Abstract
The crucial roles of Sec1/Munc18 (SM)-like proteins in membrane fusion have been evidenced in genetic and biochemical studies. SM proteins interact directly with SNAREs and contribute to SNARE pairing by a yet unclear mechanism. Here, we show that the SM protein, Sly1, interacts directly with the conserved oligomeric Golgi (COG) tethering complex. The Sly1–COG interaction is mediated by the Cog4 subunit, which also interacts with Syntaxin 5 through a different binding site. We provide evidence that disruption of Cog4–Sly1 interaction impairs pairing of SNAREs involved in intra-Golgi transport thereby markedly attenuating Golgi-to-ER retrograde transport. These results highlight the mechanism by which SM proteins link tethering to SNAREpin assembly.
Keywords: COG, Golgi, retrograde transport, SM proteins, SNARE pairing
Introduction
Membrane fusion along the secretory pathway is driven by different SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) protein complexes consisting of t-SNAREs on the target membrane and v-SNAREs on the transport vesicle (Hong, 2005). The assembly of cognate v- and t-SNAREs induces the formation of trans-SNARE complexes or SNAREpins, in which four SNARE motifs assemble as a twisted parallel four-helix bundle that brings the opposing membranes together and eventually catalyses membrane fusion (Hanson et al, 1997; Chen and Scheller, 2001; Ungar and Hughson, 2003). Pairing of distinct SNAREs and assembly of functional SNAREpins are used by different membrane-trafficking events (Hong, 2005). However, certain t-SNAREs are assembled into different SNAREpins involved in distinct intracellular transport steps (Malsam et al, 2008). Thus, the mechanisms regulating SNARE pairing and SNAREpins assembly are crucial not only for the specificity of the fusion events, but also for coordinating different membrane-trafficking pathways.
Increasing evidence suggests that the Sec1/Munc18 (SM) proteins, as well as tethering factors, drive the specificity of membrane-fusion events (Whyte and Munro, 2002; Koumandou et al, 2007). SM proteins are soluble, peripheral membrane proteins of 60–90 kDa that function in distinct intracellular transport steps, and interact directly with SNARE proteins (Malsam et al, 2008). Their ability to interact with syntaxins has been shown in yeast and mammals and has been extensively studied for different SM and syntaxin proteins. SM proteins seem to interact with three distinct conformations of syntaxins. The mammalian N-sec1/Munc18 can interact with the closed conformation of Syntaxin1, thereby precluding it from SNARE-complex assembly (Garcia et al, 1994; Pevsner et al, 1994; Dulubova et al, 1999; Yang et al, 2000), whereas the yeast Sec1p interacts with a fully assembled exocytic SNARE complex (Carr et al, 1999). On the other hand, the SM proteins Sly1p/rSly1 and Vps45p, which are involved in ER-Golgi and TGN/early endosomal transport, bind to their syntaxins, Sed5p/Syntaxin 5 and Tlg2p/Syntaxin 16, respectively, through a short N-terminal peptide (Bracher and Weissenhorn, 2002; Yamaguchi et al, 2002; Dulubova et al, 2003). Accordingly, it has been proposed that SM proteins regulate the proper folding of syntaxins and control SNARE-complex assembly. In vitro binding assays have suggested that the interaction of SM proteins with SNAREs prevents the formation of non-physiological SNARE complexes and stimulates specific SNARE pairing (Peng and Gallwitz, 2002). Consistent with these observations, reconstitution studies have shown that SM proteins strongly accelerate SNARE-mediated fusion of cognate SNAREs, thereby enhancing fusion specificity (Shen et al, 2007). Collectively, these studies suggest that SM proteins regulate both the speed and the specificity of the fusion reaction, and therefore are implicated in selective activation of cognate SNAREpins. However, those in vitro studies may not reflect their actual mode of action in intact cells, and additional components are probably involved.
Tethering factors, which mediate the physical contact between the vesicle and its target membrane, together with the small GTPases Rabs, play a critical role in determining the specificity of vesicle targeting and, therefore, fusion events (Whyte and Munro, 2002; Cai et al, 2007). Thus, Rabs and tethering factors, which act upstream of the SNAREs, must function coordinately with SM proteins. Indeed, earlier studies in yeast have shown that a single amino acid (aa) substitution in Sly1p (E532K) generates a dominant mutant, SLY1-20, which bypasses the requirement for the small Rab GTPases Ypt1p (Rab1) (Dascher et al, 1991; Ossig et al, 1991) and Ypt6p (Rab6). SLY1-20 also suppresses mutations in the tethering protein Uso1p (p115) (Sapperstein et al, 1996) and subunits of the conserved oligomeric Golgi (COG) complex (VanRheenen et al, 1998). These genetic data suggest that Sly1 protein interacts functionally with other components of the membrane-fusion machinery. However, little is known about the interacting partners of SM proteins.
In this study, we show that the COG complex interacts directly with the SM protein Sly1 in mammalian cells. The COG is a Golgi-associated tethering complex consisting of eight subunits (Cog1–Cog8), which can be divided into two functionally and structurally distinct subcomplexes; lobe A (Cog1–4) and lobe B (Cog5–8) (Ungar et al, 2002, 2005; Fotso et al, 2005; Oka et al, 2005). Subunits of the first lobe are essential for normal cell growth in yeast, and therefore, Cog1–4 are considered essential components of the complex (Whyte and Munro, 2001; Oka and Krieger, 2005). The COG complex controls multiple features of Golgi structure and function from yeast to humans and is thought to be directly involved in retrograde trafficking of Golgi-resident proteins, thereby affecting the Golgi-glycosylation machinery (Podos et al, 1994; Chatterton et al, 1999; Whyte and Munro, 2001; Ram et al, 2002; Suvorova et al, 2002; Ungar et al, 2002, 2006). Earlier studies in yeast have shown that the COG complex is a Ypt1p effector involved in retrograde intra-Golgi trafficking that interacts with several Golgi SNAREs, including Sed5p, Gos1p (GOS-28/GS28), Ykt6p and Sec22p (VanRheenen et al, 1998, 1999; Suvorova et al, 2002). More recently, a direct interaction between the COG complex and Sed5/Syntaxin 5 has been shown (Shestakova et al, 2007) and has been proposed to stabilize intra-Golgi SNARE complexes. The studies described here show that the COG complex also interacts with the SM protein Sly1, and that disruption of COG–Sly1 interaction impairs Golgi SNARE pairing and consequently, Golgi-to-ER retrograde transport. These results show the coordinated functions of tethering factors, SM proteins, and SNAREs in regulating intracellular membrane fusion.
Results
The COG complex interacts with Sly1 through its Cog4 subunit
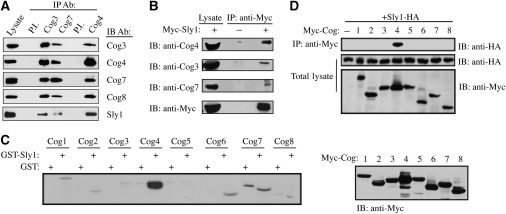
Earlier genetic studies in yeast have shown that the SLY1-20 allele, a dominant allele of Sly1p, suppresses null mutations in SEC35 (Cog2) and SEC34 (Cog3) (VanRheenen et al, 1998, 1999). These results suggest functional relations between the COG complex and Sly1 action. To explore this possibility, we first assessed whether the COG complex interacts with Sly1 in mammalian cells by coimmunoprecipitation experiments. As shown (Figure 1A), a weak but specific interaction between Sly1 and either Cog3 or Cog7, two representative subunits of the two COG lobes, was observed. On the other hand, Sly1 was strongly detected in the immunocomplex of Cog4. These results suggest that the interaction between Sly1 and the COG complex is mediated by the Cog4 subunit, and that the presence of Sly1 in either Cog3 or Cog7 immunocomplexes results from their interaction with Cog4 (Figure 1A). To further corroborate these results, a reciprocal immunoprecipitation analysis of HeLa cells expressing a Myc-tagged Sly1 was carried out using anti-Myc antibody for immunoprecipitation followed by western blotting with antibodies against Cog3, Cog7, or Cog4. As shown (Figure 1B), the endogenous Cog3 and Cog7 subunits were weakly detected in Sly1 immunocomplex as compared with Cog4, suggesting that Cog4 mediates the interaction between Sly1 and the COG complex. We consequently examined the interaction between Sly1 and the eight different subunits of the COG complex by either GST pull-down experiments using a recombinant Sly1 protein immobilized on glutathione-agarose beads (Figure 1C), or coimmunoprecipitation studies using cell lysates of HEK293 coexpressing HA-tagged Sly1 and the different Myc-tagged COG subunits (Figure 1D). In both approaches, Sly1 interacted strongly and selectively with the Cog4 subunit, suggesting that Cog4 mediates the interaction between the COG complex and Sly1 in mammalian cells.
Figure 1.
The interaction between the COG complex and Sly1 is mediated by the Cog4 subunit. (A) An endogenous COG complex interacts with Sly1. HeLa cell lysate was subjected to immunoprecipitation (IP) with anti-Cog3, anti-Cog7 or anti-Cog4 specific antibodies. Preimmune (P.I.) sera of Cog3 and Cog4 were used as a control. The presence of Sly1 in the immunocomplexes of the indicated COG subunits was determined by immunobloting (IB) with anti-Sly1 antibody. The interaction between the different COG subunits was assessed by immunobloting with the indicated anti-Cog antibodies. (B) Sly1 interacts with endogenous COG subunits. HeLa cells were transiently transfected with expression vector encoding Myc-tagged Sly1. Sly1-Myc was immunoprecipitated by anti-Myc antibody and the presence of Cog3, Cog7 or Cog4 in Sly1 immunocomplexes was determined by immunoblotting with the indicated anti-Cog antibodies. (C) Cog4 interacts with recombinant GST-Sly1. HEK293 cells were transiently transfected with expression vectors encoding the eight different subunits of the COG complex as Myc-tagged proteins. The cell lysates were incubated with either GST or GST-Sly1 bound to glutathione-agarose beads. The samples were washed and then resolved by SDS–PAGE, transferred to a nitrocellulose membrane and immunoblotted with anti-Myc antibody (left panel). The expression level of each COG subunit was determined by western blotting with anti-Myc antibody (right panel). (D) Cog4-Myc interacts with Sly1-HA. HEK293 cells were either transiently transfected with an expression vector encoding Sly1-HA or cotransfected with Sly1-HA and the different Cog-Myc subunits. The COG subunits were immunoprecipitated with anti-Myc antibody, and their association with Sly1-HA was determined by immunoblotting with anti-HA antibody. The expression level of the transfected proteins was assessed by immunoblotting of total cell lysates with the indicated antibodies (lower panels).
The N-terminal of Cog4 consists of distinct binding sites for Sly1 and Syntaxin 5
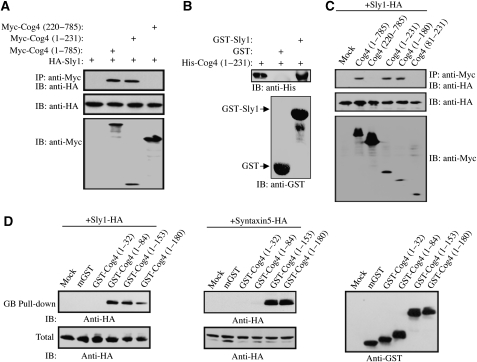
To further characterize the interaction between Sly1 and Cog4, a set of Cog4-truncated mutants was prepared as described in Figure 2A and C. These Myc-tagged truncated mutants were coexpressed with HA-tagged Sly1 in HEK293 cells, and their interaction was assessed by coimmunoprecipitation studies. As shown, deletion of the first 220 aa of Cog4 abolished the interaction, whereas a mutant consisting of only the first 231 aa interacted strongly with Sly1, suggesting that the N-terminal fragment of Cog4 contains the binding site for Sly1 (Figure 2A). This was further confirmed by in vitro binding assay using recombinant GST-tagged Sly1 and His-tagged Cog4 (aa 1–231) purified from bacteria (Figure 2B). Further deletion analysis narrowed down the interaction to the first 180 aa and showed the importance of the first 81 aa for Sly1 binding (Figure 2C). Collectively, these results suggest that Cog4 interacts directly with Sly1, and that the first N-terminal 81 aa are required for binding.
Figure 2.
Sly1 interacts with an N-terminal fragment of Cog4. HEK293 cells were transiently cotransfected with expression vectors encoding the indicated Cog4-truncated mutants (A, C) as Myc-tagged proteins together with Sly1-HA. The interaction between these truncated mutants and Sly1 was determined by immunoprecipitation with anti-Myc antibody and immunoblotting with anti-HA antibody. The numbers indicate the aa residues. The expression level of the transfected proteins was assessed by immunoblotting of total cell lysates with the indicated antibodies (A, C; lower panels). (B) A direct interaction between the N-terminal fragment of Cog4 and Sly1 was determined by binding of a recombinant His-tagged Cog4 fragment (aa 1–231) to either recombinant GST or GST-Sly1 immobilized on glutathione-agarose beads, followed by immunoblotting with anti-His antibody. (D) The N-terminal fragment of Cog4 consists of distinct binding sites for Sly1 and Syntaxin 5. The indicated Cog4-truncated mutants were expressed as GST-fusion proteins in HEK293 cells with either Sly1-HA or Syntaxin 5-HA. The cell lysates were subjected to glutathione-agarose beads (GB) pull down. Interactions between the Cog4-truncated mutants and either Sly1 or Syntaxin 5 were determined by immunoblotting with anti-HA antibody. Mammalian GST (mGST) was used as a control, whereas the expression level of each truncated mutant was determined by immunoblotting with anti-GST antibody.
Earlier studies have shown that the N-terminal fragment of Cog4 (aa 1–222) also interacts with Syntaxin 5 (Shestakova et al, 2007). We, therefore, examined whether this fragment contains distinct binding sites for Syntaxin 5 and Sly1. We prepared a new set of Cog4-truncated mutants fused to mammalian GST, coexpressed them with either HA-tagged Sly1 or HA-tagged Syntaxin 5, and assessed their interaction by GST pull-down experiments. A strong interaction was detected between the first 153 aa of Cog4 and either Sly1 or Syntaxin 5 (Figure 2D). However, a fragment consisting of the first 84 aa failed to interact with Syntaxin 5, but strongly interacted with Sly1 (Figure 2D). These results suggested that the N-terminal fragment of Cog4 (aa 1–153) consists of two distinct binding sites: an N-terminal binding site for Sly1 (aa 1–84), and a separate binding site for Syntaxin 5, probably aa 84–153.
Highly conserved residues in the N-terminal region of Cog4 mediate the interaction with Sly1
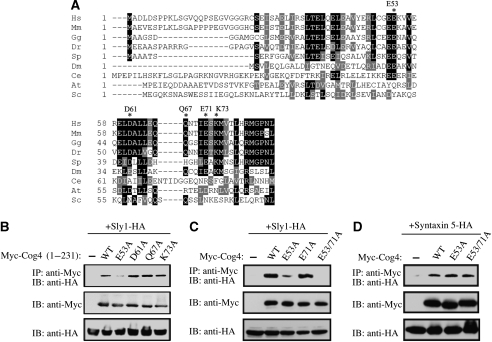
Earlier studies on Sed5p (Syntaxin 5) have shown that a single substitution of a highly conserved residue at position 10 (F10A) eliminates its interaction with Sly1 (Yamaguchi et al, 2002). Accordingly, we looked for highly conserved residues in the N-terminal region of Cog4 (aa 1–84). Multiple sequence alignment of Cog4 from different species revealed several highly conserved residues, including E52, E53, D61, Q67, E71 and K73 (Figure 3A). These residues were substituted by alanine, either individually or in combination, and their influence on Sly1 binding was examined by coimmunoprecipitation studies (Figure 3B and C). Substitution of E53 by alanine severely impaired binding to Sly1, whereas substitutions at positions 61, 67 and 73 had no apparent effect (Figure 3B). A double mutant at positions 52 and 53 (E52/53A) was very similar to E53A (Supplementary Figure S1). The E71A mutation slightly affected the binding to Sly1, whereas substitution of glutamic acids at positions 53 and 71 by alanine (E53/71A) completely abolished Cog4–Sly1 interaction (Figure 3C). Remarkably, the E53/71A mutations had no detectable effect on either Syntaxin 5 binding (Figure 3D), the ability of Cog4 to interact with Cog2, or its ability to be incorporated into the endogenous COG complex (Supplementary Figure S2). It is worth noting that the effects of these mutations were assessed by coimmunoprecipitation between Sly1 and the N-terminal fragment of Cog4 (aa 1–231; Figure 3B) and were further confirmed in the context of the full-length Cog4 (Figure 3C). Overall, this analysis suggests that the interaction between Cog4 and Sly1 is confined to the N-terminal region of Cog4 (aa 1–84), with E53 and E71 playing a crucial role, and that this region has been highly conserved throughout evolution with the exception of the Saccharomyces cerevisiae homologue. The sequence differences between the mammalian and yeast Cog4 homologues might be related to the inability of mammalian Sly1 to functionally replace Sly1p in S. cerevisiae (Li et al, 2007).
Figure 3.
Glutamic acids at positions 53 and 71 of the human Cog4 are critical for Sly1 binding. (A) Multiple sequence alignment of Cog4 from different species. The sequences were aligned using ClustalW. Black and gray background represents degree of similarity. Asterisks mark highly conserved residues that have been mutated. Hs: Homo sapiens, Mm: Mus musculus, Gg: Gallus gallus, Dr: Danio rerio, Sp: Strongylocentrotus purpuratus, Dm: Drosophila melanogaster, Ce: Caenorhabditis elegans, At: Arabidopsis thaliana, Sc: Saccharomyces cerevisiae. (B) Interaction between Sly1-HA and Myc-tagged Cog4 fragments (aa 1–231) containing the indicated point mutations was assessed by coimmunoprecipitation using anti-Myc antibody for immunoprecipitation and anti-HA antibody for immunoblotting. (C) Substitution of glutamic acids at positions 53 and 71 by alanine abolished the interaction between Sly1 and Cog4, with no detectable effect on the Syntaxin 5–Cog4 interaction (D). The interaction between the indicated Myc-tagged Cog4 mutants and either Sly1-HA or Syntaxin 5-HA was determined by coimmunoprecipitation using anti-Myc antibody for immunoprecipitation and anti-HA antibody for immunoblotting. The expression level of the transfected proteins was assessed by immunoblotting of total cell lysates with the indicated antibodies (lower panels, B–D).
The Cog4–Sly1 interaction is required for colocalization of SNAREs involved in intra-Golgi transport
To elucidate the physiological role of the Cog4–Sly1 interaction, we exploited the binding data shown in Figures 2 and 3, and ectopically expressed the N-terminal fragment of Cog4 (aa 1–84), containing the Sly1-binding site, in mammalian cells as a GFP-fusion protein. We then examined its effect on the steady-state distribution of SNARE proteins involved in intra-Golgi transport (Pelham et al, 1995; Hong, 2005; Lupashin and Sztul, 2005), including the t-SNARE Syntaxin 5 and the v-SNARE GS15. As shown, under a moderate level of expression, the GFP-Cog4 (1–84) fragment caused the redistribution of GS15 from its typical Golgi localization into diffuse punctate cytosolic structures. Concomitantly, GS15 lost its colocalization with Syntaxin 5 (Figure 4A). Remarkably, however, a GFP-Cog4 (1–84) fragment carrying the E53/71A mutations had a minor effect on GS15 distribution. Quantification of Syntaxin 5-GS15 colocalization revealed that 70%±10.5 of Syntaxin 5 is colocalized with GS15 in the control cells (n=20), whereas only 15%±7.3 in cells expressing the GFP-Cog4 (1–84) fragment. Together, these results suggest that disruption of Cog4–Sly1 binding impairs the interaction between these SNAREs.
Figure 4.
The N-terminal fragment of Cog4 disrupts the interaction between Cog4 and Sly1 and the colocalization of GS15 with Syntaxin 5. (A) GFP-tagged fragment consisting of the first 84 aa of either the wild-type Cog4 or the E53/71A double mutant was transiently transfected into HeLa cells. Two days later, the cells were fixed and double-immunostained with anti-GS15 and anti-Syntaxin 5 antibodies. Shown are representative confocal images of cells expressing the GFP-tagged Cog4 fragments (green) along with GS15 (red) and Syntaxin 5 (light blue). As shown, colocalization of GS15 and Syntaxin 5 was impaired in cells expressing moderate levels of the wild-type GFP-Cog4 (aa 1–84). Higher levels of expression caused Golgi fragmentation (C), as determined by immunostaining with anti-GRASP-65 antibody. Cells expressing a high level of these fragments are marked by arrows, whereas those expressing a low level are marked by arrowheads. Scale bar, 10 μm. (B) The GFP-Cog4 fragment (aa 1–84) inhibits the interaction between Cog4 and Sly1. Equal amounts of cell lysate of HEK293 cells expressing the Cog4-Myc were mixed with increasing amounts of cell lysates that were prepared from either HEK293 cells or HEK293 cells expressing the GFP-Cog4 (1–84) fragment or GFP. Equal volumes of the cell lysates mixtures were then incubated with recombinant GST, GST-Sly1 or GST-Syntaxin 5 immobilized on glutathione-agarose beads (GB pull-down), as indicated. The samples were washed and then resolved by SDS–PAGE, transferred to a nitrocellulose membrane and immunoblotted with the indicated antibodies. As shown, the GFP-Cog4 (1–84) fragment inhibited the binding of Cog4-Myc to GST-Sly1 in a concentration-dependent manner but had no effect on Cog4-Myc binding to GST-Syntaxin 5 (upper panels). The expression level of Cog4-Myc, the GFP-Cog4 (1–84) fragment or GFP in the lysates mixtures was determined by western blotting with either anti-Myc or anti-GFP antibodies (lower panels). (D) The Myc-tagged Cog4 fragment (aa 1–84) of wild-type Cog4 but not of the E53/71A double mutant disrupts the Golgi-targeting of GS15, as shown by double-immunostaining of fixed HeLa cells expressing the indicated Cog4 fragments with anti-Myc (green) and anti-GS15 (red) antibodies. Scale bar, 10 μm.
To show that this fragment indeed disrupts the interaction between Cog4 and Sly1, we assessed its influence on Cog4–Sly1 interaction using pull-down experiments with GST-Sly1 immobilized on glutathione-agarose beads. This fragment inhibited the binding of Cog4 to Sly1 in a concentration-dependent manner (Figure 4B), but had no effect on Cog4–Syntaxin 5 interaction. Consistent with these results, we noticed that high expression levels of GFP-Cog4 (1–84) cause Golgi fragmentation, as shown by the steady-state distribution of several Golgi markers, including GRASP-65 (Figure 4C). These observations suggest that disruption of the Cog4–Sly1 interaction can impair the steady-state structure of the Golgi complex, possibly by inhibiting SNARE pairing and therefore certain Golgi-fusion reactions. To ensure that these observations did not hinge on unusual properties of the GFP-tagged proteins, we expressed the Cog4 (1–84) fragment as a Myc-tagged protein and examined its influence on GS15 localization by indirect immunofluorescence analysis. As shown, only the wild-type Cog4 (1–84) fragment caused redistribution of GS15 from the Golgi complex (Figure 4D), consistent with the results obtained with the GFP-fusion proteins shown in Figure 4A.
To further show the importance of the Cog4–Sly1 interaction for Golgi SNARE pairing, we applied a different strategy and knocked-down Cog4 expression in mammalian cells by RNA interference (RNAi) using a hairpin siRNA expression vector. The specificity of this RNAi for Cog4 was confirmed by transient coexpression with expression vectors encoding either Cog3 or Cog4 (Figure 5A). Accordingly, stable HeLa cell lines expressing the siRNA expression vector were established and examined for Cog4 expression by immunoblotting (Figure 5B) and by inmmunofluorescence analysis (Figure 5C). As shown in Figure 5C, depletion of Cog4 expression affected the compact organization of the Golgi complex, consistent with the established role of the COG complex in maintaining Golgi structure (Chatterton et al, 1999; Ungar et al, 2002; Oka et al, 2005; Zolov and Lupashin, 2005). We next examined the effect of Cog4 depletion on the steady-state distribution and protein levels of the t-SNAREs Syntaxin 5 and GS28, and the v-SNARE GS15, by indirect immunofluorescence (Figure 5D) and immunoblotting (Figure 5E) analysis, respectively. Depletion of Cog4 had no apparent effect on the steady-state levels of Syntaxin 5 or GS28, but caused a significant reduction (∼60%) in the steady-state level of GS15. The Golgi localization of Syntaxin 5 was not affected by Cog4 knock-down. However, Cog4 depletion markedly affected the distribution of GS28 and GS15. These SNAREs lost their compact perinuclear localization and were visualized in small puncta in the cytosol (Figure 5D). They also lost their colocalization with Syntaxin 5 (Supplementary Figure S3), suggesting that Cog4 knock-down impaired the pairing of these SNAREs.
Figure 5.
Wild-type Cog4, but not the E53/71A double mutant, restores the Golgi-targeting of SNAREs in Cog4-depleted cells. (A) HeLa cells were transiently cotransfected with shRNA construct encoding siRNA of the human Cog4 together with expression vectors encoding either Myc-tagged Cog3 or Cog4. The specificity of this siRNA was determined by immunoblotting with anti-Myc antibody. Stable HeLa cell lines depleted of Cog4 (Cog4-KD) were established and assessed for Cog4 expression by immunoblotting (B) and immunofluorescence (C) using anti-Cog4 antibody. As shown (C), depletion of Cog4 impairs the compact organization of the Golgi complex. Scale bar, 10 μm. (D) Localization of the SNARE proteins GS15, GS28 and Syntaxin 5 was determined in control and Cog4-knock-down cells by immunofluorescence analysis using the corresponding antibodies. Shown are representative confocal images. Scale bar, 10 μm. (E) Expression levels of GS15, GS28, Syntaxin 5 and Sly1 in control and Cog4-depleted HeLa cells were determined by immunoblotting using the indicated antibodies. The two bands of Syntaxin 5 represent its two isoforms. (F) Silent mutations within the RNAi targeting sequence of Cog4 protect its expression from its RNAi. HeLa cells were transiently cotransfected with the shRNA construct along with Myc-tagged of either the wild-type or the silent Cog4 mutant. Expression of Cog4 was determined by western blotting with anti-Myc antibody. (G) The wild-type Cog4, but not the E53/71A double mutant, restores the targeting of GS15 and GS28 to the Golgi in Cog4-depleted cells and consequently, SNARE pairing. Cog4-depleted HeLa cells were transiently transfected with either the wild-type or the E53/71A double mutant of Myc-tagged Cog4 containing the silent mutations within the RNAi targeting sequence. Two days later, the cells were fixed and double-immunostained with anti-Myc and either anti-GS15 or anti-GS28 antibodies. Transfected cells are indicated by arrowheads. Scale bar, 10 μm.
If indeed the interaction of Cog4 with Sly1 is required for these SNAREs' pairing, as suggested by the results shown in Figure 4, then wild-type Cog4, but not the E53/71A-Cog4 double mutant, would be expected to restore SNARE pairing in Cog4-knocked-down cells. To explore this possibility, we introduced silent mutations within the RNAi targeting sequence of the wild-type and E53/71A Cog4 cDNAs. These silent mutations protected Cog4 expression from Cog4-RNAi (Figure 5F). Next, we ectopically expressed the wild-type Cog4 and the E53/71A-Cog4 double mutant in Cog4-depleted cells and examined their influence on the steady-state distribution of GS15 and GS28 by indirect immunofluorescence analysis. As shown in Figure 5G, both the wild-type and the E53/71A mutant were expressed in Cog4-depleted cells and were localized to the Golgi complex. However, only the wild-type Cog4 could restore the Golgi targeting of GS28 and GS15, and the colocalization between GS15 and Syntaxin 5 (Supplementary Figure S4), suggesting that the Cog4–Sly1 interaction is required for SNAREpin assembly.
The Cog4–Sly1 interaction is required for Golgi-to-ER retrograde transport
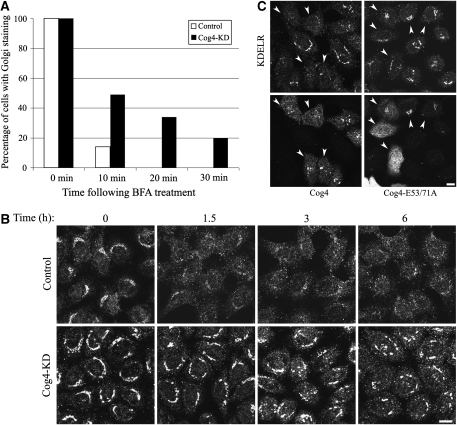
It is well established that the SNARE complex consisting of Syntaxin 5, GS28, Ykt6 and GS15 functions in intra-Golgi and Golgi-to-ER retrograde transport (Hong, 2005). This led us to further explore the role of the Cog4–Sly1 interaction in these trafficking pathways. We first characterized the effect of Cog4 depletion on protein transport to and from the Gogi complex through several approaches: treatment with brefeldin A (BFA) and washout was used to assess Golgi-to-ER and ER-to-Golgi transport, respectively. BFA is a fungal metabolite that inhibits anterograde but not retrograde vesicular transport (Lippincott-Schwartz et al, 1989). It is, therefore, commonly used to measure Golgi-to-ER retrograde transport. The effect of BFA on the localization of mannosidase II, a medial Golgi resident protein that recycles through the ER, was determined at different time points after BFA treatment by indirect immunofluorescence analysis. As shown (Figure 6A; Supplementary Figure S5A), knocking down of Cog4 substantially inhibited the transport of mannosidase II from the Golgi to the ER with no detectable effect on ER-to-Golgi anterograde transport (Supplementary Figure S5B), consistent with the established role of the COG complex in intra-Golgi and Golgi-to-ER retrograde transport (Whyte and Munro, 2001; Oka et al, 2004; Steet and Kornfeld, 2006; Ungar et al, 2006).
Figure 6.
Cog4 restores Golgi-to-ER retrograde transport in a Sly1-interaction-dependent manner. (A) Depletion of Cog4 attenuates the transport of mannosidase II from the Golgi to the ER in response to BFA treatment. Control and Cog4-depleted HeLa cells were treated with BFA (5 μg/ml) for the indicated times, fixed, immunostained with anti-mannosidase II antibody, and analysed by confocal microscopy. The percentage of cells in which mannosidase II was localized to the Golgi was calculated from 200 cells at each time point. Representative confocal images are shown in Supplementary Figure S5A. (B) Depletion of Cog4 attenuates retrograde transport of KDELR. Control and Cog4-depleted HeLa cells were incubated at 15°C for the indicated times, to block protein exit from the ERGIC. The cells were then fixed, immunostained with anti-KDELR antibody and analysed by confocal microscopy. Shown are representative confocal images of control and Cog4-depleted cells at the indicated time points. Scale bar, 10 μm. (C) Cog4-depleted HeLa cells were transiently transfected with either the wild-type or E53/71A double mutant of Myc-tagged Cog4 containing the silent mutations within the RNAi targeting sequence. Two days later, the cells were incubated for 1.5 h at 15°C, fixed and double-immunostained with anti-Myc and anti-KDELR antibodies. Transfected cells are indicated by arrowheads. As shown, the wild-type Cog4, but not the E53/71A double mutant, restored retrograde transport of KDELR. Scale bar, 10 μm.
In a complementary approach, we examined the subcellular distribution of the KDEL receptor (KDELR) at steady-state and after shifting the temperature to 15°C for different time periods by indirect immunofluorescence analysis. As shown in Figure 6B, at steady-state (time 0), KDELR was localized to the Golgi and also appeared in small puncta throughout the cytosol of the control cells. Its Golgi localization, however, was enhanced in Cog4-depleted cells. As KDELR retrieves KDEL-containing proteins from the cis-Golgi to the ER, its enhanced Golgi localization may reflect an inhibitory effect of Cog4 knock-down on Golgi-to-ER retrograde transport. This was much more pronounced after shifting the temperature to 15°C, which blocks protein exit from the ER-Golgi intermediate compartment (ERGIC) (Luna et al, 2002). At this temperature, KDELR, which cycles between the Golgi and the ER, loses its Golgi localization and is trapped in the ERGIC, as clearly observed for the control, but not for the Cog4-depleted cells. In these latter cells, KDELR was detected in the Golgi for a very long time (6 h) after the temperature shift, showing the remarkable inhibitory effect of Cog4 depletion on retrograde transport from the Golgi complex. Accordingly, we examined whether wild-type Cog4 and the E53/71A-Cog4 mutant could restore the redistribution of KDELR from the Golgi of Cog4-depleted cells after reducing the temperature to 15°C for 1.5 h. As shown in Figure 6C, the wild-type but not the E53/71A-Cog4 mutant restored the redistribution of KDELR. These results are consistent with the observations shown in Figure 5G and suggest that the Cog4–Sly1 interaction is required for the pairing of SNAREs involved in intra-Golgi transport and consequently for Golgi-to-ER retrograde transport.
Discussion
Selective SNARE pairing and assembly of SNAREpins at specific membrane-fusion sites are fundamental for every vesicular transport step. The involvement of SM proteins and various tethering factors in these processes has been shown by numerous genetic and biochemical studies. However, a direct interaction between SM proteins and tethering factors has never been described in mammalian cells. Rather, the SM protein, Vps33p, has been identified as one subunit of the homotypic fusion and vacuole protein sorting complex in yeast, which acts during the docking stage of vacuole fusion (Rieder and Emr, 1997; Sato et al, 2000; Subramanian et al, 2004). In this study, we show that the tethering complex COG interacts directly with the SM protein Sly1 through its Cog4 subunit. This interaction was confined to the first N-terminal 84 aa of Cog4 (Figure 2). Further mutagenesis studies of highly conserved residues showed the importance of the glutamic acids at positions 53 and 71 for the Cog4–Sly1 interaction (Figure 3). Strikingly, an N-terminal fragment consisting of the first 153 aa of Cog4 also interacted with Syntaxin 5 (Figure 2D). However, the E53/71A-Cog4 double mutant, which failed to interact with Sly1, interacted with Syntaxin 5 (Figure 3D), whereas a fragment consisting of the first 84 aa of Cog4, which interacted with Sly1, failed to interact with Syntaxin 5 (Figure 2D). These results suggest that the N-terminal fragment of Cog4 consists of two adjacent but distinct binding sites for Sly1 and Syntaxin 5. Interestingly, Syntaxin 5 also has two separate binding sites for Cog4 and Sly1. It interacts with Sly1 through a short N-terminal peptide consisting of its first 20 aa (Yamaguchi et al, 2002), and with Cog4 through its H3 fragment containing the SNARE domain (Shestakova et al, 2007). These observations suggest that these three proteins may interact simultaneously with each other and may function interdependently. Indeed, we found that Cog4 can interact simultaneously with Sly1 and Syntaxin 5 using coimmunoprecipitation studies (Supplementary Figure S7C). In these studies, wild-type Cog4, but not the E53/71A Cog4 mutant could restore an interaction between a Syntaxin 5 mutant (T7/F10A), which cannot bind Sly1 (Yamaguchi et al, 2002) (Supplementary Figures S7A), and the wild-type Sly1 protein. Whether the interaction of Cog4 with Sly1 enhances its binding to Syntaxin 5, or vice versa, is not currently known. Further characterization of COG, Sly1 and Syntaxin 5 interactions is expected to provide mechanistic insight into their mode of action in intra-Golgi vesicular fusion.
Earlier studies in yeast have shown that mutations in several COG subunits, including sec36/Cog1, sec35/Cog2 and sec34/Cog3, display severe synthetic growth defects with a sly1-ts mutation, and that expression of the SLY1-20 dominant allele efficiently suppresses the growth defect of the sec36-1, sec34-1 and sec35-1 mutants (VanRheenen et al, 1998, 1999; Ram et al, 2002). SLY1-20 also suppresses ypt1 mutant, a small GTPase that is required for tethering of ER-derived vesicles to Golgi membranes (Ossig et al, 1991; Cao et al, 1998). It was proposed that Sly1-20p represents an activated form of Sly1 that bypasses the tethering requirement (Sapperstein et al, 1996; Cao et al, 1998), thereby suppressing ypt1, uso1 and sec34/35 mutants. Structural studies have revealed that the Sly1-20p point mutation resides on the surface of a short helix (α20), which together with α21 may act as a Rab-regulated lid to control Sly1p activity (Bracher and Weissenhorn, 2002). Thus, Sly1-20p (E532A) may have a permanently open lid representing a constitutive active form. Conformational changes in Sly1 have also been observed on Syntaxin 5 binding. The Syntaxin 5–Sly1 interaction has been shown to induce a significant alteration in the overall shape of the full-length rSly1 that could be critical for its function (Arac et al, 2005). Whether the Sly1–COG interaction also induces a conformational change in Sly1 is currently unknown. Nevertheless, we provide evidence of the physiological importance of this interaction.
Overexpressing the N-terminal fragment of Cog4 containing the Sly1-binding site (aa 1–84) induced redistribution of GS15 from its typical Golgi localization to vesicular-like structures, and concomitantly abolished its colocalization with Syntaxin 5. These results were obtained with the wild-type Cog4, but not with its E53/71A mutant (Figure 4), suggesting that inhibition of the Cog4–Sly1 interaction markedly affects the pairing of these SNAREs. A similar strategy was used earlier to inhibit the Syntaxin 5–Sly1 interaction in mammalian cells by applying either the N-terminal fragment of Syntaxin 5 containing the Sly1-binding site, or the N-terminal fragment of Sly1 (aa 1–147) containing the Syntaxin 5-binding site. Those studies suggested that the Syntaxin 5–Sly1 interaction is required for both ER-to-Golgi transport and maintenance of the Golgi morphology (Dascher and Balch, 1996; Yamaguchi et al, 2002; Dulubova et al, 2003; Williams et al, 2004). These observations are consistent with the essential role of Sly1 in the fusion of ER-derived vesicles with the Golgi membranes (Kosodo et al, 2002; Peng and Gallwitz, 2002, 2004). However, Sly1 has also been implicated in regulating Golgi-to-ER retrograde transport as evidenced by the transport defects of a sly1-5 mutant harbouring a single aa substitution, R452A, in domain III of Sly1p (Li et al, 2005).
Consistent with these findings, we show here that the Cog4–Sly1 interaction is required for Golgi-derived retrograde transport (Figure 6C), as evidenced by the ability of the wild-type Cog4, but not the E53/71A double mutant, to restore the transport defect of KDELR from the Golgi of Cog4-depleted cells, after shifting the temperature to 15°C. We also show that depletion of Cog4 subunit by RNAi affects the Golgi structure (Figure 5C), and substantially inhibits retrograde transport from the Golgi, as determined by BFA treatment (Figure 6A; Supplementary Figure S5A) and by the redistribution of KDELR in response to a 15°C temperature shift (Figure 6B), with no detectable effect on ER-to-Golgi anterograde transport (Supplementary Figures S5B and S6). These results are consistent with the established roles of the COG complex in maintaining Golgi structure and regulating retrograde vesicular trafficking within the Golgi complex (Chatterton et al, 1999; Oka et al, 2004, 2005; Wu et al, 2004; Zolov and Lupashin, 2005; Shestakova et al, 2006; Steet and Kornfeld, 2006; Ungar et al, 2006; Kranz et al, 2007; Ng et al, 2007). Cog4 depletion had no apparent effect on the Golgi localization of either Syntaxin 5 (Figure 5D) or Sly1 (not shown), but markedly affected the steady-state distribution of the Golgi SNAREs GS15 and GS28 (Figure 5D), and consequently Syntaxin 5-GS15 colocalization (Supplementary Figure S3). The specificity of these effects was evidenced by the ability of wild-type Cog4 to restore the Golgi structure, as well as the Golgi localization of these SNARE proteins (Figure 5G). Strikingly, however, the E53/71A double mutant failed to restore the Golgi localization of GS15 and GS28 (Figure 5G), the colocalization between GS15 and Syntaxin 5 (Supplementary Figure S4), or retrograde transport of KDELR (Figure 6C). Together, these results suggest that the Cog4–Sly1 interaction is required for pairing of SNAREs involving in intra-Golgi transport.
How are the interactions of COG with Sly1 and Syntaxin 5 coordinately operated in intra-Golgi membrane fusion? The COG is considered to act as a tethering complex that mediates the first point of contact between a vesicle and its target membrane, before SNARE pairing. As such, it might mediate the initial interaction between Syntaxin 5 on the Golgi membrane and the cognate v-SNARE GS15 on the vesicle. This potential role is consistent with the impaired pairing of intra-Golgi SNAREs obtained in cells that have been depleted or mutated in certain COG subunits (Ungar et al, 2002; Oka et al, 2004, 2005; Zolov and Lupashin, 2005), including Cog4 (Supplementary Figure S3). However, in a recent in vitro reconstitution study, COG failed to stimulate trans-SNARE complex formation (Shestakova et al, 2007). On the other hand, the SM proteins were found to markedly accelerate the rate of membrane fusion in vitro (Scott et al, 2004; Shen et al, 2007). SM proteins seem to act on a transient, partially assembled intermediate of the SNARE complex and promote progression of the fusion event by direct interaction with both t- and v-SNAREs (Shen et al, 2007). Thus, it could be that the COG complex is involved in the initial assembly of these intermediate species by stabilizing the transient v- or t-SNARE interaction, as has been recently proposed (Shestakova et al, 2007). These intermediate species could then provide a platform for SM protein action. Subsequently, COG may act coordinately on the assembled complex with the SM protein, by direct interaction with Sly1. Consistent with this hypothesis, recent studies have shown that the COG complex displays a high affinity for Sed5p-containing SNARE complex (Shestakova et al, 2007), and our studies indicate that the COG–Sly1 interaction is required for intra-Golgi membrane fusion (Figures 5 and 6). It is currently unclear whether the COG–Sly1 interaction is involved in the activation of Sly1 through a conformational change, or simply provides a scaffold for the assembly of multiprotein complexes involved in the membrane-fusion machinery, thereby increasing the local concentration of these components to facilitate SNARE pairing and eventually, membrane fusion.
Materials and methods
Antibodies, reagents and chemicals
Polyclonal antibodies against Cog3 (aa 1–274), Cog4 (aa 1–337) and Cog7 (aa 1–329) were raised in rabbits immunized with recombinant GST-fusion proteins consisting of the indicated aa residues. The antibodies were affinity purified by two sequential steps; the antiserum was first purified on a GST-bound agarose column to remove the anti-GST antibodies, and subsequently on a GST-Cog-bound agarose column. Antiserum against Cog8 was kindly provided by D Ungar (York University, York, UK), whereas polyclonal antibodies against GS28 and Syntaxin 5 and monoclonal anti-VSV-G antibody were a generous gift of Z Elazar (Weizmann Institute of Science, Rehovot, Israel). Polyclonal antibodies against GRASP-65 (Peretti et al, 2008) and GS15 (Xu et al, 1997), and monclonal anti-GS28 antibody (Subramaniam et al, 1995) were described earlier. Monoclonal anti-GS15 and anti-KDELR antibodies were purchased from BD Biosciences (San Jose, CA) and from Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Polyclonal antibodies against mannosidase II and Sly1 (SCFD1) were purchased from the University of Georgia (Athens, GA) and from Proteintech Group, Inc. (Chicago, USA), respectively. Monoclonal antibody against p115 was kindly provided by the late D Shields (Albert Einstein College of Medicine, New York, NY). Alexa-488 donkey anti-mouse and anti-rabbit immunoglobulin Gs (IgGs) were purchased from Invitrogen (Carlsbad, CA). Cyanine (Cy)3-conjugated goat anti-rabbit and goat anti-mouse IgGs, as well as Cy5-conjugated goat anti-rabbit IgG were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
ProteinA-agarose beads were purchased from Repligen Corp. (Waltham, MA). Ni-NTA agarose was from Qiagen, whereas BFA, puromycin, and anti-mouse IgG conjugated to agarose beads were purchased from Sigma-Aldrich.
DNA constructs
Bacterial expression vector encoding rSly1 fused to GST was kindly provided by JC Hay (University of Montana, Missoula, Montana). The cDNA of rSly1 was subcloned into the mammalian expression vector pCMV-neo-HA. The cDNA of the human Golgi isoform of Syntaxin 5 was kindly provided by J Gerst (Weizmann Institute of Science, Rehovot, Israel) and subcloned into pCMV-neo-HA. Threonine at position 7 and phenylalanine at position 10 of Syntaxin 5 were substituted by alanine to produce the Syntaxin 5(T7/F10A) mutant using the QuickChange site directed mutagenesis kit and the following sense and anti-sense primers: (5′-CCTGCCGGGATCGCGCTCAAGAAGCTCTGTCTGCCTGCAAGTCGC-3′) and (5′-GCGACTTGCAGGCAGACAGAGCTTCTTGAGCGCGATCCCGGCAGG-3′). The DNA constructs encoding the different Myc-tagged COG subunits have been described earlier (Loh and Hong, 2002, 2004). Truncated Cog4 mutants were produced by subcloning of the corresponding PCR products into either pCMV-neo-Myc, pEGFP-C1 (Clontech), or mammalian-GST expression vectors. The following sense and anti-sense primers have been used for PCR amplifications: Cog4 (1–32); (5′-TTCGGATCCTTGGCGGACCTTGATTCGCC-3′) and (5′-AAAGCGGCCGCTCAGAGCTCAGCGGAGATTTC-3′); Cog4 (1–84); (5′-TTCGGATCCTTGGCGGACCTTGATTCGCC-3′) and (5′-AAGGCGGCCGCTCACAGATTAGGACCCATTCG-3′); Cog4 (1–153); (5′-TTCGGATCCTTGGCGGACCTTGATTCGCC-3′) and (5′-AAAGCGGCCGCCTACTGCTCATAATCTTCACTC-3′); Cog4 (1–180); (5′-AGAGGATCCAATGGCGGACCTTGATTCG-3′) and (5′-AAAGGTACCCTACATGCTCCCCTCTTTGCC-3′); Cog4 (1–231); (5′-TTCGGATCCTTGGCGGACCTTGATTCGCC-3′) and (5′-AAACTCGAGCTATCCCTCCTCATGCAAACCC-3′); Cog4 (81–231); (5′-AAAGGATCCAATGGGTCCTAATCTGCAG-3′) and (5′-AAACTCGAGCTATCCCTCCTCATGCAAACCC-3′).
A BglII-NotI fragment of Cog4 cDNA was subcloned into pCMV-neo-Myc vector to produce a Myc-tagged Cog4 (220–785) truncated mutant. Point mutations within Cog4 cDNA were introduced by the QuickChange site directed mutagenesis kit according to the manufacturer's instructions (Stratagene), and confirmed by DNA sequencing. The following oligonucleotide primers have been used for the mutagenesis (only the sense primer is specified): E53A (5′-GCTCTGCGGCGAAGCCAAAGTGGTGGAGAGAG-3′); E52A/E53A (5′-GCTCTGCGGCGCAGCCAAAGTGGTGGAG-3′); D61A (5′-GGTGGAGAGAGAGTTGGCCGCTCTTTTGGAACAGC-3′); Q67A (5′-GCTCTTTTGGAACAGGCTAACACCATTGAAAG-3′); E71A (5′-GGAACAGCAAAACACTATAGCAAGTAAGATGGTCACTCTCC-3′); K73A (5′-GGAACAGCAAAACACAATTGAAAGTGCGATGGTCACTCTCC-3′).
Silent mutations within the RNAi targeting sequence of Cog4 were introduced by the same procedure using the following sense and anti-sense primers: (5′-GTACCATGCAGGAGCTCATCGGGTTGTATGTTACCATGGAGG-3′) and (5′-CCTCCATGGTAACATACAACCCGATGAGCTCCTGCATGGTAC-3′).
Cell culture, transfection and immunofluorescence microscopy
HEK293 and HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, 100 μg/ml penicillin and 100 μg/ml streptomycin. The cells were transfected using the calcium–phosphate method. Transfected HeLa cells were grown on coverslips, washed with phosphate-buffered saline (PBS), and fixed either in methanol or methanol-acetone (50%/50%) for 1 h at −20°C, or in 1% paraformaldehyde in KM buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.2, 10 mM NaCl, 1.5 mM MgCl2 and 2.5% glycerol) for 20 min at room temperature. Cells were immunostained as described earlier (Litvak et al, 2002). The specimens were analysed by a confocal laser scanning microscope (Zeiss 510; Carl Zeiss, Jena, Germany) by using the 488-, 543-nm and either 405- or 633-nm excitation for fluorescein, Cy3 epifluorescence and either 4,6-diamidino-2-phenylindole (Hoechst) or Cy5, respectively. Quantification of colocalization on confocal sections was performed by the LSM510 colocalization software. Standard deviation was calculated from the quantification of 20 cells.
Establishment of Cog4 knock-down stable cell lines
The mammalian pSUPER-puro vector was used for expression of a short hairpin (sh) RNA corresponding to nucleotides 1242–1260 (5′-GGAGCTAATTGGCTTATAT-3′) of the human Cog4 cDNA. HeLa cells were transfected with the Cog4 shRNA construct, and 36 h later were splitted into selection medium containing 0.5 μg/ml puromycin. Depletion of Cog4 expression in puromycin-resistant clones was verified by both immunoblot and immunofluorescence analysis. Stable cell-line harbouring an empty pSUPER vector was established and used as control.
Cell extracts, immunoprecipitations and pull-down experiments
Cells were lysed in lysis buffer containing: 1% triton X-100, 20 mM Hepes pH 7.5, 100 mM NaCl, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 1 mM PMSF, 10 μg/ml leupeptin and 10 μg/ml aprotinin. The lysate was centrifuged at 15 000 g for 15 min at 4°C, and the supernatant was used in either pull-down or immunoprecipitation experiments. For pull-down assays, GST and GST-rSly1 were expressed in bacteria, purified by standard procedures (Amersham Biosciences), and incubated with cell lysates expressing the indicated protein for 2 h at 4°C. The samples were then washed twice in buffer containing; 20 mM Hepes pH 7.5, 250 mM NaCl and 1 mM DTT, followed by three washes in buffer containing; 20 mM Hepes pH 7.5, 250 mM NaCl, 1% triton X-100, and 1 mM DTT, and finally with buffer containing; 20 mM Hepes pH 7.5, 100 mM NaCl and 1 mM DTT. The bound proteins were analysed by western blotting. For direct binding assays, His-Cog4 (1–231) was purified from bacteria on a nickel column according to the manufacturer's instructions (Qiagen), incubated with GST or GST-rSly1-bound to glutathione agarose beads, washed as described above, and analysed by western blotting. Immunoprecipitations were performed essentially as described earlier (Amarilio et al, 2005).
Transport assays
Control and Cog4 knocked-down cell-lines grown in 24-well plates were either treated with 5 μg/ml BFA or incubated at 15°C for the indicted time points. The cells were then fixed either in acetone-methanol (50%/50%) or in methanol for 1 h at −20°C, immunostained with either anti-mannosidase II or anti-KDELR antibodies, respectively, and analysed by confocal microscopy. For the BFA wash-out experiments, cells were treated with 0.25 μg/ml BFA for 1 h, extensively washed in PBS, allowed to recover in regular medium for the indicated time points, fixed and immunostained with anti-mannosidase II antibody, as described above. VSV-G transport assay was performed essentially as described earlier (Litvak et al, 2005).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure Legends
Review Process File
Acknowledgments
Sima Lev is the incumbent of the Joyce and Ben B Eisenberg Chair of Molecular Biology and Cancer Research. This work was supported by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel's Ministry of Science and Technology (MOST). We thank Daniel Unger, York University, UK, Jesse C Hay, University of Montana, Montana, USA, Monty Krieger, Massachusetts Institute of Technology, Cambridge, USA, and Zvulun Elazar, Weizmann Institute of Science, Israel, for generously providing reagents and for productive discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amarilio R, Ramachandran S, Sabanay H, Lev S (2005) Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem 280: 5934–5944 [DOI] [PubMed] [Google Scholar]
- Arac D, Dulubova I, Pei J, Huryeva I, Grishin NV, Rizo J (2005) Three-dimensional structure of the rSly1 N-terminal domain reveals a conformational change induced by binding to syntaxin 5. J Mol Biol 346: 589–601 [DOI] [PubMed] [Google Scholar]
- Bracher A, Weissenhorn W (2002) Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J 21: 6114–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S (2007) Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Cao X, Ballew N, Barlowe C (1998) Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J 17: 2156–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ (1999) Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol 146: 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterton JE, Hirsch D, Schwartz JJ, Bickel PE, Rosenberg RD, Lodish HF, Krieger M (1999) Expression cloning of LDLB, a gene essential for normal Golgi function and assembly of the ldlCp complex. Proc Natl Acad Sci USA 96: 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH (2001) SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol 2: 98–106 [DOI] [PubMed] [Google Scholar]
- Dascher C, Balch WE (1996) Mammalian Sly1 regulates syntaxin 5 function in endoplasmic reticulum to Golgi transport. J Biol Chem 271: 15866–15869 [DOI] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD (1991) Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol 11: 872–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J (1999) A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J 18: 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Arac D, Li H, Huryeva I, Min SW, Rizo J, Sudhof TC (2003) Convergence and divergence in the mechanism of SNARE binding by Sec1/Munc18-like proteins. Proc Natl Acad Sci USA 100: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV (2005) Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem 280: 27613–27623 [DOI] [PubMed] [Google Scholar]
- Garcia EP, Gatti E, Butler M, Burton J, De Camilli P (1994) A rat brain Sec1 homologue related to Rop and UNC18 interacts with syntaxin. Proc Natl Acad Sci USA 91: 2003–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Heuser JE, Jahn R (1997) Neurotransmitter release—four years of SNARE complexes. Curr Opin Neurobiol 7: 310–315 [DOI] [PubMed] [Google Scholar]
- Hong W (2005) SNAREs and traffic. Biochim Biophys Acta 1744: 120–144 [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Noda Y, Adachi H, Yoda K (2002) Binding of Sly1 to Sed5 enhances formation of the yeast early Golgi SNARE complex. J Cell Sci 115: 3683–3691 [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RM, Field MC (2007) Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol 7: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Winkel RD, Cipollo JF, Costello CE, Loh E, Hong W, Freeze HH (2007) COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet 16: 731–741 [DOI] [PubMed] [Google Scholar]
- Li Y, Gallwitz D, Peng R (2005) Structure-based functional analysis reveals a role for the SM protein Sly1p in retrograde transport to the endoplasmic reticulum. Mol Biol Cell 16: 3951–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schmitt HD, Gallwitz D, Peng RW (2007) Mutations of the SM protein Sly1 resulting in bypass of GTPase requirement in vesicular transport are confined to a short helical region. FEBS Lett 581: 5698–5702 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan LC, Bonifacino JS, Klausner RD (1989) Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S (2005) Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat Cell Biol 7: 225–234 [DOI] [PubMed] [Google Scholar]
- Litvak V, Tian D, Carmon S, Lev S (2002) Nir2, a human homolog of Drosophila melanogaster retinal degeneration B protein, is essential for cytokinesis. Mol Cell Biol 22: 5064–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Hong W (2002) Sec34 is implicated in traffic from the endoplasmic reticulum to the Golgi and exists in a complex with GTC-90 and ldlBp. J Biol Chem 277: 21955–21961 [DOI] [PubMed] [Google Scholar]
- Loh E, Hong W (2004) The binary interacting network of the conserved oligomeric Golgi tethering complex. J Biol Chem 279: 24640–24648 [DOI] [PubMed] [Google Scholar]
- Luna A, Matas OB, Martinez-Menarguez JA, Mato E, Duran JM, Ballesta J, Way M, Egea G (2002) Regulation of protein transport from the Golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol Biol Cell 13: 866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin V, Sztul E (2005) Golgi tethering factors. Biochim Biophys Acta 1744: 325–339 [DOI] [PubMed] [Google Scholar]
- Malsam J, Kreye S, Sollner TH (2008) Membrane fusion: SNAREs and regulation. Cell Mol Life Sci 65: 2814–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, Kranz C, Hagebeuk EE, Duran M, Abeling NG, Wuyts B, Ungar D, Lupashin V, Hartdorff CM, Poll-The BT, Freeze HH (2007) Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol Genet Metab 91: 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Krieger M (2005) Multi-component protein complexes and Golgi membrane trafficking. J Biochem (Tokyo) 137: 109–114 [DOI] [PubMed] [Google Scholar]
- Oka T, Ungar D, Hughson FM, Krieger M (2004) The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol Biol Cell 15: 2423–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Vasile E, Penman M, Novina CD, Dykxhoorn DM, Ungar D, Hughson FM, Krieger M (2005) Genetic analysis of the subunit organization and function of the conserved oligomeric golgi (COG) complex: studies of COG5- and COG7-deficient mammalian cells. J Biol Chem 280: 32736–32745 [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D (1991) The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol 11: 2980–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR, Banfield DK, Lewis MJ (1995) SNAREs involved in traffic through the Golgi complex. Cold Spring Harb Symp Quant Biol 60: 105–111 [DOI] [PubMed] [Google Scholar]
- Peng R, Gallwitz D (2002) Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J Cell Biol 157: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Gallwitz D (2004) Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J 23: 3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S (2008) Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol Biol Cell 19: 3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Scheller RH (1994) n-Sec1: a neural-specific syntaxin-binding protein. Proc Natl Acad Sci USA 91: 1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podos SD, Reddy P, Ashkenas J, Krieger M (1994) LDLC encodes a brefeldin A-sensitive, peripheral Golgi protein required for normal Golgi function. J Cell Biol 127: 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram RJ, Li B, Kaiser CA (2002) Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol Biol Cell 13: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD (1997) A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell 8: 2307–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG (1996) Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol 132: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Rehling P, Peterson MR, Emr SD (2000) Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell 6: 661–671 [DOI] [PubMed] [Google Scholar]
- Scott BL, Van Komen JS, Irshad H, Liu S, Wilson KA, McNew JA (2004) Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol 167: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ (2007) Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell 128: 183–195 [DOI] [PubMed] [Google Scholar]
- Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V (2007) Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol 179: 1179–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Zolov S, Lupashin V (2006) COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic 7: 191–204 [DOI] [PubMed] [Google Scholar]
- Steet R, Kornfeld S (2006) COG-7-deficient human fibroblasts exhibit altered recycling of Golgi proteins. Mol Biol Cell 17: 2312–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam VN, Krijnse-Locker J, Tang BL, Ericsson M, Yusoff AR, Griffiths G, Hong W (1995) Monoclonal antibody HFD9 identifies a novel 28 kDa integral membrane protein on the cis-Golgi. J Cell Sci 108 (Part 6): 2405–2414 [DOI] [PubMed] [Google Scholar]
- Subramanian S, Woolford CA, Jones EW (2004) The Sec1/Munc18 protein, Vps33p, functions at the endosome and the vacuole of Saccharomyces cerevisiae. Mol Biol Cell 15: 2593–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV (2002) The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol 157: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Hughson FM (2003) SNARE protein structure and function. Annu Rev Cell Dev Biol 19: 493–517 [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG (2002) Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol 157: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Krieger M, Hughson FM (2006) Retrograde transport on the COG railway. Trends Cell Biol 16: 113–120 [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Vasile E, Krieger M, Hughson FM (2005) Subunit architecture of the conserved oligomeric Golgi complex. J Biol Chem 280: 32729–32735 [DOI] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Lupashin VV, Barlowe C, Waters MG (1998) Sec35p, a novel peripheral membrane protein, is required for ER to Golgi vesicle docking. J Cell Biol 141: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Cao X, Sapperstein SK, Chiang EC, Lupashin VV, Barlowe C, Waters MG (1999) Sec34p, a protein required for vesicle tethering to the yeast Golgi apparatus, is in a complex with Sec35p. J Cell Biol 147: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JR, Munro S (2001) The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell 1: 527–537 [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S (2002) Vesicle tethering complexes in membrane traffic. J Cell Sci 115: 2627–2637 [DOI] [PubMed] [Google Scholar]
- Williams AL, Ehm S, Jacobson NC, Xu D, Hay JC (2004) rsly1 binding to syntaxin 5 is required for endoplasmic reticulum-to-Golgi transport but does not promote SNARE motif accessibility. Mol Biol Cell 15: 162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH (2004) Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med 10: 518–523 [DOI] [PubMed] [Google Scholar]
- Xu Y, Wong SH, Zhang T, Subramaniam VN, Hong W (1997) GS15, a 15-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) homologous to rbet1. J Biol Chem 272: 20162–20166 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Sudhof TC (2002) Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell 2: 295–305 [DOI] [PubMed] [Google Scholar]
- Yang B, Steegmaier M, Gonzalez LC Jr, Scheller RH (2000) nSec1 binds a closed conformation of syntaxin1A. J Cell Biol 148: 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov SN, Lupashin VV (2005) Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol 168: 747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure Legends
Review Process File