zimmerman et al, 2004), whereas CG-NAP is essential for mature centrosomes to show microtubule-organizing activity. During this process, Kiz, recruited by Cep72, strengthens the integrity of mitotic centrosomes in a Plk1-mediated phosphorylation-dependent manner (Oshimori et al, 2006). It is important to note that among the several centrosome proteins examined, Plk1-mediated phosphorylation specifically strengthened the association between Kiz and pericentrin (Oshimori et al, 2006).
Our previous study shows that during spindle formation, forces generated by chromosome movement are converged on the centrosome by spindle microtubule-mediated mechanisms. The Kiz-mediated centrosome stabilization is essential to protect the PCM from the forces and prevent its fragmentation (Oshimori et al, 2006). Although Cep72 contributes to this process by recruiting Kiz, it also facilitates the tethering of the minus ends of non-centrosomal microtubules to the PCM. Accordingly, it seems that microtubule-mediated forces do not converge well on the Cep72-depleted centrosomes, which would explain the partial relief of PCM fragmentation in Cep72-depleted cells, a condition of Kiz deprivation from the centrosomes.
The pulling and pushing forces exerted by mitotic spindle are important for chromosomes to align at metaphase plate and segregate towards spindle poles. These forces create tension across the kinetochore and are required for silencing the spindle checkpoint (Pinsky and Biggins, 2005). The bipolar spindles assembled in the absence of Cep72 can neither align chromosomes efficiently (Figure 6E) nor generate sufficient tension between kinetochores to shut off the spindle-assembly checkpoint (Figure 6F). The data suggest that bundling and focusing of minus ends of spindle microtubules by non-centrosomal proteins, such as NuMA, alone are not sufficient for generating forces that are required for proper alignment and segregation of chromosomes. The microtubule minus-ends need to be focused and in contact with the rigid PCM structure at poles to accept and endure the forces generated during chromosomal movement. The Cep72 has key roles in both constructing rigid PCM structure and anchoring spindle poles to the PCM. We speculate that even in the acentrosomal spindles, which do not have a centriole but seem to contain PCM proteins at poles (Dictenberg et al, 1998), Cep72 may contribute to the functional spindle-pole formation. Further studies on the mechanisms of Cep72-mediated recruitment and anchoring of centrosomal proteins will provide much-needed insights into the formation of the spindle pole.
Materials and methods
Cep72 cDNA, plasmids, and antibodies
The human Cep72 cDNA clone (KIAA1519; GenBank accession number, NM_018140) was obtained from the Kazusa DNA Research Institute (Chiba, Japan). In this study, cDNAs encoding Cep72 were subcloned into pcDNA3-FLAG and pcDNA3-HA vectors. The wild-type Kiz or Kiz (T379A) was subcloned into the pME18S-Myc vector. The anti-Cep72 rabbit polyclonal antibodies were generated against amino-acids (a.a.) 1–107 and 321–427 of human Cep72 and affinity-purified. The antibodies against kendrin/pericentrin, CG-NAP, and GCP2 (gifts from Y Ono), α-tubulin, γ-tubulin (monoclonal antibody), centrin, and FLAG (Sigma, St Louis, MO), γ-tubulin (polyclonal antibodies; Santa Cruz Biotechnology, Santa Cruz, CA), NuMA (Monosan, Uden, The Netherlands), Myc (Cell Signaling Technology, Danvers, MA), and golgin-97 (Invitrogen, Carlsbad, CA) were also used.
Cell culture and synchronization
HeLa and HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) and U2OS cells were grown in RPMI-1640, both supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2 mM L-glutamine. For the RNAi experiments, HeLa or U2OS cells were transfected with siRNA and then synchronized using a double thymidine block. Approximately 10–12 h after release from the second thymidine block, cells were harvested for further analyses.
Plasmid DNA transfection
HeLa cells were plated on culture dishes with or without coverslips 1 day before transfection with FuGENE6 (Roche, Basel, Switzerland) using 0.5–5 μg of total DNA, depending on the experiments. HEK293T cells were plated on 60-mm dishes 1 day before calcium phosphate transfection with 5 μg total DNA. At 24–48 h after transfection, the cells were harvested and used for further experiments.
Microtubule depolymerization and microtubule-re-growth assay
HeLa cells were seeded on coverslips, transfected with siRNAs, and grown for 60 h as described above. For microtubule depolymerization, the cells were transferred into ice-cold medium containing 1 μM nocodazole, incubated for 30 min, and fixed with −20°C methanol for 5 min. For the microtubule re-growth assay, these cells were transferred into warm (37°C) medium to allow them to recover microtubule nucleation and fixed with –20°C methanol 5 or 15 min after re-growth.
RNA interference
RNA oligonucleotides targeting the following cDNA sequences were synthesized (JBioS, Saitama, Japan) and annealed using standard protocols: 5′-AAGCGATTTGAGCGTGTCCAA-3′ for Kiz siRNA, 5′-TTGCAGATCGCTGGACTTCAA-3′ for Cep72 siRNA, 5′-CAGGTGATGAAGGAAAGCCTT-3′ for CG-NAP siRNA, and 5′-AATTCTCCGAACGTGTCACGT-3′ for control siRNA.
These RNA duplexes (50 nM) were transfected into HeLa cells using LipofectAMINE RNAiMAX reagent (Invitrogen, Carlsbad, CA).
Indirect immunofluorescence microscopy
HeLa cells were grown on a glass coverslip, fixed with –20°C methanol for 5 min, and stained with primary antibodies and secondary antibodies coupled to Alexa488, Alexa555, or Cy3 (Invitrogen, Carlsbad, CA). DNA was stained with Hoechst33342 (Invitrogen). Immunofluorescence image stacks were captured on an Olympus IX-70 inverted microscope (Olympus, Melville, NY) controlled by Delta Vision Softworx (Applied Precision, Issaquah, WA) using × 60 or × 100 objective lenses. Deconvolution was performed, when necessary. The image stacks were quick projected and saved as Photoshop files. Data analysis was carried out using ImageJ software.
Immunoprecipitation and western blotting analysis
For immunoprecipitation experiments, the cells were washed once with cold phosphate-buffered saline (PBS) and lysed in TNE buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 12 mM β-glycerophosphate, 1 mM Na3VO4, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF)) for 1 h at 4°C on ice. The lysates were clarified by centrifugation and incubated with the appropriate antibodies (2 μg) for 2 h at 4°C. A total of 400–600 μg of lysates was used for immunoprecipitation and 5% of them were applied as input samples. After incubation, 20 μl of protein G Sepharose (GE Healthcare, Buckinghamshire, UK) were added and the mixtures were rotated for 1 h at 4°C. The beads were then washed five times in TNE buffer and re-suspended in gel sample buffer. The bound proteins were resolved by SDS–PAGE and transferred to an Immobilon-P membrane (Millipore, Billerica, MA). The membrane was blocked with 4% BSA and probed with primary antibodies. The immunoreactive proteins were detected using secondary antibodies conjugated with horseradish peroxidase (GE Healthcare) using the chemiluminescent reagents Western Lightning (PerkinElmer, Waltham, MA) or Immobilon Western (Millipore).
Live-cell time-lapse microscopy
U2OS cells stably expressing α-tubulin fused to the C-terminal portion of the Venus fluorescent protein (Supplementary Movies 1 and 2) were cultured on glass-bottomed dishes. During observation, the cells were maintained on the IX-70 microscope stage (Olympus) in warm, humid air supplemented with 5% CO2. Time-lapse image sequences were recorded at 30-s (Supplementary Movies 1 and 2) intervals.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary Figure 10
Supplementary Movie 1
Supplementary Movie 2
Supplemental Figures and Movies Legends
Acknowledgments
We thank Y Ono and M Takahashi for antibodies against kendrin/pericentrin, CG-NAP and GCP2; A Miyawaki for Venus fluorescent-protein expression plasmid; and N Tokai-Nishizumi, K Tanaka, and RF Whittier for fruitful discussions. This study was supported by the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (JSPS) to NO, grants-in-aid from JSPS and the Ministry of Education, Cultures, Sports, Science and Technology, Japan to MO and TY, and Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), MEXT, Japan.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM (2007) Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol 8: 451–463 [DOI] [PubMed] [Google Scholar]
- Blagden SP, Glover DM (2003) Polar expeditions—provisioning the centrosome for mitosis. Nat Cell Biol 5: 505–511 [DOI] [PubMed] [Google Scholar]
- Bornens M (2002) Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 14: 25–34 [DOI] [PubMed] [Google Scholar]
- Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R, Nigg EA (2003) Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell 5: 113–125 [DOI] [PubMed] [Google Scholar]
- Compton DA, Cleveland DW (1994) NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr Opin Cell Biol 6: 343–346 [DOI] [PubMed] [Google Scholar]
- Dammermann A, Merdes A (2002) Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol 159: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ (1998) Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Choi YK, Rattner JB, Qi RZ (2008) CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the γ-tubulin ring complex. Mol Biol Cell 19: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ (2007) Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol 17: 1960–1966 [DOI] [PubMed] [Google Scholar]
- Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A (2006) NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol 172: 505–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M, Tassin AM (2003) Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol Cell 14: 4260–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160: 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocca MC, Jin M, Goldenring JR (2006) AKAP350 modulates microtubule dynamics. Eur J Cell Biol 85: 611–619 [DOI] [PubMed] [Google Scholar]
- Luders J, Patel UK, Stearns T (2006) GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol 8: 137–147 [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167: 831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW (2000) Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A, Ramyar K, Vechio JD, Cleveland DW (1996) A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87: 447–458 [DOI] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M (2000) Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 113: 3013–3023 [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA (1995) Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature 378: 638–640 [DOI] [PubMed] [Google Scholar]
- Nigg EA (2002) Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2: 815–825 [DOI] [PubMed] [Google Scholar]
- Nigg EA (2007) Centrosome duplication: of rules and licenses. Trends Cell Biol 17: 215–221 [DOI] [PubMed] [Google Scholar]
- O′Connell CB, Khodjakov AL (2007) Cooperative mechanisms of mitotic spindle formation. J Cell Sci 120: 1717–1722 [DOI] [PubMed] [Google Scholar]
- Oshimori N, Ohsugi M, Yamamoto T (2006) The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X (2000) Centrosome maturation. Curr Top Dev Biol 49: 449–470 [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M (1996) Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci 109: 3089–3102 [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Biggins S (2005) The spindle checkpoint: tension versus attachment. Trends Cell Biol 15: 486–493 [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Andreassen PR, Lacroix FB, Wilson L, Margolis RL (2001) Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc Natl Acad Sci USA 98: 4492–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ (2004) The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol 16: 49–54 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y (2002) Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring γ-tubulin ring complex. Mol Biol Cell 13: 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Rusan NM, Wadsworth P (2003) Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol 13: 1894–1899 [DOI] [PubMed] [Google Scholar]
- Wadsworth P, Khodjakov A (2004) E pluribus unum: towards a universal mechanism for spindle assembly. Trends Cell Biol 14: 413–419 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T (1995) Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378: 578–583 [DOI] [PubMed] [Google Scholar]
- Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L (2008) The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol 18: 136–141 [DOI] [PubMed] [Google Scholar]
- Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ (2004) Mitosis-specific anchoring of γ tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell 15: 3642–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure 9
Supplementary Figure 10
Supplementary Movie 1
Supplementary Movie 2
Supplemental Figures and Movies Legends