Abstract
Chorismate mutase catalyzes a key step in the shikimate biosynthetic pathway towards phenylalanine and tyrosine. Curiously, the intracellular chorismate mutase of Mycobacterium tuberculosis (MtCM; Rv0948c) has poor activity and lacks prominent active-site residues. However, its catalytic efficiency increases >100-fold on addition of DAHP synthase (MtDS; Rv2178c), another shikimate-pathway enzyme. The 2.35 Å crystal structure of the MtCM–MtDS complex bound to a transition-state analogue shows a central core formed by four MtDS subunits sandwiched between two MtCM dimers. Structural comparisons imply catalytic activation to be a consequence of the repositioning of MtCM active-site residues on binding to MtDS. The mutagenesis of the C-terminal extrusion of MtCM establishes conserved residues as part of the activation machinery. The chorismate-mutase activity of the complex, but not of MtCM alone, is inhibited synergistically by phenylalanine and tyrosine. The complex formation thus endows the shikimate pathway of M. tuberculosis with an important regulatory feature. Experimental evidence suggests that such non-covalent enzyme complexes comprising an AroQδ subclass chorismate mutase like MtCM are abundant in the bacterial order Actinomycetales.
Keywords: enzyme catalysis, multi-enzyme complex, Mycobacterium tuberculosis Rv0948c, shikimate pathway, X-ray crystal structure
Introduction
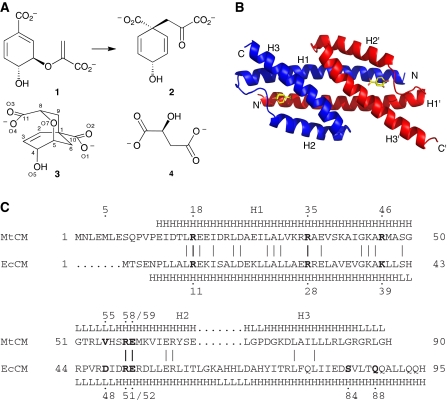
The shikimate pathway serves the biosynthesis of aromatic compounds in prokaryotes, ascomycete fungi, apicomplexans, and plants (Haslam, 1993; Richards et al, 2006). Its absence in mammals makes it a potential antibiotic drug target. The initial step of the shikimate pathway is the synthesis of 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) from phosphoenolpyruvate and D-erythrose-4-phosphate by DAHP synthase. Six subsequent enzymatic steps lead to the central metabolite chorismate, at which the metabolic flux diverges into several branches. Chorismate mutase (CM) accelerates the rearrangement of chorismate to prephenate (Figure 1A), the last common biosynthetic precursor of phenylalanine and tyrosine, typically by more than a millionfold (Andrews et al, 1973).
Figure 1.
Catalytic reaction, structure, and sequence of AroQ chorismate mutases (CMs). (A) CMs convert chorismate (1) to prephenate (2) through a transition state that resembles inhibitor 3 (Bartlett and Johnson, 1985), and to some extent L-malate (4). (B) Structure of the AroQ prototype EcCM with transition-state analogue 3 bound to its active sites (Lee et al, 1995a). (C) Sequence alignment and secondary structure (H=helix; L=loop) of MtCM (subclass AroQδ) and EcCM (AroQα), according to the crystal structures of MtCM (PDB ID: 2VKL) and EcCM (1ECM) (Lee et al, 1995a). Sequence identity is 25.3%; bold, active-site residues in EcCM and their homologues in MtCM.
Structurally, CMs can be classified into two unrelated groups (Lee et al, 1995b; MacBeath et al, 1998). The rare AroH class has a trimeric pseudo α/β barrel structure, as exemplified by the monofunctional CM from Bacillus subtilis (Chook et al, 1993). The largely α-helical AroQ class is more widespread and has recently been divided into the subclasses AroQα through to AroQδ (Ökvist et al, 2006). The AroQα subclass is represented by the structurally characterized CM domain of the bifunctional Escherichia coli chorismate mutase-prephenate dehydratase (EcCM; Figure 1B) (Lee et al, 1995a), whereas the more sophisticated allosteric CM from Saccharomyces cerevisiae (Xue et al, 1994) represents the AroQβ subclass.
In Mycobacterium tuberculosis, the causative agent of tuberculosis, a highly active CM is encoded by the open reading frame Rv1885c (Prakash et al, 2005; Sasso et al, 2005; Kim et al, 2006). The structure of this protein (*MtCM) was solved and assigned to the AroQγ subclass of CMs (Ökvist et al, 2006). However, this CM is secreted and thus unavailable to support the cytoplasmic biosynthesis of phenylalanine and tyrosine in M. tuberculosis (Sasso et al, 2005; Kim et al, 2006). There is another open reading frame, Rv0948c, which encodes a protein with homology to AroQ CMs. The primary sequence of this protein, which we refer to as MtCM, shows features that are quite unusual for AroQ CMs, prompting its assignment to a new AroQδ subclass (Ökvist et al, 2006). For instance, MtCM seemed to lack the counterpart of the second half of helix H3 carrying the catalytically crucial Gln 88 in EcCM (Liu et al, 1996), and it has an arginine residue (Arg 46) at the position of the most prominent active-site lysine typical for AroQ CMs (Liu et al, 1996; MacBeath et al, 1998) (Figure 1C). The aroQδ gene from M. tuberculosis was investigated by several groups (Prakash et al, 2005; Sasso et al, 2005; Ökvist et al, 2006; Kim et al, 2008), and the purified MtCM was reported to possess unusually poor CM activity in vitro (Prakash et al, 2005; Kim et al, 2008).
Here, we show that MtCM forms a complex with the first enzyme of the shikimate pathway and that complex formation both increases the CM activity by more than two orders of magnitude and endows MtCM with regulatory features. Furthermore, we report the first crystal structure of this non-covalent protein complex at high resolution, with and without a bound transition-state analogue, and provide a detailed mechanistic explanation for the observed catalytic activation of MtCM.
Results
Cloning of the M. tuberculosis aroQδ gene
To characterize the intracellular CM from M. tuberculosis, we cloned the open reading frame Rv0948c from chromosomal DNA. Rv0948c, predicted from the genome sequence of strain H37Rv (Cole et al, 1998), has three methionine codons that could possibly initiate translation of the native MtCM, with the second one being the most likely site (Figure 2) (Kim et al, 2008). On a high-copy expression plasmid, the corresponding gene encoding a 90-residue protein (Figure 1C) complemented the CM-deficiency of E. coli mutant strain KA12/pKIMP-UAUC (Kast et al, 1996) (see Supplementary Table SI). This established that this MtCM version functions as an active CM.
Figure 2.
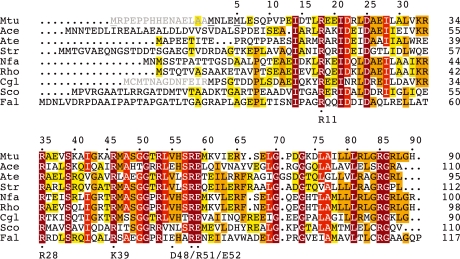
Multiple sequence alignment of representative AroQδ proteins. A PSI-BLAST search (Altschul et al, 1997) was carried out to identify close MtCM homologues. Hit sequences were retained if they, compared with EcCM, had a shortened C-terminus and an arginine at the position corresponding to EcCM Lys 39. The alignment shows an arbitrarily selected sequence from each genus. The sequences were directly adopted from database entries and may not necessarily identify the correct start position. In this work, shorter versions were used for Mtu (MtCM) and Cgl (CgCM), as indicated by an N-terminal extension in grey and adjustments in the numbering. Three possible start methionines for MtCM are underlined. Although circumstantial evidence from frameshift experiments in Mycobacterium smegmatis was interpreted in favour of the originally predicted (Cole et al, 1998) 105-amino acid MtCM variant (Rv0948c) (Schneider et al, 2008), this study focused on the 90-residue version starting with the second methionine (residue numbers on top; also corresponding to MtCM in Figure 1C). This decision was based on the inspection of possible Shine–Dalgarno sequences (Doran et al, 1997), lack of sequence homology in the N-terminal region of predicted AroQδ proteins (even observed when comparing M. smegmatis and M. tuberculosis variants), and the similarity of the catalytic parameters (unpublished results; Kim et al, 2008) for the longest version and MtCM used here. Important active-site residues in EcCM with counterparts in AroQδ enzymes are identified below the alignment. Residues are boxed according to sequence identity within the AroQδ subgroup from yellow (⩾33%), orange (⩾50%), bright red (⩾75%), to dark red (100% identity). Mtu, M. tuberculosis; Ace, Acidothermus cellulolyticus; Ate, Actinoplanes teichomyceticus; Str, Salinospora tropica; Nfa, Nocardia farcinica; Rho, Rhodococcus sp.; Cgl, Corynebacterium glutamicum; Sco, Streptomyces coelicolor; Fal, Frankia alni.
Crystal structure of MtCM containing L-malate
MtCM, the first representative identified as an AroQδ-subclass CM (Ökvist et al, 2006), was overproduced in the CM-deficient E. coli strain KA13 (MacBeath et al, 1998) in its native form and crystallized. The crystal structure of MtCM was solved at 1.65 Å resolution in complex with L-malate, a component of the crystallization buffer (Table I). The dicarboxylic acid malate may have promoted crystallization by its virtue of resembling the normal ligands of MtCM (Figure 1A) and thus being able to bind to and stabilize the active-site region. While this work was in progress, Kim et al (2008) deposited the 2.0-Å structure of ligand-free MtCM in the Protein Data Bank (PDB) (PDB ID: 2QBV).
Table 1.
Summary of data collection and refinement statistics
| MtCM with bound L-malate | MtCM–MtDS complex | MtCM–MtDS–TSA (3) ternary complex | |
|---|---|---|---|
| Data collection | |||
| Space group | P43212 | P3221 | P3221 |
| Cell dimensions | |||
| a, b, c (Å) | 62.3 62.3 37.5 | 204.0, 204.0, 66.5 | 205.9, 205.9, 67.2 |
| α,β,γ (deg) | 90.0 90.0 90.0 | 90.0 90.0 120.0 | 90.0 90.0 120.0 |
| Resolution (Å) | 20.7–1.65 (1.74–1.65) | 66.8–2.15 (2.27–2.15) | 40.93-2.35 (2.48–2.35) |
| Rsym or Rmerge | 0.046 (0.68) | 0.109 (0.53) | 0.105 (0.45) |
| I/σ(I) | 23.8 (2.2) | 14.2 (2.5) | 9.8 (2.7) |
| Completeness (%) | 99.8 (100.0) | 96.6 (81.0) | 98.9 (95.4) |
| Redundancy | 6.6 (6.4) | 6.8 (3.8) | 3.6 (3.2) |
| Refinement | |||
| Resolution (Å) | 19.0–1.65 | 66.8–2.15 | 39.84–2.35 |
| Number of unique reflections | 8844 | 79086 | 63874 |
| Rwork/Rfree | 0.180/0.235 | 0.186/0.221 | 0.193/0.230 |
| Number of atoms | |||
| Protein | 659 | 7519 | 8127 |
| Ligand/ion | 9 | 81 | 105 |
| Water | 37 | 351 | 214 |
| B-factors (Å2) | |||
| Protein | 23.8 | 32.6 | 32.0 |
| Ligand/ion | 26.6 | 36.5 | 41.3 |
| Water | 35.0 | 31.5 | 26.1 |
| r.m.s.d. values from ideal geometry | |||
| Bond lengths (Å) | 0.019 | 0.017 | 0.014 |
| Bond angles (deg) | 1.6 | 1.6 | 1.6 |
| PDB entry code | 2VKL | 2W19 | 2W1A |
| Residues not modelled | |||
| MtCM | A1–12, A90 | C1–18, D1–15 | C1–12, D1–11 |
| MtDS | A[-9]-A2, A264–266, A372–380, A414–442, B[-9]-B2, B10–14, B372–380, B414–442 | A[-9]-A2, A234–240, A264–265, B[-9]-B2, B11–14, B234–240 | |
| One crystal was used per structure. Values in parentheses are for highest-resolution shell. | |||
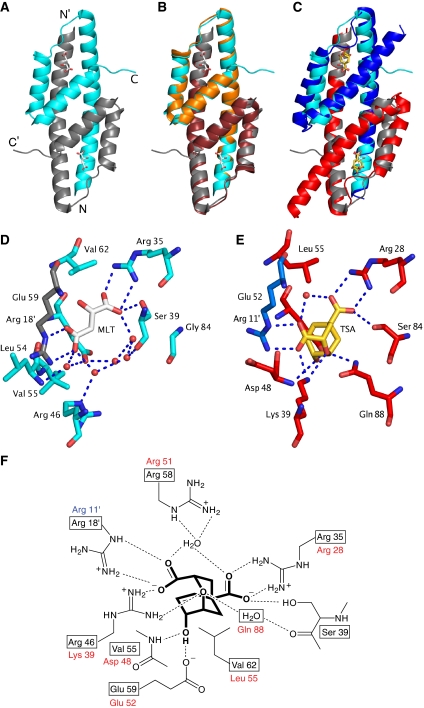
As expected from the sequence similarity to EcCM (Figure 1C), MtCM adopts the same overall fold consisting of an intertwined dimer with three helices per protomer, forming two active sites, each embedded in a four-helix bundle (Figure 3A–C). The most eye-catching difference between MtCM and EcCM is the truncation of helix H3. In fact, the C-terminal residues Gly 86 to His 90 of MtCM take on a structure, stabilized by extensive crystal contacts, that extends away from the active site rather than being part of H3 (Figure 3C).
Figure 3.
Structure of MtCM and active-site details. Ligands are shown in stick representation (carbons in white for malate, in gold for inhibitor 3). (A) Overall structure of the malate-binding MtCM dimer (PDB ID: 2VKL). The two protomers are represented in cyan and grey, respectively; N- and C-termini are labelled. (B) Superimposition of MtCM as in (A) with unliganded MtCM dimer in orange/ruby (2QBV) (Kim et al, 2008). (C) Superimposition of MtCM as in (A) with EcCM liganded to 3 (1ECM) (Lee et al, 1995a). EcCM protomers are highlighted in blue and red. (D) Active-site details of malate (MLT)-bound MtCM. An apostrophe identifies residues contributed by the other protomer. Red spheres and broken lines represent water molecules and key hydrogen bonds, respectively. Arg 58 was omitted for clarity. (E) Active-site details of EcCM with bound 3 (TSA). Arg 51, which coordinates to the water molecule bridging the carboxylates of 3, was omitted for clarity. (F) Scheme of MtCM key active-site residues (boxed) with bound 3 (heavy lines; interactions according to 2W1A, see below). Homologous EcCM residues are listed next to the boxes.
Active-site residues of MtCM
One malate ion is bound to each active site of the MtCM dimer in a manner mimicking the binding of the transition-state analogue 3 in EcCM (Figure 3D–F). Furthermore, most features of EcCM's active site are conserved in MtCM. For instance, residues Arg 35 and Arg 18′, corresponding to Arg 28 and Arg 11′ in EcCM, are in place to coordinate both carboxylate groups of the ligand. Glu 59 of MtCM is found in a position corresponding to EcCM's Glu 52, which binds the hydroxyl group of 3.
As discussed above, one striking difference between the active sites is the replacement of the otherwise strictly conserved (MacBeath et al, 1998) lysine residue of EcCM (Lys 39; Figure 3E) by an arginine in MtCM (Arg 46; Figure 3D). Lys 39 hydrogen bonds to the ether oxygen of the transition-state analogue 3 (Lee et al, 1995a), and is believed to be required for electrostatic stabilization of the transition state in AroQ CMs (MacBeath et al, 1998). In fact, substitution in EcCM of Lys 39 by Ala, Gln or Arg reduces the catalytic efficiency (kcat/Km) by five orders of magnitude to essentially background levels (Liu et al, 1996).
Moreover, also the second residue that interacts with the ether oxygen of 3 in EcCM, Gln 88 (Figure 3E), lacks a similar counterpart in MtCM (Figure 1C). Instead, the void is filled with water molecules. The importance of Gln 88 was shown by Q88A or Q88K mutation in EcCM that lowered the catalytic efficiency by 2 × 104-fold (Liu et al, 1996). The truncation of helix H3 of MtCM leaves the active site open in this region and accessible to bulk solvent, in sharp contrast to being buried in the protein, as is the case for EcCM (Lee et al, 1995a), *MtCM (Ökvist et al, 2006), and mMjCM (Pervushin et al, 2007). Another, more subtle consequence of the shorter H3 in MtCM is the lack of a homologous counterpart to EcCM's Ser 84, which hydrogen bonds to O1 of 3. Instead, its role is assumed by Ser 39 of helix H2 in MtCM (Figure 3D–F).
Enzymatic activity of MtCM
The CM activity of MtCM follows Michaelis–Menten kinetics, with a catalytic efficiency kcat/Km of 1.7 × 103 M−1 s−1. This is two orders of magnitude lower than usual for wild-type CMs (Schnappauf et al, 1997; Mattei et al, 1999; Sasso et al, 2005) (Table II). The rather poor activity of MtCM was already noticed in earlier reports (Prakash et al, 2005; Kim et al, 2008). To examine whether alternative MtCM start sites (described in Figure 2) could influence activity, two additional MtCM variants were constructed that were either 15 amino acids longer or 4 residues shorter than MtCM. While both N-terminal variants were able to complement the CM deficiency of KA12/pKIMP-UAUC in vivo (Supplementary Table SI), kinetic studies indicated similarly low catalytic efficiencies as found for the 90-residue MtCM variant, independent of the start site (kcat/Km values roughly within a factor of two; unpublished data, 2007), in agreement with a recent report (Kim et al, 2008).
Table 2.
Catalytic parameters for the chorismate mutase reaction catalyzed by different enzymesa
| Proteinb | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) | Fold activation by MtDS | Relative specific activity when complexed with MtDSc |
|---|---|---|---|---|---|
| MtCMd | 2.0±0.1 | 1140±20 | 1750±90 | 140±35 | NA |
| MtDSe | ND | ND | < 20 | NA | NA |
| MtCM+MtDSd | 8.1±1.9 | 34±3 | (2.4±0.6) × 105 | NA | NA |
| H6-CgCMd | ND | ≫2000 | 390±60 | 12±2 | NA |
| H6-MtCM | ND | >1400 | 920 | 67±12f | 100% |
| H6-MtCM R46K | ND | >1000 | 17±1 | 300±80f | 8% |
| H6-MtCM G86A | 0.11 | 570 | 190 | 7±1f | 3% |
| H6-MtCM R87A | ND | >1500 | 670 | 33±1f | 36% |
| H6-MtCM L88A | ND | >1400 | 1040 | 7.6±0.6f | 14% |
| H6-MtCM L88* | ND | >1700 | 1100 | 2.8±0.2f | 6% |
| BsCM | 50 | 85 | 5.9 × 105 | NA | NA |
| *MtCM | 50 | 180 | 2.7 × 105 | NA | NA |
| ScCMg | 180 | 3800 | 4.7 × 104 | NA | NA |
| ScCM+10 μM Trpg | 170 | 400 | 4.4 × 105 | NA | NA |
| aStandard CM assays were carried out at 30°C in 50 mM potassium phosphate (pH 7.5), unless stated otherwise. kcat (calculated per CM protomer) and Km were derived from Michaelis–Menten plots with ⩾5 different substrate concentrations. The s.d. values given (σn−1) generally refer to assays carried out with two independent protein preparations. ND indicates that Km was too high for accurate determination of individual catalytic parameters at experimentally accessible chorismate concentrations (typically up to ≈2 mM). NA, not applicable. The data for BsCM (Mattei et al, 1999), *MtCM (Sasso et al, 2005), and ScCM (Schnappauf et al, 1997) were taken from the literature. | |||||
| bMtCM is the untagged and H6-MtCM the His-tagged version of the Mycobacterium tuberculosis CM (AroQδ subclass). MtDS is the His-tagged M. tuberculosis DAHP synthase. H6-CgCM is a proteolytically stable AroQδ CM derived from open reading frame Cgl0853 from Corynebacterium glutamicum by replacing the 13 N-terminal amino acids with a His tag. BsCM, *MtCM, and ScCM refer to CMs from Bacillus subtilis (monofunctional AroH), M. tuberculosis (secreted AroQγ), and Saccharomyces cerevisiae (allosteric AroQβ), respectively. | |||||
| cDetermined as the relative activity of the His-tagged MtCM variants in the presence of MtDS compared to wild-type H6-MtCM complexed with MtDS (=100%). The percentage reflects the ratio of averaged initial velocities (v0 (MtDS+MtCM)), normalized by MtCM-variant and chorismate concentrations. v0 (MtDS+MtCM) was measured at least twice for each variant and was obtained with 30 nM MtCM, 300 nM MtDS at ≈23 μM chorismate in BTP+ buffer, which is 50 mM BTP (1,3-bis[tris(hydroxymethyl)methylamino]propane), pH 7.5, 0.5 mM TCEP [tris(2-carboxyethyl)phosphine hydrochloride], 0.2 mM phosphoenolpyruvate, and 0.1 mM MnCl2. BTP+ contains buffer components recommended in some protocols to stabilize MtDS (Webby et al, 2005). | |||||
| dMeasured in 50 mM BTP, pH 7.5 (except for H6-CgCM+MtDS, which was measured in BTP+). When determined in 50 mM potassium phosphate, pH 7.5, the kcat for MtCM was ≈40% lower. The activation factor (=ratio of kcat/Km(MtDS+CM) over kcat/Km(CM)) was determined at 2 μM MtDS, corresponding to a 200-fold molar excess over the CM. | |||||
| eNo CM activity above background could be detected for 2 μM MtDS in the presence of ≈50 μM chorismate in BTP+ buffer (two independent measurements). | |||||
| fAn apparent activation factor was estimated as the ratio of ⩾2 averaged initial velocities (v0(MtDS+MtCM)), normalized by MtCM-variant and chorismate concentrations, over kcat/Km (MtCM). The velocities v0(MtDS+MtCM) were obtained using 30 nM MtCM variant, 300 nM MtDS at ≈23 μM chorismate in BTP+. | |||||
| gThe assay buffer was 100 mM Tris–HCl, pH 7.6, 2 mM EDTA, and 20 mM DTT (Schnappauf et al, 1997). | |||||
We tested whether the unusual Arg 46 in the active site of MtCM (Figure 3) was responsible for the reduced activity by changing it to the strictly conserved Lys of all other AroQ subclasses. Although the charge-conservative K39R mutation in EcCM essentially killed CM activity (Liu et al, 1996), the reverse R46K mutation in MtCM did not improve activity; on the contrary, kcat/Km dropped by another 50-fold (Table II).
It was further probed whether the mediocre activity of MtCM could be altered by changing assay conditions or by adding potential co-factors or regulators. However, neither pH variation between 5.9 and 8.7, nor provision of 0.1 mg/ml BSA, 2 mM Ca2+, 10 mM Mg2+, 1 mM EDTA, 1 mM EGTA, 1 mM 1,10-phenanthroline, 1 mM phenylalanine, 1 mM tyrosine, 1 mM tryptophan, or 0.6 mM salicylate affected catalytic activity by more than a factor of 2. As CMs sometimes occur as covalently fused domains to other shikimate-pathway enzymes (MacBeath et al, 1998), we wondered whether such enzymes could non-covalently influence MtCM activity. Therefore, M. tuberculosis open reading frames Rv2178c and Rv3838c, encoding DAHP synthase (MtDS) and prephenate dehydratase (MtPDT), respectively, were cloned and overexpressed in E. coli KA13. While addition of purified MtPDT to a crude MtCM preparation had no effect on its CM activity, a dialyzed lysate of cells producing MtDS increased the chorismate consumption of MtCM by up to a factor of 25 (data not shown). The MtDS lysate did not have any detectable CM activity over background on its own. This result suggested that MtCM productively interacts with MtDS.
Complex formation with MtDS strongly activates MtCM
To characterize the interaction between the two enzymes, we produced N-terminally His-tagged MtDS (472 residues) in E. coli strain KA13. The CM activity assays with purified MtDS corroborated the observations with the crude lysates. While not showing significant CM activity on its own, an excess of purified MtDS was capable of activating MtCM by more than two orders of magnitude to a catalytic efficiency (kcat/Km) typical for wild-type CMs (Table II). This activation is primarily caused by a more than 30-fold-decreased Km value, but also by a four-fold increase in kcat. In contrast, MtDS activity was not affected by addition of an up to 100-fold molar excess of pure MtCM. The measured kcat/Km values for phosphoenolpyruvate were identical within experimental error (6%), and they were within a factor of two of the kcat/Km (8.4 × 104 M−1 s−1) determined previously for MtDS (Webby et al, 2005).
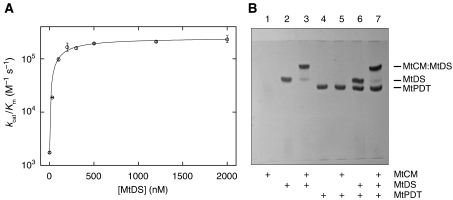
Assuming that a physical association of MtCM with MtDS correlates with the boost in MtCM's catalytic activity, the interaction between the two proteins was examined by titration of MtCM with different concentrations of MtDS. Under all conditions, CM activity followed hyperbolic (Michaelis–Menten) kinetics. From the titration data shown in Figure 4A, an apparent affinity constant Kd,app of 140±30 nM was calculated for a hypothetical 1:1 stoichiometry. It should be noted that this experiment necessitated the presence of chorismate, which probably affects the Kd,app value. This assessment is supported by preliminary gel-filtration experiments, in which we could observe co-elution of MtCM with the MtDS fraction after Superdex 75 size-exclusion chromatography, but exclusively in the presence of chorismate (≈500 μM).
Figure 4.
Interaction between MtCM and MtDS. (A) Titration of MtCM with MtDS. The catalytic efficiency (kcat/Km) of the CM reaction was determined at different MtDS concentrations in 50 mM BTP (pH 7.5) at 30°C. The MtCM concentration was held constant at 10 nM (100 nM in the absence of MtDS). At each MtDS concentration, Michaelis–Menten kinetics with ⩾5 different chorismate concentrations were measured. Error bars correspond to the s.d. of ⩾2 independent Michaelis–Menten kinetics. (B) Band-shift assay. Purified proteins were mixed according to the scheme below the gel (at 10 μM final concentration each) and separated on a 12.5% native polyacrylamide gel at 15°C with 0.25 M Tris and 0.88 M L-alanine (pH 8.8) as the running buffer (further details in the Supplementary data). Proteins were stained with Coomassie Blue. The band shifts in lanes 3 and 7 compared with lanes 2 and 6 indicate an interaction between MtCM and MtDS. MtPDT is prephenate dehydratase from M. tuberculosis. The absence of an MtCM band when loaded alone (lane 1) is possibly due to a lack of a net charge at the pH of the running buffer, resulting in diffusion of MtCM at the site of sample application.
Native polyacrylamide gel electrophoresis (PAGE) of purified proteins provided additional and complementary evidence for the existence of the MtCM–MtDS complex in solution. When MtCM was mixed with MtDS before native PAGE, a band shift was observed relative to the band of MtDS alone (Figure 4B, lanes 3 and 2, respectively), visualizing the interaction of the two proteins. In contrast, MtPDT does not seem to associate with MtCM or MtDS, as it did not induce a band shift (Figure 4B, lanes 4–7).
Crystal structure of the non-covalent MtCM–MtDS complex
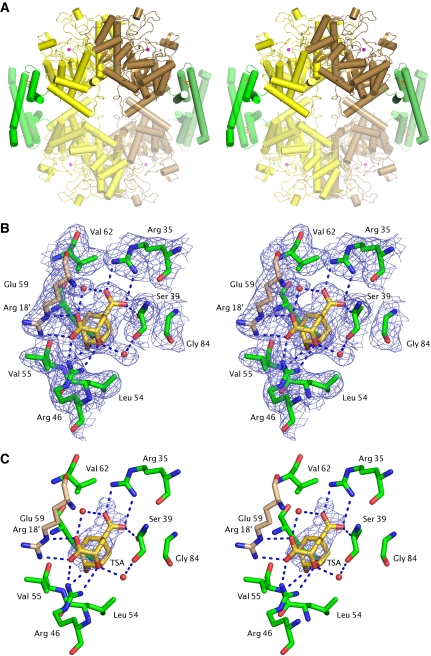
To investigate the nature of the non-covalent interaction between MtCM and MtDS and gain insight into the mechanism of MtCM activation, we determined the crystal structure of the MtCM–MtDS complex in the presence and absence of ligand 3. The crystals of the complex were found to be isomorphous to the previously determined MtDS structure (PDB ID: 2B7O) (Webby et al, 2005), which was used to solve the complex structure by molecular replacement (Table I and Supplementary data). The MtDS protomers adopt TIM barrel folds, essentially identical to the crystal structure of the uncomplexed protein (Webby et al, 2005). The asymmetric unit contains a homodimer (buried interface ca. 1600 Å2 per protomer) held together by exchanging N-terminal strands that form an antiparallel β-ribbon. In addition, in the complex each MtDS protomer contacts one protomer of MtCM. Exploiting two-fold crystallographic symmetry completes the CM active sites by generating MtCM homodimers strongly resembling functional EcCM. Simultaneously, an MtDS homotetramer is formed (average contact area 1062 Å2 per MtDS protomer), which results in an MtCM–MtDS hetero-octamer consisting of a central core of four identical MtDS subunits adorned on opposite peripheral sides by two MtCM dimers (Figure 5A). A stereo picture containing the backbone trace of the complex is provided as Supplementary Figure S1.
Figure 5.
Structure of the MtCM–MtDS complex. (A) Stereo cartoon of the entire MtCM–MtDS hetero-octameric complex. Green, MtCM; yellow/brown, MtDS dimer; red spheres, manganese atoms; sticks, inhibitor 3. Protomers placed by exploiting two-fold crystallographic symmetry are shown in faded colour. (B) Stereo figure showing the active site of MtCM with ligand 3 (gold carbons) of the MtCM–MtDS complex (simulated annealed composite OMIT map contoured at 1.5 σ). Arg 18′ (wheat carbons) is provided from the second MtCM protomer. For comparison, the electron density for the binary MtCM–MtDS complex is shown as Supplementary Figure S3. (C) Stereo figure as in (B), showing changes in the active site on binding of 3 with an Fo–Fo map (contoured at 2 σ). The electron density is carved out around 3. Additional density differences caused by a 1 Å shift in crystal cell parameters are not shown for clarity.
According to a prediction from the EBI PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html; Krissinel and Henrick, 2007), this octameric quaternary structure is stable, with a free energy ΔGdiss of 23.7 kcal mol−1 (at 27°C) for the dissociation of the hetero-octamer into an MtDS tetramer and two MtCM dimers. Moreover, the MtDS homotetramer is predicted to be stable (ΔGdiss=23.3 kcal mol−1 for the dissociation into dimers; derived from 2B7O (Webby et al, 2005)), as is the MtCM dimer (ΔGdiss=27.1 kcal mol−1; structure with bound malate; 2VKL). Although dissociation of the MtCM–MtDS complex into stable components qualitatively agrees with the observed activity titration results (Figure 4A), the free energy calculated from the experimental Kd,app value (ΔGdiss=9.5 kcal mol−1; 30°C) may not be directly comparable with the ΔGdiss calculated from the structure, as it still needs to be verified, which oligomeric states actually occur in solution or are relevant for CM activity.
The MtDS tetramerization interface comprises helices α2a and α2b protruding from the TIM barrel, with Phe 227 and Cys 231 from one protomer fitting into a pocket in the opposite protrusion formed by the base of the α2a/α2b helix hairpin and the C-terminal residues of helix α1. The MtCM–MtDS interface on average buries 855 Å2 per subunit and involves residues from helices H2 and H3 as well as the loop between H1 and H2 of MtCM and MtDS helices α7, α8, α2b, and the loop preceding α2b. The interface contains numerous non-polar and polar interactions, including the close packing of MtCM's C-terminal residues Leu 88, Gly 89, and His 90 into a shallow groove on the MtDS surface between helices α7 and α8 (Figures 5A and 7B), which otherwise forms a solvent-exposed hydrophobic patch. The C-terminal His 90 of MtCM also shows contacts (<4 Å) with MtDS through the side chain of Glu 396 and the C-terminal carboxylate group of MtDS.
Active site of MtCM with bound transition-state analogue in the MtCM–MtDS complex
The CM active site of the MtCM–MtDS complex containing compound 3 (Figure 5C and list of protein–ligand interactions in Supplementary Table SII) shows clear differences to the MtCM-malate structure shown in Figure 3D. In particular, the positioning/interactions of the three active-site residues Arg 46, Val 55, and Glu 59 (and to a lesser extent Arg 18′) are distinct (Figures 6B and 7B).
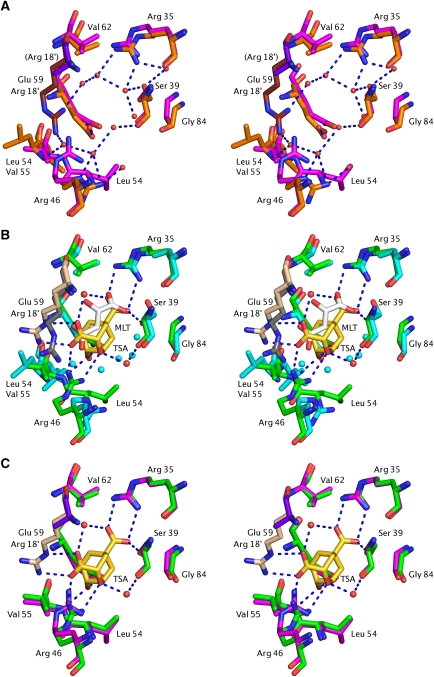
Figure 6.
Conformational changes in MtCM active site on MtDS binding. (A) Unliganded MtCM (PDB ID 2QBV, orange/ruby carbons) (Kim et al, 2008) and MtCM–MtDS complex without ligand (2W19, magenta/purple carbons). Arg 18′ in parentheses indicates that this residue (purple) has been modelled as an alanine in the unliganded MtCM–MtDS complex due to lack of electron density. (B) Ternary complex of MtCM–MtDS (PDB ID 2W1A, green/wheat carbons) with 3 (TSA, carbons in gold) and MtCM binding malate (2VKL; cyan/grey carbons). Key hydrogen bonds between MtDS-complexed MtCM and inhibitor 3 are shown by broken blue lines; red and cyan spheres represent water molecules in the ternary and the malate complex, respectively. Glu 59, which stretches from helix H2 into the active site (behind the TSA), hydrogen bonds to the hydroxyl group of inhibitor 3 in the ternary complex. (C) Ligand-free MtCM–MtDS complex superimposed onto the ternary complex, represented as in panels (A) and (B), respectively.
In the MtCM–MtDS complex, the side chain of Arg 46 interacts with both the ether oxygen of 3 and one of its carboxylate groups, in a manner similar to Lys 39 in EcCM (compare Figures 3E–F and 5B–C). Val 55 in the ternary complex is involved in a hydrogen bond to the hydroxyl group of 3 through its main chain amide group (similar to Asp 48 of EcCM), whereas the MtCM-malate structure features Leu 54 at the equivalent position in an orientation unsuitable to provide such a hydrogen bond (Figure 3D–F and 6B). Glu 59 (positioned behind 3 in Figure 6B–C) superimposes rather well in the structures with and without MtDS (and also superimposes well with EcCM residue Glu 52; Figure 3), but it has different interaction partners in the two structures. While this residue interacts with Arg 53 (from the H1–H2 loop) in the MtCM-malate structure, it hydrogen bonds to Arg 85 (positioned in H3) in the ternary complex (Figure 7D). Arg 58 in the active site of the MtCM–MtDS ternary complex coordinates a water molecule that bridges both carboxylate groups of 3 (Figure 7C), in the same way as Arg 51 in EcCM (Figure 3F), while interacting with the hydroxyl group of malate in an analogous position in the MtCM structure (Figure 7B). A second tightly bound water molecule coordinates to the ether oxygen of 3, mimicking the interaction provided by EcCM's Gln 88 (Figure 3F).
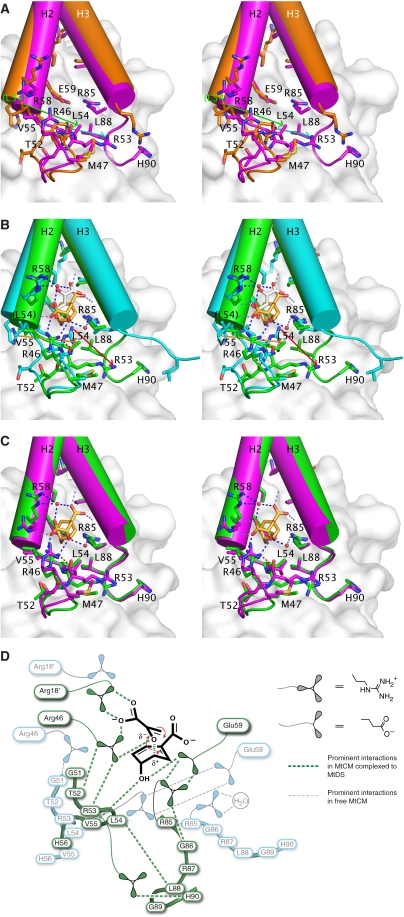
Figure 7.
Global conformational changes in MtCM on binding to MtDS. (A) Stereo figure of ligand-free MtCM (2QBV, orange carbons) superimposed onto the ligand-free MtCM–MtDS complex (2W19, magenta carbons). Residue labels concern the MtCM–MtDS complex (except for Leu 54 in parentheses belonging to the MtCM structure). Green and cyan arrows highlight the difference in position for Leu 54 and Arg 53, respectively (MtCM → MtCM–MtDS). (B) Stereo figure of superimposed ligand complexes of MtCM (2VKL, cyan carbons; malate, white carbons) and the MtCM–MtDS complex (2W1A, green carbons; inhibitor 3, gold carbons). Residue labelling and arrows (orange and red for shifts of Leu 54 and Arg 53, respectively) are as in (A). (C) Stereo figure of the superimposed complexes of ligand-free MtCM–MtDS (2W19, magenta carbons) and MtCM–MtDS with bound inhibitor 3 (2W1A, green carbons). In panels (A) to (C), MtDS is shown in the background in faded surface representation. Residues in helix H1 (i.e., preceding Met 47) and part of the loop to H2 have been omitted from MtCM for clarity. Dotted lines (in B and C) mark hydrogen bonds, with those between the Arg 46 side chain and the backbone carbonyls of Thr 52 and Arg 53 highlighted in red. (D) Simplified representation of prominent and catalytically relevant conformational changes observed on complex formation (free MtCM in cyan, MtDS-complexed MtCM in green). Important residues and the reacting substrate chorismate are depicted together with the network of critical interactions (broken lines). Except for Arg 53 and Arg 85, the residues in the 50's loop and at the C-terminus (Arg 85–His 90) are shown without side chains.
Structural changes upon complex formation
In contrast to the small differences found when comparing MtDS in the three available crystal structures (mostly consisting of minor side-chain rearrangements at the interface with MtCM), MtCM shows a markedly distinct structure in the MtCM–MtDS complex (with or without ligand 3) when compared either with the malate-bound MtCM (2VKL) or the quite similar MtCM apo structure (2QBV) (Figures 6 and 7). These differences are not a consequence of ligand binding, as a superimposition shows very similar structures of both MtCM–MtDS complexes, independent of the presence of 3 (Figures 6C and 7C). As summarized schematically in Figure 7D, conformational differences on MtDS binding are particularly large at the C-terminus and for the 50's loop between H1 and H2, regions that are involved in contacts with MtDS and, in addition, are close to the active site (Figure 7).
When the MtCM-malate structure is superimposed onto the ternary complex, the Cα coordinates of Leu 88 are ≈14 Å apart, whereas Cα distances decrease for the residues preceding Leu 88 to just 0.6 Å for Gly 84 (Figure 7B). Arg 85 shows a Cα-Cα difference of 1.8 Å, and while its side chain roughly occupies the same space in both structures, its hydrogen-bonding partners are distinct. In the absence of MtDS, it interacts with a water molecule whereas in the complex it forms two hydrogen bonds with the active-site residue Glu 59 and another hydrogen bond with the carbonyl oxygen of Gly 86, which adopts a cis-conformation in the complex. Thus, there is a direct link, through the strongly conserved residues Arg 85 and Gly 86 (Figure 2), between the conformation of the C-terminus and the positioning of the active-site residue Glu 59 (Figure 7).
Furthermore, the loop between H1 and H2 of MtCM shows large deviations when comparing structures with and without MtDS. In particular, Arg 53 exhibits a dramatic shift (over 10 Å for some atoms) in the position of its guanidinium group, which can hydrogen bond to Glu 59 only in the MtCM structures without MtDS, whereas it is next to the C-terminus of MtCM in the MtDS complexes (Figure 7A and B, highlighted by cyan and red arrows, respectively). The differential positioning of the 50's loop also affects Arg 46 in the active site. In the complex, the carbonyl oxygens of Thr 52 and Arg 53 coordinate the side chain of Arg 46 through its Nɛ and Nη2 atoms, respectively, whereas in the absence of MtDS, the Nη1 atom of Arg 46 hydrogen bonds to the backbone oxygen of Gly 51. These two coordination modes result in different conformations of the catalytically important Arg 46, with up to 6 Å deviation in positions of side-chain atoms (Figures 6 and 7). Furthermore, a shift in the position of MtCM residues Leu 54, Val 55, and His 56 by one peptide unit in the presence of MtDS (2.8–4.7 Å differences in Cα positions), combined with the alterations at the C-terminus, result in a contact between Leu 54 and Leu 88, whereas these residues are 20 Å apart in the absence of MtDS (Figure 7A and B, Leu 54 shift emphasized by green and orange arrows, respectively). In the latter structure, Leu 54 packs against the active-site residue Arg 18′ (Figure 3D), which is slightly bent rather than fully extended as in the complex with MtDS, in which the shorter Val 55 occupies the equivalent position (Figure 6B).
The CM activity of the MtCM–MtDS complex is regulated by phenylalanine and tyrosine
A rationale for MtCM–MtDS complex formation would be the acquisition of regulatory behaviour as it occurs, for instance, in ScCM, the CM from S. cerevisiae, in which tyrosine is an inhibitor and tryptophan an activator (Schnappauf et al, 1997; Sträter et al, 1997). Here, we conducted an inhibition study on MtCM and the MtCM–MtDS complex (Table III). Addition of 200 μM phenylalanine, tyrosine or tryptophan did not affect the CM activity of MtCM alone. In contrast, when these amino acids were added to a mixture of MtCM and MtDS, phenylalanine and tyrosine inhibited the CM activity by about 70 and 40%, respectively, whereas the addition of tryptophan still had no effect. When phenylalanine and tyrosine were tested simultaneously at the same total concentration, synergistic inhibition by a factor of 25 was observed.
Table 3.
Influence of aromatic amino acids on the CM activity of MtCM and the MtCM–MtDS complex
| Amino-acid added | Remaining specific activity v0/[MtCM] (s−1)a | |
|---|---|---|
| 100 nM MtCM | 30 nM MtCM+300 nM MtDS | |
| — | 0.041±0.001 | 3.16±0.29 (100%) |
| 200 μM Phe | 0.042±0.002 | 0.90±0.13 (28%)* |
| 200 μM Tyr | 0.043±0.001 | 2.00±0.10 (63%)* |
| 100 μM Tyr+100 μM Phe | 0.041±0.003 | 0.13±0.01 (4.1%)* |
| 200 μM Trp | 0.038±0.002 | 3.03±0.43 (96%) |
| aActivity was assayed by observing substrate (initially ≈23 μM) consumption at 30°C and 274 nm in 50 mM BTP, pH 7.5, (for MtCM) or BTP+ (for the complex). Mean and s.d. values were calculated from duplicate (for MtCM alone) or triplicate measurements (both enzymes present). An asterisk denotes a statistically significant deviation (two-tailed t-test; P<0.05; for details, see Supplementary Table SIII) from the corresponding control measurement without addition. Note that MtCM may be only partially complexed with MtDS at the concentrations used; thus, the inhibitory effects may actually be larger. | ||
The MtCM homologue from Corynebacterium glutamicum can be activated by MtDS
Sequence comparisons showed that close homologues of MtCM exist in the bacterial order Actinomycetales (Figure 2). These proteins, which we refer to as forming the AroQδ subclass of CMs, show a strong conservation of residues that are found in the active sites of EcCM (Lee et al, 1995a) and MtCM (Figure 3), such as Arg 11′, Arg 28, Arg 51, and Glu 52 (EcCM numbering). Instead of the catalytic lysine at position 39 in EcCM, all AroQδ members have in common an arginine residue (Figure 2) corresponding to Arg 46 that interacts with the ether oxygen of transition-state analogue 3 in MtCM (Figure 5). Furthermore, they all lack a homologue of EcCM residue Gln 88 able to provide the second hydrogen bond to the ether oxygen of 3. Instead, they possess a shortened C-terminal helix H3 and a strictly conserved Arg–Gly dyad (Figures 1 and 2), which in MtCM is needed to allow the bending of the C-terminus towards MtDS and to link it (through Arg 85) to the active site (to Glu 59) (Figure 7).
The consensus features of the AroQδ subclass, combined with the structural insights obtained from the MtCM structures presented here, suggest that all AroQδ enzymes engage in complex formation and activation by their cognate DAHP synthase. To experimentally explore this idea, the corresponding aroQδ gene from C. glutamicum (derived from Cgl0853, encoding Cgl in Figure 2) was cloned and overexpressed in E. coli. The catalytic efficiency of the purified protein CgCM was ≈103-fold lower than typical for CMs, but could be increased by more than an order of magnitude by addition of the heterologous MtDS (Table II). Native PAGE confirmed an MtDS band shift by CgCM (Supplementary Figure S2).
Importance of C-terminal residues for the activation mechanism
The overlays of uncomplexed and MtDS-complexed MtCM structures suggest that on complex formation, the C-terminal residues and the 50's loop are brought into a catalytically beneficial conformation, particularly as these segments directly contact active-site residues (Figure 7). If complex formation and activation are general features of all AroQδ subclass CMs, conserved residues at the C-terminus of AroQδ enzymes are likely to be involved in the activation mechanism. From the sequence alignment in Figure 2 and the structural observations (Figure 7), the strictly conserved residues Arg 85 and Gly 86, the moderately conserved Arg 87, and the last common hydrophobic residue Leu 88 seem particularly interesting in this respect. The activities of site-directed C-terminal mutants, in comparison with the otherwise identical (N-terminally tagged) wild-type H6-MtCM, are summarized in Table II.
Three of the four MtCM mutants, R87A, L88A, and L88* (lacking the last three residues), preserved wild-type-like CM activity in the absence of MtDS. Thus, in contrast to Arg 46, residues Arg 87 and Leu 88 are not involved in basal MtCM activity. However, the ability to become activated by MtDS is impaired by 10- and 20-fold for MtCM variants L88A and L88*, respectively, supporting the structure-based hypothesis that Leu 88 is involved in the activation switch (Figure 7D). Even though kcat/Km for the G86A version was five-fold lower than wild type, the participation of the strictly conserved Gly 86 in the activation mechanism is evident from the 10-fold reduced activation factor (Table II). Not all manipulations at the C-terminus destroy this mechanism, as exemplified by the R87A mutation, for which the activation factor was affected only two-fold. Remarkably, the catalytically catastrophic R46K variant is at least as strongly activated by MtDS as wild-type MtCM, suggesting that the activation and catalytic machineries do not fully overlap.
Discussion
The M. tuberculosis genome encodes two chorismate mutases. The highly active *MtCM of the AroQγ subclass is secreted by this Gram-positive bacterium and thus not part of the intracellular shikimate pathway (Sasso et al, 2005; Kim et al, 2006), but instead might have a function in intra- or inter-species interactions (Sasso et al, 2005). The intracellular MtCM was reported to possess only modest activity (Prakash et al, 2005; Kim et al, 2008). This is untypical for natural enzymes, which generally evolved to operate at maximum catalytic efficiencies (Radzicka and Wolfenden, 1995). The poor kcat/Km, which is approximately two orders of magnitude lower than usual for CMs, was explained by the absence in MtCM of several active-site residues that are conserved in the AroQ class (Kim et al, 2008). In fact, in place of a Lys and a Gln/Glu residue, which presumably serve to stabilize the developing negative charge in the transition state (Lee et al, 1995a; Liu et al, 1996; Künzler et al, 2005), MtCM shows only Arg 46 (Figure 3F). Interestingly, arginine is the crucial transition-state stabilizing residue in the evolutionarily unrelated AroH class of CMs (Chook et al, 1994; Kast et al, 1996), and a combinatorial mutagenesis study has shown that lysine can functionally replace it (Kast et al, 1996), albeit with a drop in activity (Kast et al, 2000). In analogy, a corresponding change of Arg 46 to the AroQ-consensus lysine was detrimental for MtCM activity (Table II). The distinct catalytic-site features of MtCM had led to the proposition that it is a member of a new subclass (termed AroQδ) (Ökvist et al, 2006) (Figure 8). Here, we show that this δ-subclass CM has yet another peculiarity. Using kinetic analysis, band-shift experiments, and high-resolution crystallography, it was established for the first time that MtCM forms a protein complex with MtDS, another enzyme of the same biosynthetic pathway. Although the exact nature of the native complex under physiological conditions still remains to be determined, the crystal structure shows a hetero-octamer of 243 kDa. On complex formation, the AroQδ CM experiences a marked activity boost to a level at which it can actually serve as the regular housekeeping CM for phenylalanine and tyrosine biosynthesis in M. tuberculosis.
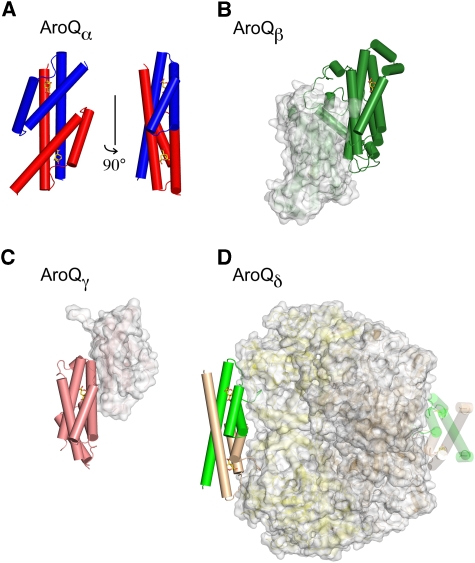
Figure 8.
Overview of known AroQ subclasses and their quaternary structure. (A) EcCM dimer [PDB ID 1ECM (Lee et al, 1995a), AroQα]. (B) ScCM dimer [4CSM (Xue et al, 1994), AroQβ]. (C) *MtCM dimer [2FP2 (Ökvist et al, 2006), AroQγ]. (D) MtCM–MtDS hetero-octamer [2W1A, AroQδ]. Representative CM active sites in (B), (C), and (D) are shown from the same angle as the right-hand view in (A). These sites are formed either by one or two protomers, as indicated by the colour of the solid helices that pack around the bound transition-state analogue 3 (sticks with golden carbons). AroQ protomers that do not contribute residues to these active sites are shown in faded surface representation in (B) and (C), like MtDS in (D). All structures are drawn to scale.
How can this activity enhancement by more than two orders of magnitude be attained? The crystal structures show that MtCM interacts with MtDS mainly through its C-terminus and the loop between H1 and H2. Notably, MtDS does not provide residues (e.g., a Gln substitute) to MtCM's active site, but instead achieves activation by inducing an MtCM conformation that is more favourable for catalysis. To this end, several MtCM residues, including Arg 53 and the three C-terminal residues Leu 88, Gly 89, and His 90, experience displacements by ⩾10 Å on complexation with MtDS, with concomitant conformational changes in the active site (Figures 6 and 7). The kinetic data for MtCM mutants G86A, L88A, and the C-terminally truncated L88* support a model in which the residues of the C-terminal ‘hook' are part of the activation mechanism, but are not directly involved in catalysis. The catalytic residues that undergo significant conformational changes during activation include the presumably transition-state-stabilizing Arg 46, which becomes pre-organized to engage in a bidentate interaction with the ligand, and Glu 59 and Val 55, which in the MtDS complex form critical hydrogen bonds to the hydroxyl group of 3 (Figure 7D). It is particularly noteworthy that the backbone amide interaction (by Val 55) to 3, probably conserved in all AroQ CMs, is only possible in the presence of MtDS. This entire reorganization of the active site is a consequence of extensive conformational changes induced at the MtCM–MtDS interface on MtDS binding, which are transmitted to the catalytic centre of MtCM.
The benefit of such an activity switch is probably the establishment of a sensible regulation regime for the costly biosynthetic pathway towards the aromatic compounds Phe and Tyr, allowing cells to conserve resources (Braus, 1991). For this reason, AroQα enzymes are often fused to protein domains enabling end-product feedback inhibition at the enzyme level (Ahmad and Jensen, 1986; Gu et al, 1997; MacBeath et al, 1998; Kleeb et al, 2006). As exemplified in structural detail for ScCM, AroQβ enzymes undergo large ligand-induced conformational changes to modulate activity by a sophisticated allosteric regulation mechanism. The binding of the inhibitor tyrosine or the activator tryptophan to the allosteric binding site results in rotation of one protomer of the dimer relative to the other, and thereby in an inter-conversion between an inactive T-state and an active R-state (Schnappauf et al, 1997; Sträter et al, 1997). For instance, as a consequence of tryptophan binding, affinity for the substrate chorismate (and thereby catalytic efficiency) increases by an order of magnitude (Table II). Whereas the secreted *MtCM is not regulated by Phe or Tyr (Sasso et al, 2005; Kim et al, 2006), we have shown that on interaction with MtDS the intracellular MtCM becomes regulatable by these shikimate-pathway end products. Possibly, such feedback inhibition could result in a shift in the equilibrium between complexed MtCM and the free form, which binds to chorismate 30-fold less tightly (Table II). The regulation by reversible MtCM–MtDS complex formation on interaction with metabolites from the shikimate pathway would allow for meaningful immediate and effective tuning of total CM activity within M. tuberculosis. Such a mechanism is supported by the relatively high dissociation constant of the complex and by observations from size-exclusion chromatography. In fact, association of CM with the larger complex partner was only detectable by gel filtration of either purified MtDS and MtCM from M. tuberculosis (our preliminary experiments), or of crude fractions containing a presumably similar complex in Brevibacterium flavum (Shiio and Sugimoto, 1979), if chorismate was added to the running buffer.
Sequence database searches retrieved MtCM homologues distinguished by their unique set of active-site residues, the ‘AroQδ signature', exclusively from the bacterial order Actinomycetales. A representative sequence alignment (Figure 2) shows, for instance, that in place of a homologue of the catalytic Gln 88 of EcCM (Figures 1 and 3), the C-termini of AroQδ sequences include a strictly conserved Arg–Gly dyad (at MtCM positions 85–86) and a hydrophobic residue at position 88. In MtCM, these residues were implicated in the MtDS-promoted structural rearrangements leading to activity enhancement. The conservation of these sequence motifs, together with the observation that also the C. glutamicum homologue could be activated by the heterologous MtDS strongly suggests that formation of a regulatory complex with DAHP synthase is a general feature of all AroQδ CMs. Our findings thus provide the basis to generalize and explain on a molecular level earlier reports about DAHP synthase-associated ‘split CMs' as described, for instance, for the actinomycetes Amycolatopsis methanolica (Euverink et al, 1995) and B. flavum (Shiio and Sugimoto, 1979; Sugimoto and Shiio, 1980), which is closely related or even identical to C. glutamicum (Liebl, 2006).
Like AroQδ CMs, also the DAHP synthase partner in the complex can be identified at the sequence level. DAHP synthases were previously divided into classes AroAI or AroAII based on sequence homology (Gosset et al, 2001). AroAII DAHP synthases, to which MtDS belongs (Webby et al, 2005), are well represented among actinomycetes, and moreover, an organism with an AroQδ sequence always seems to possess an AroAII sequence, suggesting that complexes containing AroQδ CMs generally involve AroAII class DAHP synthases. Very interestingly, members of the AroAIβ subclass of the other class (AroAI) of DAHP synthases were found to occur as covalent fusions with AroQα domains (Wu and Woodard, 2006). In these fusions, the CM domain is either on the N- or on the C-terminal side, as found, for instance, in the bacterial phyla Firmicutes or Bacteroidetes, respectively, but not in Actinobacteria, which includes the order Actinomycetales. Like the non-covalent AroQδ CM-AroAII DAHP synthase complexes, which were explored here at the molecular level, the covalent protein fusions were also shown to have acquired regulatory properties through domain interactions (Wu and Woodard, 2006). Thus, the main driving force for both the covalent and non-covalent association of CMs and DAHP synthases, which evolved independently at least three times, may have been the gain of regulatory potential. Whereas prephenate binding to the AroQ domain in the case of the covalent associations provides feedback inhibition to DAHP synthase (Wu and Woodard, 2006), we have shown that the non-covalent complex enables feedback regulation of intracellular CM activity. For M. tuberculosis, this adds metabolic control also at the chorismate branch point, in addition to the feedback regulation of the first enzyme MtDS by the shikimate-pathway products Tyr, Phe, Trp, and chorismate (Webby et al, 2005).
The CM–DAHP synthase complex from M. tuberculosis is the first non-covalent complex involving an enzyme of the common shikimate pathway described at high resolution. Given that many dreaded pathogens, such as M. tuberculosis and Corynebacterium diphtheriae belong to the Actinomycetales (Ventura et al, 2007), disrupting the formation of this protein complex, which presumably is specific for this group of bacteria, may be a viable intervention strategy against diseases caused by these organisms. The structures and mechanisms determined in this work may thus also be useful to guide efforts for rational drug design.
Materials and methods
Materials and general procedures
Chromosomal DNA from M. tuberculosis strain H37Rv and the C. glutamicum strain ATCC 13032 were provided by Roland Brosch (Institut Pasteur, Paris, France) and Tobias Fuhrer (Institute of Molecular Systems Biology, ETH Zurich), respectively. All oligonucleotides were custom-synthesized by Microsynth (Balgach, Switzerland). DNA sequencing was carried out on an ABI PRIZM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) by the chain-termination method (Sanger et al, 1977), using the BigDye Sequencing Kit from the same company. Chorismate was prepared as described by Grisostomi et al (1997). The cloning techniques and general media preparations were according to standard procedures (Sambrook and Russell, 2001).
Bacterial strains and plasmids
General cloning was carried out in E. coli strain XL1-Blue (Stratagene, La Jolla, CA, USA). E. coli KA12/pKIMP-UAUC (Kast et al, 1996) and KA13 (MacBeath and Kast, 1998; MacBeath et al, 1998) have been described. The construction of plasmids pKTCM-H, pKTCMM-H, pKTCMM-HN, pKTCM-HC, pMG246, pKTDS-H, pKTDS-HN, pKIMP-UM, pMG213H-W, pMG213H-Mtu, pKCGCM-H, pKCGCM-HC, and pKCGCM-HN, needed, for example, for expressing the various wild-type formats of the aroQδ, aroG, and pheA genes from M. tuberculosis and aroQδ from C. glutamicum, is detailed in the Supplementary data. There, details are also provided for plasmids pKTCMM-HN-R46K, pKTCMM-HN-G86A, pKTCMM-HN-R87A, pKTCMM-HN-L88A, and pKTCMM-HN-L88* needed for the production of the corresponding set of MtCM mutants (tagged with Met–His6–Ser–Ser–Gly fused to Met 1 in Figure 1C) in strain KA13.
Overproduction and purification of MtCM, MtDS, CgCM, and MtPDT
Wild-type MtCM (no tag), MtDS, CgCM, and MtPDT (the latter three with an N-terminal Met–His6–Ser–Ser–Gly sequence) were overproduced from plasmids pKTCMM-H, pKTDS-HN, pKCGCM-HN, and pMG213H-Mtu, respectively, in the CM-deficient E. coli strain KA13, which carries an IPTG-inducible chromosomal T7 RNA polymerase gene. The gene expression by IPTG exploits a lac operator-controlled T7 promoter on the plasmids (which also harbour the lacI gene).
MtCM was purified using a three-step strategy consisting of cation and anion exchange chromatography (at, respectively, 1.5 pH units below and above the experimental isoelectric point of 8.3), followed by gel filtration. The final yield of SDS gel-electrophoretically pure MtCM was 5 mg MtCM per litre of bacterial culture. The integrity of the purified MtCM was confirmed using mass spectrometry (expected [M-H]+, 10 091 Da; observedMALDI−MS, 10 093 Da).
MtDS was produced in a bioreactor and purified using Ni–NTA affinity chromatography followed by gel filtration. To preserve MtDS enzyme integrity, it was essential to base all MtDS purification buffers on BTP++, which consists of 20 mM BTP (1,3-bis[tris(hydroxymethyl)methylamino]propane), pH 7.5, 150 mM NaCl, 0.5 mM TCEP [tris(2-carboxyethyl)phosphine hydrochloride], 0.2 mM phosphoenolpyruvate, and 0.1 mM MnCl2 (Webby et al, 2005). On average, 1 mg of pure MtDS having the correct mass (expected, 51 826 Da; observedESI−MS, 51 827 Da) was obtained per litre of bacterial culture. When necessary, MtCM and MtDS were concentrated using ultrafiltration with Macrosep centrifugal devices (Pall, Basel, Switzerland).
Further details on cell culturing and protein purification are provided in the Supplementary data, except for wild-type MtCM and MtDS, for which the procedure will be published elsewhere (M Ökvist, S Sasso, K Roderer, P Kast and U Krengel, in preparation).
Electrophoresis, mass spectrometry, and quantification of purified proteins
The protein preparations were checked for electrophoretic homogeneity by SDS–PAGE using the PhastSystem (GE Healthcare, Otelfingen, Switzerland) and their correct Mr using mass spectrometry (for details on sample workup and Mr determination, see the Supplementary data). The isoelectric point was determined using an IEF 3–9 PhastGel and the Isoelectric Focusing Calibration Kit (GE Healthcare).
The concentration of purified proteins was determined using the Bradford assay (Bradford, 1976). The assay was calibrated for both wild-type MtCM and MtDS with the results from quantitative total protein hydrolysis to correct for differential colouration properties (see Supplementary data). Compared to the standard protein bovine serum albumin, MtCM is stained roughly 1.5 times stronger whereas MtDS is stained ca. 1.2 times weaker by the Coomassie Blue G-250 dye used in the Bradford assay. For the MtCM mutants, the correction factor determined for wild-type MtCM was applied. CgCM was directly compared with the bovine serum albumin standard.
Crystal structure analysis
The diffraction-quality crystals for malate-bound MtCM (concentrated to 2.45 mg ml−1 in BTP++ buffer) were obtained with the hanging-drop vapour diffusion technique at 20°C. Final crystallization conditions contained 15% PEG 1500 and 0.1 M of the L-malate-containing MMT buffer system (Newman, 2004), pH 8–9.
The diffraction-quality crystals of the MtCM–MtDS complexes were obtained by streak-seeding into conditions containing a 1:1 mixture of MtCM (0.28 mg/ml) and MtDS (1.7 mg/ml), both in BTP++ buffer, with or without a 2:1 excess of 3, equilibrated against a reservoir containing 0.9 M ammonium sulfate, 100 mM Tris (pH 7.9–8.0), and 1–5% PEG 400 (M Ökvist, S Sasso, K Roderer, P Kast, and U Krengel, in preparation).
Diffraction and refinement statistics are provided in Table I; further information on crystallization and structure determination is provided in the Supplementary data. All structures show good geometry. The real-space correlation coefficients are 0.964, 0.947, and 0.940 for the structures of MtCM (with bound malate), the MtCM–MtDS complex (without ligand), and the ternary MtCM–MtDS complex with 3, respectively, consistent with an overall good fit of the structures to the electron density (e.g., Figure 5B and Supplementary Figure S3). Between 11 and 18 N-terminal residues of MtCM showed disorder in individual protomers and were thus excluded from the models, as were some MtDS loop residues (e.g., 372–380 and 414–442 near the damaged manganese site in the unliganded MtCM–MtDS complex; for a detailed listing, see Table I). Residues Ser 49, Gly 50, and Gly 51 in the unliganded MtCM structure (loop region connecting helices H1 and H2) show weaker electron density.
CM activity assays and determination of the apparent affinity constant
In vitro CM activity in the presence of MtPDT was assessed with a discontinuous CM assay (Cotton and Gibson, 1965). In all other cases, CMs were characterized using a continuous assay at 274 nm or 310 nm. Initial velocities were fitted to the Michaelis–Menten equation (Segel, 1976), and the catalytic rate constant kcat was calculated per CM active site. The relatively high apparent dissociation constant (Kd,app) of the complex, and the need to keep MtCM concentrations low enough for monitoring the initial portion of the reaction, necessitated a large excess of MtDS for assaying the complexes. The Kd,app for MtCM–MtDS was calculated for a 1:1 stoichiometry using equations derived in the Supplementary data, in which further experimental details on the CM activity assays are provided.
DAHP synthase activity assay
MtDS activity was monitored in 50 mM BTP (pH 7.5), containing 100 μM MnCl2 at 30°C by a continuous assay following phosphoenolpyruvate consumption (ɛ232=2800 M−1 cm−1) over time, according to a previously described procedure (Schoner and Herrmann, 1976). The kcat/Km for phosphoenolpyruvate was derived under saturating D-erythrose-4-phosphate (240 μM) and low (<10 μM) phosphoenolpyruvate concentrations.
Accession codes
Protein Data Bank: atomic coordinates and structure-factor amplitudes have been deposited under accession codes 2VKL, 2W19, and 2W1A.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank R Brosch (Institut Pasteur, Paris) for the provision of M. tuberculosis chromosomal DNA, T Fuhrer for C. glutamicum, J Haugstetter for the construction of pKIMP-UM and pMG213H-Mtu, B Orelli for pKTDS-HN and initial experiments, and D Künzler for the preparation of chorismate. This study was supported by funds from Novartis Pharma, the ETH Zurich, the University of Oslo, and the Swiss National Science Foundation (grant no. 3100A0-116475/1).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmad S, Jensen RA (1986) The evolutionary history of two bifunctional proteins that emerged in the purple bacteria. Trends Biochem Sci 11: 108–112 [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews PR, Smith GD, Young IG (1973) Transition-state stabilization and enzymic catalysis. Kinetic and molecular orbital studies of the rearrangement of chorismate to prephenate. Biochemistry 12: 3492–3498 [DOI] [PubMed] [Google Scholar]
- Bartlett PA, Johnson CR (1985) An inhibitor of chorismate mutase resembling the transition-state conformation. J Am Chem Soc 107: 7792–7793 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Braus GH (1991) Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev 55: 349–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Gray JV, Ke H, Lipscomb WN (1994) The monofunctional chorismate mutase from Bacillus subtilis: structure determination of chorismate mutase and its complexes with a transition state analog and prephenate, and implications for the mechanism of the enzymatic reaction. J Mol Biol 240: 476–500 [DOI] [PubMed] [Google Scholar]
- Chook YM, Ke H, Lipscomb WN (1993) Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc Natl Acad Sci USA 90: 8600–8603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544, Erratum (1998) Nature 396: 190–198 [DOI] [PubMed] [Google Scholar]
- Cotton RGH, Gibson F (1965) The biosynthesis of phenylalanine and tyrosine: enzymes converting chorismic acid into prephenic acid and their relationships to prephenate dehydratase and prephenate dehydrogenase. Biochim Biophys Acta 100: 76–88 [DOI] [PubMed] [Google Scholar]
- Doran T, Tizard M, Millar D, Ford J, Sumar N, Loughlin M, Hermon-Taylor J (1997) IS900 targets translation initiation signals in Mycobacterium avium subsp. paratuberculosis to facilitate expression of its hed gene. Microbiology 143: 547–552 [DOI] [PubMed] [Google Scholar]
- Euverink GJW, Hessels GI, Franke C, Dijkhuizen L (1995) Chorismate mutase and 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase of the methylotrophic actinomycete Amycolatopsis methanolica. Appl Environ Microbiol 61: 3796–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosset G, Bonner CA, Jensen RA (2001) Microbial origin of plant-type 2-keto-3-deoxy-D-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J Bacteriol 183: 4061–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisostomi C, Kast P, Pulido R, Huynh J, Hilvert D (1997) Efficient in vivo synthesis and rapid purification of chorismic acid using an engineered Escherichia coli strain. Bioorg Chem 25: 297–305 [Google Scholar]
- Gu W, Williams DS, Aldrich HC, Xie G, Gabriel DW, Jensen RA (1997) The AroQ and PheA domains of the bifunctional P-protein from Xanthomonas campestris in a context of genomic comparison. Microb Comp Genomics 2: 141–158 [DOI] [PubMed] [Google Scholar]
- Haslam E (1993) Shikimic Acid: Metabolism and Metabolites. New York: John Wiley & Sons [Google Scholar]
- Kast P, Asif-Ullah M, Jiang N, Hilvert D (1996) Exploring the active site of chorismate mutase by combinatorial mutagenesis and selection: the importance of electrostatic catalysis. Proc Natl Acad Sci USA 93: 5043–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast P, Grisostomi C, Chen IA, Li S, Krengel U, Xue Y, Hilvert D (2000) A strategically positioned cation is crucial for efficient catalysis by chorismate mutase. J Biol Chem 275: 36832–36838 [DOI] [PubMed] [Google Scholar]
- Kim S-K, Reddy SK, Nelson BC, Robinson H, Reddy PT, Ladner JE (2008) A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H37Rv and the secreted chorismate mutase (y2828) from Yersinia pestis. FEBS J 275: 4824–4835 [DOI] [PubMed] [Google Scholar]
- Kim S-K, Reddy SK, Nelson BC, Vasquez GB, Davis A, Howard AJ, Patterson S, Gilliland GL, Ladner JE, Reddy PT (2006) Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids. J Bacteriol 188: 8638–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeb AC, Kast P, Hilvert D (2006) A monofunctional and thermostable prephenate dehydratase from the archaeon Methanocaldococcus jannaschii. Biochemistry 45: 14101–14110, Erratum (2007) Biochemistry 46: 2552–2552 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Künzler DE, Sasso S, Gamper M, Hilvert D, Kast P (2005) Mechanistic insights into the isochorismate pyruvate lyase activity of the catalytically promiscuous PchB from combinatorial mutagenesis and selection. J Biol Chem 280: 32827–32834 [DOI] [PubMed] [Google Scholar]
- Lee AY, Karplus PA, Ganem B, Clardy J (1995a) Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J Am Chem Soc 117: 3627–3628 [Google Scholar]
- Lee AY, Stewart JD, Clardy J, Ganem B (1995b) New insight into the catalytic mechanism of chorismate mutases from structural studies. Chem Biol 2: 195–203 [DOI] [PubMed] [Google Scholar]
- Liebl W (2006) Corynebacterium—Nonmedical. In The Prokaryotes, Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds), Vol. 3, 3rd edn, pp 796–818. New York: Springer [Google Scholar]
- Liu DR, Cload ST, Pastor RM, Schultz PG (1996) Analysis of active site residues in Escherichia coli chorismate mutase by site-directed mutagenesis. J Am Chem Soc 118: 1789–1790 [Google Scholar]
- MacBeath G, Kast P (1998) UGA read-through artifacts—when popular gene expression systems need a pATCH. Biotechniques 24: 789–794 [DOI] [PubMed] [Google Scholar]
- MacBeath G, Kast P, Hilvert D (1998) A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii. Biochemistry 37: 10062–10073 [DOI] [PubMed] [Google Scholar]
- Mattei P, Kast P, Hilvert D (1999) Bacillus subtilis chorismate mutase is partially diffusion-controlled. Eur J Biochem 261: 25–32 [DOI] [PubMed] [Google Scholar]
- Newman J (2004) Novel buffer systems for macromolecular crystallization. Acta Crystallogr D Biol Crystallogr 60: 610–612 [DOI] [PubMed] [Google Scholar]
- Ökvist M, Dey R, Sasso S, Grahn E, Kast P, Krengel U (2006) 1.6 Å Crystal structure of the secreted chorismate mutase from Mycobacterium tuberculosis: novel fold topology revealed. J Mol Biol 357: 1483–1499 [DOI] [PubMed] [Google Scholar]
- Pervushin K, Vamvaca K, Vögeli B, Hilvert D (2007) Structure and dynamics of a molten globular enzyme. Nat Struct Mol Biol 14: 1202–1206 [DOI] [PubMed] [Google Scholar]
- Prakash P, Aruna B, Sardesai AA, Hasnain SE (2005) Purified recombinant hypothetical protein coded by open reading frame Rv1885c of Mycobacterium tuberculosis exhibits a monofunctional AroQ class of periplasmic chorismate mutase activity. J Biol Chem 280: 19641–19648 [DOI] [PubMed] [Google Scholar]
- Radzicka A, Wolfenden R (1995) A proficient enzyme. Science 267: 90–93 [DOI] [PubMed] [Google Scholar]
- Richards TA, Dacks JB, Campbell SA, Blanchard JL, Foster PG, McLeod R, Roberts CW (2006) Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot Cell 5: 1517–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74: 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso S, Ramakrishnan C, Gamper M, Hilvert D, Kast P (2005) Characterization of the secreted chorismate mutase from the pathogen Mycobacterium tuberculosis. FEBS J 272: 375–389 [DOI] [PubMed] [Google Scholar]
- Schnappauf G, Sträter N, Lipscomb WN, Braus GH (1997) A glutamate residue in the catalytic center of the yeast chorismate mutase restricts enzyme activity to acidic conditions. Proc Natl Acad Sci USA 94: 8491–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CZ, Parish T, Basso LA, Santos DS (2008) The two chorismate mutases from both Mycobacterium tuberculosis and Mycobacterium smegmatis: biochemical analysis and limited regulation of promoter activity by aromatic amino acids. J Bacteriol 190: 122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner R, Herrmann KM (1976) 3-Deoxy-D-arabino-heptulosonate 7-phosphate synthase. Purification, properties, and kinetics of the tyrosine-sensitive isoenzyme from Escherichia coli. J Biol Chem 251: 5440–5447 [PubMed] [Google Scholar]
- Segel IH (1976) Biochemical Calculations: How to Solve Mathematical Problems in General Biochemistry. New York: John Wiley & Sons [Google Scholar]
- Shiio I, Sugimoto S (1979) Two components of chorismate mutase in Brevibacterium flavum. J Biochem 86: 17–25 [PubMed] [Google Scholar]
- Sträter N, Schnappauf G, Braus G, Lipscomb WN (1997) Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure 5: 1437–1452 [DOI] [PubMed] [Google Scholar]
- Sugimoto S, Shiio I (1980) Purification and properties of dissociable chorismate mutase from Brevibacterium flavum. J Biochem 88: 167–176 [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D (2007) Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71: 495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby CJ, Baker HM, Lott JS, Baker EN, Parker EJ (2005) The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis reveals a common catalytic scaffold and ancestry for type I and type II enzymes. J Mol Biol 354: 927–939 [DOI] [PubMed] [Google Scholar]
- Wu J, Woodard RW (2006) New insights into the evolutionary links relating to the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase subfamilies. J Biol Chem 281: 4042–4048 [DOI] [PubMed] [Google Scholar]
- Xue Y, Lipscomb WN, Graf R, Schnappauf G, Braus G (1994) The crystal structure of allosteric chorismate mutase at 2.2 Å resolution. Proc Natl Acad Sci USA 91: 10814–10818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File