Abstract
We have trapped elongation factor G (EF-G) from Escherichia coli in six, functionally defined states, representing intermediates in its unidirectional catalytic cycle, which couples GTP hydrolysis to tRNA–mRNA translocation in the ribosome. By probing EF-G with trypsin in each state, we identified a substantial conformational change involving its conserved switch I (sw1) element, which contacts the GTP substrate. By attaching FeBABE (a hydroxyl radical generating probe) to sw1, we could monitor sw1 movement (by ∼20 Å), relative to the 70S ribosome, during the EF-G cycle. In free EF-G, sw1 is disordered, particularly in GDP-bound and nucleotide-free states. On EF-G•GTP binding to the ribosome, sw1 becomes structured and tucked inside the ribosome, thereby locking GTP onto EF-G. After hydrolysis and translocation, sw1 flips out from the ribosome, greatly accelerating release of GDP and EF-G from the ribosome. Collectively, our results support a central role of sw1 in driving the EF-G cycle during protein synthesis.
Keywords: elongation factor, GTP hydrolysis, ribosome, translation
Introduction
The high fidelity and speed of cellular protein synthesis are achieved, in part, through an array of GTP hydrolytic proteins (GTPases), which sequentially and transiently associate with the ribosome. Prototypical examples are the two bacterial elongation factors: EF-Tu, which delivers correct aminoacyl-tRNA substrates to the ribosome; and EF-G, which promotes precise translocation of tRNAs and mRNA in the ribosome. With homologues in all cells, these protein factors share a number of intriguing similarities and contrasts.
EF-Tu and EF-G bind to a common site within the cavity between the 30S and 50S subunits of the bacterial 70S ribosome. However, these factors cycle alternately on and off the ribosome, in a cooperative manner, 6 to 20 times per second in vivo (Sørensen and Pedersen, 1991; Mesters et al, 1994). They can be conceptualized as going through unidirectional catalytic cycles, which couple steps of GTP binding, hydrolysis, and release of products (GDP and inorganic phosphate (Pi)) to their specific functions on the ribosome. In their GTP-bound (active) states, they bind to the ribosome. Although GTP hydrolysis is catalysed by residues in the G domain (domain I) shared by both factors, this reaction is intrinsically slow and activated through specific interactions of each factor with the ribosome (Mohr et al, 2002; Kothe et al, 2004; Nechifor et al, 2007). Similar to other GTPases, GTP hydrolysis and subsequent Pi release induces a cascade of conformational changes, starting in two ‘switch' elements (sw1 and sw2) in the G domain (Vetter and Wittinghofer, 2001). In their GDP-bound (inactive) states, the factors dissociate from the ribosome, and they are converted back to their GTP-bound states off the ribosome.
In the EF-Tu cycle (reviewed in Rodnina and Wintermeyer, 2001; Ogle and Ramakrishnan, 2005), a ternary complex of EF-Tu•GTP•aminoacyl-tRNA binds to a ‘posttranslocational' ribosome that displays an unpaired mRNA codon in its 30S A site. The acceptor (aminoacyl) end of the tRNA is held in a cleft between the three domains of EF-Tu, whereas its anticodon end pairs with the codon in the A site. If codon•anticodon pairing is correct, GTP hydrolysis is activated. Crystal structures of free EF-Tu, with GDPNP (nonhydrolysable GTP analogue: guanosine 5′-[β,γ-imido]triphosphate) or GDP, showed a dramatic conformational change induced by its switches (Berchtold et al, 1993; Abel et al, 1996). Sw1 converted from two α-helices, adjacent to GDPNP, to a β-hairpin projected away from GDP. The α-helix and loop of sw2 rotated as a rigid body. In combination, the switches open the interdomain cleft of EF-Tu. GTP hydrolysis is thought to release the aminoacyl-tRNA from EF-Tu to the 50S A site, allowing for peptide bond formation with peptidyl-tRNA in the P site, and release of EF-Tu•GDP from the ribosome (Rodnina and Wintermeyer, 2001). Off the ribosome, EF-Tu•GDP is kinetically stable and requires EF-Ts to exchange its bound GDP for a fresh GTP (Gromadski et al, 2002).
In the EF-G cycle (reviewed in Frank et al, 2007; Shoji et al, 2009), EF-G•GTP binds to a ‘pretranslocational' ribosome that results after peptide bond formation and contains two tRNAs in the A and P sites in both ribosomal subunits. EF-G•GTP binding to the ribosome stabilizes the first (50S) step of translocation, transferring the acceptor ends of the two tRNAs to 50S P and E sites, whereas their anticodon ends remain anchored in 30S A and P sites (Moazed and Noller, 1989; Spiegel et al, 2007). 50S translocation is accompanied by rotation of the two ribosomal subunits, relative to one other (Frank and Agrawal, 2000; Valle et al, 2003; Agirrezabala et al, 2008). Through GTP hydrolysis, EF-G accelerates the second (30S) step of translocation, transferring the tRNA anticodons and their paired mRNA codons to the 30S P and E sites (Rodnina et al, 1997; Katunin et al, 2002; Wilson and Nechifor, 2004; Pan et al, 2007). GTP hydrolysis is thought to induce conformational changes within EF-G, which are propagated into the ribosome, and somehow ‘unlock' the ribosome (Spirin, 1985, 2009). These conformational changes lead to 30S translocation and Pi release (in random order), followed by EF-G and GDP release (in unknown order) from the ribosome (Savelsbergh et al, 2003). Off the ribosome, EF-G exchanges nucleotides rapidly, in contrast to EF-Tu (Wilden et al, 2006).
Many structural approaches have been taken to characterize conformational changes in bacterial EF-G proteins in various functional states. X-ray crystallography first showed that Thermus thermophilus EF-G is folded into five domains (I–V), organized into two supradomain units (I–II and III–V; Ævarsson et al, 1994; Czworkowski et al, 1994; Laurberg et al, 2000). From small-angle X-ray scattering techniques, the global conformations of free EF-G•GTP and EF-G•GDP could not be distinguished in solution (Czworkowski and Moore, 1997). This was corroborated by additional crystal structures of free EF-G, with various mutations and bound nucleotides, which showed similar conformations (Hansson et al, 2005a, 2005b). Only minor changes in sw2 were observed (involving rotations of amino-acid side chains), whereas sw1 was mostly invisible (disordered). Cryo-electron microscopy (cryo-EM) showed a very different conformation of EF-G•GDPNP on the ribosome, in which the two supradomains rotate as rigid bodies, relative to crystalline-free EF-G (Valle et al, 2003). Finally, two EF-G paralogs of T. thermophilus showed a striking resemblance on and off the ribosome (Connell et al, 2007): a cryo-EM structure of ribosome-bound EF-G•GDPNP, and a crystal structure of free EF-G-2•GDPNP. Both structures included an ordered sw1, adjacent to the bound GDPNP, similar to EF-Tu.
On the other hand, parallel structural studies on Saccharomyces cerevisiae eEF2, a eukaryotic homologue of bacterial EF-G, yielded somewhat different observations. Two crystal structures of free eEF2, with or without the fungicide sordarin, showed large conformational differences, involving rotations of three supradomain units (I–II, III, and IV–V; Jørgensen et al, 2003). Sw1 was invisible, whereas sw2 underwent more substantial movements. Cryo-EM structures of ribosome-bound eEF2, trapped with GDPNP or sordarin after GTP hydrolysis, adopted similar conformations, intermediate between the two crystal structures, with subtle changes attributed to GTP hydrolysis (Spahn et al, 2004; Taylor et al, 2007). Assuming a common catalytic cycle for EF-G and eEF2, the earlier studies raise a few key questions: What conformational changes occur in each factor as it cycles on and off the ribosome? When do they occur during the catalytic cycle? What functions do they serve?
Here, using a combination of biochemical and biophysical methods, we have probed the conformations of Escherichia coli EF-G trapped in six functional states, on and off the ribosome. These experiments led us to uncover a substantial movement of sw1, which we could interpret in the context of earlier structural studies. In particular, sw1 oscillates between ordered and disordered conformations, dependent on GTP or GDP, in both free and ribosome-bound EF-G. On EF-G•GTP binding to a pretranslocational ribosome, sw1 becomes structured and tucked inside the ribosome cavity, which locks the GTP substrate onto EF-G. After GTP hydrolysis and both steps of translocation, sw1 flips out from the cavity, assuming a disordered conformation, which greatly accelerates release of GDP and EF-G from the posttranslocational ribosome. On the basis of our findings, we present a model in which sw1 conformational changes play central roles in driving the unidirectional cycle of EF-G during protein synthesis.
Results
Six states of EF-G, free or ribosome-bound
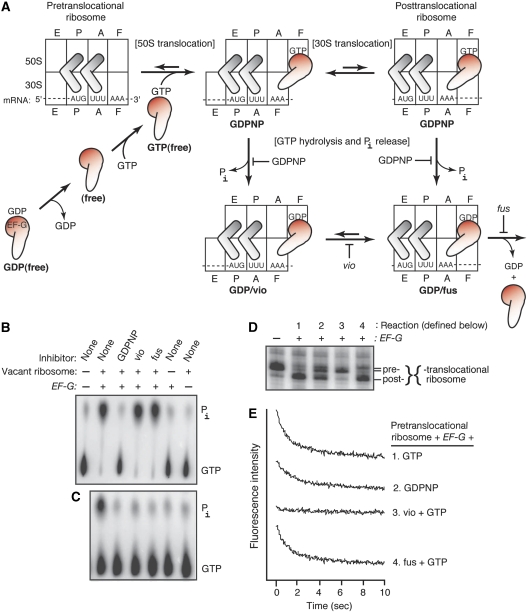
We began this study by defining six states of EF-G, stalled at different points in its catalytic cycle (Figure 1A). Three states of free EF-G were formed with GTP, GDP, or without nucleotide, which we call GTP(free), GDP(free), and (free), respectively. Three states of ribosome-bound EF-G were formed by binding EF-G to a pretranslocational ribosome, in the presence of GDPNP or GTP and antibiotic (viomycin or fusidic acid).
Figure 1.
Six states of EF-G, free or ribosome-bound. (A) The EF-G catalytic cycle. Boxes represent tRNA interaction sites (A, P, and E) on the 30S and 50S subunits of the ribosome. The EF-G interaction sites are designated by F. Rotation of the ribosomal subunits is represented by the shift in boxes. The six trapped states are highlighted by the red EF-G cartoons and are identified by bold lettering. (B, C) GTP hydrolysis assays. Panel B: single-turnover conditions: EF-G (0.8 μM), GTP (0.5 μM, with trace radioactive [γ32-P]GTP), and vacant ribosome (1.0 μM). Panel C: multiple-turnover conditions: EF-G (0.04 μM), GTP (50 μM), and vacant ribosome (0.5 μM). Inhibitors: GDPNP (100 μM), vio (250 μM), or fus (200 μM). Reactions (37°C, 20 min) were quenched with formic acid (30%) and resolved by thin layer chromatography (Nechifor and Wilson, 2007). (D) Extent of ribosome translocation, monitored by the toeprinting assay (Wilson and Nechifor, 2004). (E) Kinetics of ribosome translocation, measured by using 3′-pyrene-mRNA (Studer et al, 2003; Nechifor et al, 2007). Fluorescence traces (offset for clarity) monitor translocation kinetics. In Panels D and E, reactions contained: pretranslocational ribosome (0.5 μM; see Materials and methods), EF-G (1.0 μM), and GTP (1 mM). Inhibitors were GDPNP (1 mM), vio (0.5 mM), or fus (1 mM).
The latter complexes are abbreviated as GDPNP, GDP/vio, and GDP/fus, respectively. They were characterized in several assays for GTP hydrolysis, ribosomal translocation, and EF-G binding to the ribosome. First, we examined GTP hydrolysis under single- and multiple-round conditions; that is stoichiometric and excess GTP, relative to EF-G (Figure 1B and C). As expected, we observed that ribosome-activated GTP hydrolysis by EF-G was competitively inhibited by GDPNP. Fusidic acid (fus) and viomycin (vio) had no effect on GTP hydrolysis under single (but not multiple) turnover conditions, suggesting that both antibiotics trap EF-G on the ribosome after hydrolysing a single GTP molecule. Second, we monitored 30S translocation by two complementary assays. The extent of 30S translocation was determined by toeprinting, which tracks ribosome movement along a defined mRNA (Figure 1D). The kinetics of 30S translocation was followed by using mRNA 3′-labelled with pyrene, whose fluorescence becomes quenched on its entry into the ribosome (Figure 1E). EF-G•GTP catalysed rapid and efficient translocation, whereas EF-G•GDPNP catalysed partial translocation, as we reported earlier (Wilson and Nechifor, 2004). However, GDPNP slowed the kinetics by only three-fold, a much smaller effect than reported earlier (Rodnina et al, 1997; Katunin et al, 2002) but in agreement with another study (Pan et al, 2007). Fus did not change the kinetics, but slightly inhibited the extent, of translocation. Vio completely blocked translocation in both assays. Third, we assessed binding of EF-G to the ribosome by filtration, which separated free and ribosome-bound EF-G (Materials and methods). The GDPNP and GDP/fus complexes were both very stable. The GDP/vio complex was less stable, but it still could be purified for probing experiments (described below).
Our results, taken in conjunction with earlier studies, suggest that GTP hydrolysis and translocation are partially coupled mechanisms, as diagrammed in Figure 1A. In particular, our GDPNP complex represents a heterogeneous translocation state and presumably a homogeneous GTP hydrolysis state (Wilson and Nechifor, 2004). Earlier studies have shown that EF-G•GDPNP stabilizes the rotated conformation of the ribosome associated with 50S translocation (Valle et al, 2003; Spiegel et al, 2007). Our GDP/vio complex is trapped homogeneously after GTP hydrolysis and before 30S translocation. Earlier studies have shown that vio allows GTP hydrolysis, 50S translocation, and ribosomal subunit rotation, but prevents 30S translocation (Rodnina et al, 1997; Ermolenko et al, 2007). Vio is believed to bind across the ribosomal subunit interface, near the 30S A site (Johansen et al, 2006). Finally, our GDP/fus complex is trapped relatively homogeneously after GTP hydrolysis and after 30S translocation (Wilson and Nechifor, 2004). Fus targets ribosome-bound EF-G•GDP, possibly binding in a crevice that opens between domains G, III, and V (Laurberg et al, 2000).
Trypsin preferentially cleaves sw1 in EF-GKGDP, free or ribosome-bound
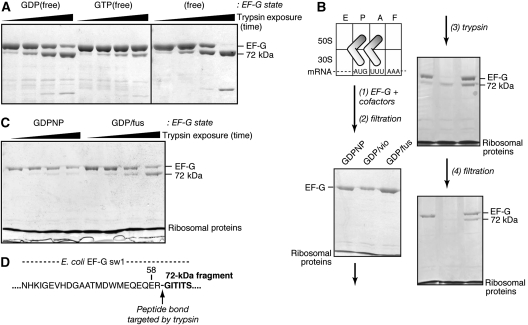
As an initial probe of EF-G conformation in solution, we exposed the three free states of EF-G to a small amount of the protease trypsin, and monitored the appearance of cleaved fragments of EF-G over time (Figure 2A). A prominent fragment with a molecular mass of ∼72 kDa appeared rapidly, whereas other smaller fragments could be detected after the majority of intact EF-G had disappeared. In the GDP(free) and (free) states, the 72-kDa fragment was formed about equally rapidly, whereas in the GTP(free) state this fragment was formed significantly more slowly. An early study observed the same 72-kDa fragment (Skar et al, 1975), but that study was restricted to the (free) state and it did not identify the site in EF-G targeted by trypsin that generated this fragment.
Figure 2.
Trypsin preferentially cleaves sw1 in EF-G•GDP, free or ribosome-bound. (A) Free EF-G states, probed with trypsin. Complexes were formed with EF-G (2 μM) containing GDP (0.5 mM), GTP (0.5 mM), or no nucleotide. Trypsin (4.5 μg/ml) was added, and reactions were incubated (20°C). Samples of each reaction were removed and denatured (after 1, 4, 16, and 64 min), and analysed by SDS–PAGE (Materials and methods). (B) Stability of ribosome-bound EF-G complexes. Step (1): EF-G (1.5 μM) was bound to a pretranslocational ribosome (1.5 μM) in the GDPNP, GDP/fus, or GDP/vio states (Materials and methods). Step (2): free EF-G was removed from ribosome-bound EF-G by filtration (Materials and methods). Step (3): complexes were treated with trypsin (4.5 μg/ml; 20°C; 10 min). Step (4): reactions were quenched with trypsin inhibitor (340 μg/ml), and the digested complexes were re-purified by filtration. (C) Ribosome-bound EF-G states, probed with trypsin. Complexes were formed with EF-G (1.7 μM) bound to a pretranslocational ribosome (2.0 μM) in GDPNP, GDP/vio, and GDP/fus states (Figure 1). After removing free EF-G, complexes were probed with trypsin and analysed by SDS–PAGE as in panel A. (D) Peptide bond cleaved by trypsin in the sequence of sw1 of E. coli EF-G, deduced from the N-terminal sequence of the 72-kDa fragment (Supplementary Table 1).
Next, we examined the three ribosome-bound states of EF-G. To ensure that we probed only ribosome-bound EF-G, we formed the complexes with a substoichiometric amount of EF-G, relative to the ribosome, and purified the resulting complexes from any remaining free EF-G by filtration (Figure 2B). After exposing the purified complexes to trypsin, we observed the same 72-kDa fragment. The relative portion of this fragment in the three complexes was different, in the order GDPNP<GDP/fus<GDP/vio. To assess whether trypsin indeed cleaved ribosome-bound EF-G, rather than EF-G that may have dissociated from the ribosome, we again filtered the three trypsin-cleaved samples. The gel analysis showed that the 72-kDa fragment remained in only the GDP/fus sample. The GDPNP sample contained only intact EF-G, whereas no EF-G was detected in the GDP/vio sample, consistent with the latter's lower stability.
We repeated the experiments on the two more stable complexes, and examined the formation rate of the 72-kDa fragment (Figure 2C). Strikingly, this fragment was formed very slowly in the purified GDPNP complex. By contrast, it was formed much faster in the GDP/fus complex, at a similar rate as in the GDP(free) state. As a control, we analysed the proteins of the ribosome on a higher percentage gel, which indicated that the ribosome remained intact during the time the complexes were exposed to trypsin (data not shown).
We isolated the 72-kDa fragment from the gel containing the digested GDP/fus sample, and determined its N-terminal amino-acid sequence by Edman degradation (Supplementary Table S1). The sequence we obtained, GITITS…, identified the peptide bond linking Arg59 and Gly60 of E. coli EF-G, which was cleaved by trypsin (Figure 2D). This site is located in the C-terminal portion of sw1, whose sequence is most highly conserved among the translational GTPases (Supplementary Figure S1). The same site of cleavage was identified in eEF2, but was purportedly similarly exposed to trypsin in its free states and protected in its ribosome-bound GDPCP state (Nilsson and Nygard, 1991). Trypsin also targets the conserved Arg in EF-Tu, as well as nonconserved Arg and Lys residues in N-terminal part of sw1 (Wittinghofer et al, 1980).
With this information, we modelled the possible interactions of trypsin with ribosome-bound EF-G, by using a cryo-EM structure of T. thermophilus EF-G in the GDPNP state (Connell et al, 2007). In this structure, sw1 is tucked inside the ribosomal cavity, and its electron density approximately fits the α-helix of sw1 in the crystal structure of T. thermophilus EF-G-2 in the GTP(free) state. We could plausibly dock a trypsin molecule onto the entrance to the cavity between the ribosomal subunits, just below the bound EF-G (Supplementary Figure S2). However, in this position the active site of trypsin cannot access the scissile peptide bond in the α-helix of sw1. This modelling, together with the above results, suggested that a conformational change in EF-G and/or the ribosome is required for trypsin to have access to sw1 in the GDP/fus state.
Sw1 moves relative to ribosome-bound EF-G, after GTP hydrolysis
As sw1 in the GDPNP state is situated just inside the ribosome cavity, between the ribosomal subunits, we wondered whether trypsin might have access to sw1 as a consequence of rotation of the ribosomal subunits (Frank and Agrawal, 2000). By modelling the counterclockwise rotation of the subunits (as viewed from the 30S subunit), we imagined that the region around sw1 might open up sufficiently for trypsin attack. Earlier studies have correlated subunit rotation with peptide bond formation (Valle et al, 2003) and 50S translocation (Ermolenko et al, 2007; Cornish et al, 2008). The former removes the peptide chain from tRNA in the P/P state, and the latter moves the resulting deacylated tRNA into the P/E state (Moazed and Noller, 1989). EF-G binding in the GDPNP or GDP/fus state stabilizes both subunit rotation and 50S translocation (Valle et al, 2003; Spiegel et al, 2007).
Thus, we could interpret the differential accessibility of sw1 in two ways: (i) sw1 may remain stationary and become accessible to trypsin after the ribosomal subunits have rotated; or (ii) sw1 may itself move from a protected location inside the ribosome cavity to the exterior where sw1 could be easily attacked by trypsin. To distinguish these possibilities, we assembled ribosome complexes in the unrotated or rotated conformation. Rotation was monitored indirectly by its associated 50S translocation. EF-G was bound to both ribosome conformers in the GDP/fus state (Zavialov and Ehrenberg, 2003). The complexes were probed again with trypsin, to ask whether sw1 accessibility depends on GTP hydrolysis and/or subunit rotation.
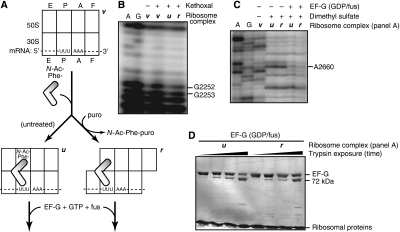
First, we bound N-acetyl-Phe-tRNAPhe (peptidyl-tRNA analogue) to the P site of a ribosome that contained mRNA and a vacant A site (Figure 3A). The resulting ribosome complex was divided into two parts: one was left untreated, whereas the other was treated with puromycin (aminoacyl-tRNA analogue), forming N-acetyl-Phe-puromycin (released from the ribosome) and deacylated tRNAPhe. By chemically probing tRNA interactions in the 50S P site, we confirmed that the former ribosome contained tRNA in the P/P state, whereas the latter ribosome underwent 50S translocation, corresponding to tRNA movement to the P/E state (Figure 3B). Then, we bound EF-G to both ribosomes in the GDP/fus state. Likewise, chemical probing experiments confirmed that EF-G was bound to both ribosomes (Figure 3C; Moazed et al, 1988). Finally, the two EF-G-containing ribosomes were purified by filtration, and probed with trypsin. In both ribosomes, sw1 was equally sensitive to trypsin (Figure 3D). Taken together, these results favour the interpretation (ii, above) that GTP hydrolysis induces a conformational change in sw1, making it more accessible to cleavage by trypsin in the GDP/fus state, rather than an indirect effect of subunit rotation.
Figure 3.
Sw1 moves relative to ribosome-bound EF-G, after GTP hydrolysis. (A) Formation of ribosome complexes. N-acetyl-Phe-tRNAPhe (2.4 μM) was bound to the P site of a vacant (v) ribosome (2.0 μM), containing T4 gene 32 mRNA. The resulting complex was split into two equal volumes: one was left untreated; the other was treated with puromycin (1 mM; 20°C, 30 min; 37°C, 8 m), forming N-acetyl-Phe-puromycin (released from the ribosome) and deacylated tRNAPhe (remaining on the ribosome). Before treatment, the ribosomal subunits are expected to adopt their unrotated (u) conformation (Valle et al, 2003), and the tRNA binds in the P/P state (Moazed and Noller, 1989). After treatment, the subunits can convert to their rotated (r) conformation, and the deacylated tRNAPhe can move to its P/E state. (B) Chemical probing of tRNA movement from P/P to P/E states in the ribosome. The three ribosomes (v, u, and r; panel A) were probed with kethoxal, which chemically modifies G2252 and G2253 of 23S rRNA in the 50S P site. Their modification was monitored by primer extension (Stern et al, 1988). tRNA occupation of the P/P state and movement to the P/E state are indicated by the relative protection and exposure (respectively) of G2252 and G2253 (Samaha et al, 1995). (C) Chemical probing of EF-G binding to the ribosome. EF-G (1.7 μM) was bound to complexes u and r in the GDP/fus state. The resulting complexes were purified from free EF-G by filtration. EF-G interaction with the 50S F site was probed with dimethyl sulfate, which chemically modifies nucleotide A2660 in 23S rRNA (Moazed et al, 1988). EF-G binding is indicated by the protection of A2660. (D) Enzymatic probing of EF-G. EF-G-containing ribosome complexes were exposed to trypsin, and analysed by SDS–PAGE (as in Figure 2C).
Sw1 flips out from the ribosome cavity, after GTP hydrolysis and 30S translocation
To examine sw1 movements more specifically, we chose a chemical probe, bromoacetamidobenzyl-EDTA•iron(II) (abbreviated as FeBABE), which can be covalently attached to the thiol group of a cysteine residue (Cys), engineered at a desired location in a Cys-free version of EF-G (Wilson and Noller, 1998). After binding EF-G to the ribosome, FeBABE generates diffusible hydroxyl radicals that cause strand cleavages in the rRNA components of the ribosome. The short-lived hydroxyl radicals are generally restricted to a localized sphere (radius<25 Å from the iron atom), which can vary somewhat by the microenvironment where FeBABE is attached (Meares et al, 2003).
We engineered Cys at residue 58 of E. coli EF-G, a partially conserved residue just upstream from the peptide bond cleaved by trypsin (Figure 2C). The 58C-FeBABE protein was bound to the ribosome in the GDP/fus state. The 16S and 23S rRNAs (of the 30S and 50S subunits, respectively) were extracted and scanned for strand cleavages by primer extension analysis. In preliminary experiments, we detected very strong cleavages in helix 95 (Sarcin-Ricin loop) of 23S rRNA. The rRNA cleavages from 58C-FeBABE were similar to those observed earlier from 196C-FeBABE (Wilson and Noller, 1998). This was unexpected, because residues 58 and 196 are separated by ∼40 Å in free EF-G-2•GDPNP, and suggested that the two residues approach each other in the GDP/fus complex.
We therefore focused our attention on comparing two proteins (58C-FeBABE and 196C-FeBABE) in the three ribosome-bound states. These proteins catalysed complete GTP hydrolysis that was activated by the ribosome, strongly inhibited by GDPNP, unaffected by vio, and slightly inhibited by fus (Supplementary Figure S3A). With GTP, they promoted efficient 30S translocation that was partially inhibited by GDPNP, completely inhibited by vio, and slightly inhibited by fus (Supplementary Figure S3B). Thus, they functioned similarly to wild-type EF-G (Figure 1B and D). They were bound to the ribosome in all three states, and the rRNAs were extracted and analysed for cleavages. As a negative control, we also analysed FeBABE-treated Cys-free EF-G in parallel.
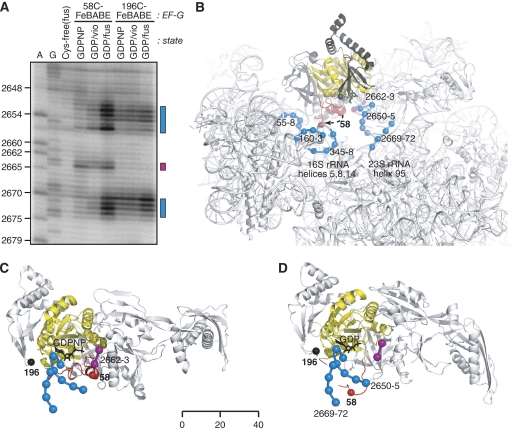
The primer extension gel in Figure 4A showed a striking difference in cleavage pattern for 58C-FeBABE. In the GDPNP state, this protein cleaved two nucleotides (2662, 2663) at the tip of helix 95 of 23S rRNA. In the GDP/vio state, the same two cleaved nucleotides were seen, along with weak cleavages in the stem of helix 95. In the GDP/fus state, the same two nucleotides were targeted again. In addition, in the latter state, this protein strongly cleaved nucleotides on both strands near the base of helix 95. In contrast, 196C-FeBABE generated cleavages in the stem of helix 95 that were similar in all three states, but somewhat stronger in GDP/fus. Further analysis showed additional cleavages by 58C-FeBABE in 16S rRNA helices 5, 8, and 14 (Supplementary Figure S4). These cleavages were consistently strongest in the GDP/fus state, weaker but still detected in the GDP/vio state, and undetected in the GDPNP state.
Figure 4.
Sw1 flips out from the ribosome cavity, after GTP hydrolysis and 30S translocatiuon. (A) Tracking sw1 movement with FeBABE. Three derivatives of E. coli EF-G (single Cys mutants 58C and 196C, and Cys-free) were treated with FeBABE (Supplementary data). Proteins 58C-FeBABE and 196C-FeBABE (2 μM) were bound to a pretranslocational ribosome (0.5 μM) in GDPNP, GDP/fus, and GDP/vio states (Figure 1; Materials and methods). The control FeBABE-treated Cys-free protein was bound in the GDP/fus state. All seven complexes were treated with hydrogen peroxide and ascorbate (Materials and methods) to generate localized hydroxyl radicals from the iron atom of the FeBABE probe (Wilson and Noller, 1998). After quenching radicals with thiourea, rRNAs were extracted from the complexes. Strand cleavages in 16S and 23S rRNA were analysed by primer extension (Stern et al, 1988; Wilson and Nechifor, 2004). Shown here is a primer extension analysis of the region including helix 95 of 23S rRNA. Lanes A, G: sequencing reactions. Numbers (left) refer to E. coli 23S rRNA nucleotides. Rectangles (right) identify strongly cleaved rRNA nucleotides, and are colour-coded as follows: magenta: 58C-FeBABE in GDPNP and GDP/vio states; blue: 58C-FeBABE in GDP/fus state. (B) rRNA nucleotides targeted by 58C-FeBABE. Magenta and blue spheres are centred on Pα atoms of cleaved rRNA nucleotides (panel A; Supplementary Figure 5), and are colour-coded as in panel A. Red and black spheres are centred on Cα atoms of EF-G residues 58 and 196, respectively. Sw1 and G domain are coloured red and yellow, respectively. The overall structural framework is based on T. thermophilus ribosome-bound EF-G•GDPNP (Connell et al, 2007). Superpositioned onto it are the structures of EF-G-2•GDPNP (Connell et al, 2007) and EF-G(G16V)•GDP (Hansson et al, 2005b). Arrow shows the movement of residue 58 between the superpositioned structures. (C) Sw1 conformation in the GDPNP state. The view is orthogonal to that of panel B. The ribosome is invisible except for nucleotides in helix 95 cleaved by 58C-FeBABE. (D) Sw1 conformation in the GDP/fus state.
We mapped the nucleotides cleaved by 58C-FeBABE onto the cryo-EM structure mentioned above (Connell et al, 2007). In this context, EF-G residue 58 is buried inside the ribosome cavity, near the γ-phosphate of GDPNP, whereas residue 196 is outside of the cavity. Residue 58 rests against helix 95, just below nucleotides 2662 and 2663 (∼16 and 9 Å away) cleaved in all three states (Figure 4B). In contrast, the nucleotides in the stem of helix 95, strongly cleaved in the GDP/fus state, are located outside of the ribosome cavity, up to 35 Å away from residue 58, but in close proximity to residue 196 (Figure 4C). These data are incompatible, from both steric and distance considerations; that is residue 58 is positioned behind helix 95, which would block hydroxyl radicals diffusing from this location to around the base of helix 95. In contrast, hydroxyl radicals would have a direct line of attack from residue 196. The cleavages in 16S rRNA are even further removed from residue 58 in the GDPNP state, but are directly across the cavity from the cleavages in the stem of helix 95.
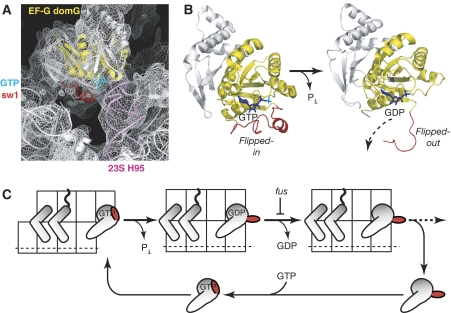
These data can be reconciled if residue 58 occupies an initial position inside the ribosome cavity before GTP hydrolysis and 30S translocation. After 30S translocation, residue 58 then moves out of the cavity in the vicinity of residue 196 after GTP hydrolysis. The movement implies that the entire sw1 element is initially tucked inside the cavity, and then flips out from the cavity in which it becomes disordered at a late stage in the EF-G cycle. The disordered, flipped-out sw1 would account for the dispersed cleavage in 16S and 23S rRNA, as well as the rapid cleavage by trypsin. Curiously, the external location of residue 58 agrees with a crystal structure of T. thermophilus EF-G(G16V)•GDP (Hansson et al, 2005b), whose sw1 was partially ordered and projected away from the G domain (Figure 4D).
Flipped-out sw1 correlates with accelerated release of GDP and EF-G from the ribosome
In its flipped-in conformation, sw1 wraps around the bound GDPNP and appears to ‘lock' the nucleotide between EF-G and helix 95 (Connell et al, 2007). Indeed, the ribosome strongly increases the binding equilibrium constant (kd) of EF-G for both GTP and GDP (Baca et al, 1976). For GDPNP in particular, kd increases by 260-fold, and the dissociation rate constant (kd) decreases by ∼104-fold (Wilden et al, 2006).
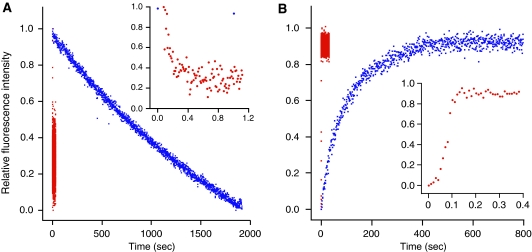
Thus, our finding that sw1 oscillates between flipped-in and flipped-out conformations suggests the possibility that sw1 may regulate nucleotide binding and release from EF-G on the ribosome. To address these issues, we measured rates of nucleotide release from ribosome-bound EF-G. We took advantage of fluorescent mant-nucleotides, which become substantially more fluorescent on binding to EF-G and the ribosome. We bound E. coli wild-type EF-G to the vacant ribosome with mant-GDPNP or mant-GDP, in the absence or presence of fus or vio. Each complex was rapidly mixed with excess unlabelled nucleotide, so that the irreversible dissociation rate of mant-nucleotide from the complex could be monitored by the fluorescence decrease over time. We used this simplified system because the experiments consumed larger quantities of materials, and earlier studies have established that the GTPase activity of EF-G does not depend on the presence of tRNA and mRNA in the ribosome (Nechifor et al, 2007). After formation of the complex with mant-GDPNP, the fluorescence intensity of the sample decreased slowly with time (Figure 5A), indicating that the bound mant-GDPNP was released gradually into solution (kd=0.0016 s−1). In contrast, mant-GDP dissociated very rapidly from ribosome-bound EF-G (7500-fold increase in kd; Table I). Both nucleotides dissociated from free EF-G at rates that were too fast to measure.
Figure 5.
Kinetics of release of nucleotide and EF-G from the ribosome. (A) Release of fluorescent nucleotide from ribosome-bound EF-G. Two complexes were formed with wild-type E. coli EF-G (2 μM), vacant ribosome (3 μM), and either mant-GDPNP (2 μM; blue dots) or mant-GDP (2 μM; red dots). These complexes were rapidly mixed with excess, unlabelled GDPNP or GDP (2.4 mM). See ‘Materials and methods'. The inserted graph focuses on the first second of the two reactions after mixing, in order to show more clearly the rapid release of mant-GDP. (B) Release of fluorescent EF-G from the ribosome. Two complexes were formed with EF-G(OG) (2 μM), vacant ribosome (3 μM), and either GDPNP (200 μM; blue dots) or GDP (200 μM; red dots). These complexes were rapidly mixed with excess, wild-type E. coli EF-G (20 μM). See ‘Materials and methods'. Likewise to panel A, the inserted graph focuses on the first 0.4 sec of the reactions.
Table 1.
Release of fluorescently labelled nucleotide and EF-G from functionally distinct ribosome complexesa
| Ribosome complex | Chase | Dissociation rate constants (kd sec−1)b | |
|---|---|---|---|
| mant-nucleotide | EF-G(OG) | ||
| mant-GDPNP•EF-G | GDPNP | 0.0016±0.0001 | |
| mant-GDP•EF-G | GDP | 12±2 | |
| mant-GDP•vio•EF-G | GDP | 0.80±0.09 | |
| mant-GDP•fus•EF-G | GDP | 0.016±0.005c | |
| mant-GDP•EF-G | GDP+fus | 0.019±0.002 | |
| GDPNP•EF-G(OG) | EF-G | 0.0093±0.0005 | |
| GDP•EF-G(OG) | EF-G | 8±2 | |
| GDP•vio•EF-G(OG) | EF-G | 0.56±0.15 | |
| GDP•fus•EF-G(OG) | EF-G | 0.043±0.001 | |
| GDP•EF-G(OG) | EF-G+fus | 0.42±0.05 | |
aSee Materials and methods.
bValues represent the averages (±s.d.), determined from time-resolved fluorescence measurements (Figure 5; Supplementary Figure S6) from three to five independent reactions.
cBiphasic kinetics (rate constant represents the fast phase).
These observations stimulated us to wonder how the bound nucleotide may affect EF-G release from the ribosome. To monitor EF-G release, we attached fluorescent Oregon Green (OG) to EF-G(426C), chosen because its fluorescence was significantly quenched on its binding to the ribosome (B Nguyen, unpublished data). The fluorescent EF-G(OG) protein was bound to the ribosome with unlabelled GDPNP, and the resulting complex was rapidly mixed with excess unlabelled EF-G. The fluorescent intensity of the mixture increased exponentially with time, reaching a plateau after 400 s, indicating EF-G(OG) release from the complex, with kd=0.0093 s−1 (Figure 5B). With GDP in the complex instead, EF-G(OG) release was accelerated by 860-fold. Without nucleotide, the fluorescence did not change, indicating that EF-G(OG) did not bind to the ribosome (data not shown).
Thus, both nucleotide and EF-G release from the ribosome strongly correlate to the predicted effects of sw1 conformational changes. Release of mant-GDP and EF-G(OG), with GDP in the complex, occurred at similar rates. The 5.8-fold difference between kd values for mant-GDPNP and EF-G(OG), with GDPNP in the complex, may be attributed to the stabilizing effect of the mant group (∼three-fold; Wilden et al, 2006). We should note, however, that we could not monitor release of both nucleotide and EF-G in the same experiment, because of technical issues of fluorescence interference between the mant and OG probes.
Finally, we examined effects of fus and vio (Supplementary Figure S5). Fluorescent complexes were formed with fus or vio already bound, and chased with unlabelled nucleotide competitor. Fus strongly suppressed mant-GDP release, whereas vio had more modest effect (750- and 15-fold, respectively). Despite these stabilizing effects, mant-GDP was released considerably faster from the fus- and vio-containing complexes, relative to mant-GDPNP (22- and 500-fold faster, respectively). Similarly, fus suppressed EF-G(OG) release much more strongly than vio (186- and 14-fold, respectively). To investigate the fus effects further, complexes were formed without fus, and were rapidly mixed with fus and unlabelled competitor. Remarkably, fus could still exert its strong effects, even when it was present initially in solution, suggesting that fus can rapidly bind to the ribosome before nucleotide or EF-G dissociate. Despite sw1 being in its flipped-out conformation in the GDP/fus complex, fus suppresses GDP release and thereby stabilizes EF-G on the ribosome.
Discussion
Structural dynamics of sw1 in EF-G and EF-Tu
GTPases share a common G domain with a universal ‘spring-loaded' switch mechanism, in which their switch elements are pulled together through hydrogen bonds from the γ-phosphate of GTP to conserved Thr (in sw1) and Gly (in sw2) residues. Consequently, their GTP-bound states are similar, whereas their GDP-bound states relax into different conformations after Pi release, as originally observed in Ras proteins (Vetter and Wittinghofer, 2001).
This principle can be illustrated by comparing crystal structures of EF-Tu and EF-G, off the ribosome (Supplementary Figure S6). In particular, sw1 adopts similar helical conformations in EF-Tu•GDPNP and EF-G-2•GDPNP (Berchtold et al, 1993; Connell et al, 2007). In EF-Tu•GDP, sw1 converts to β-hairpin structure, which projects into the interdomain core of EF-Tu (Abel et al, 1996). In EF-G-GDP structures, sw1 is usually disordered. However, in the structure of T. thermophilus EF-G(G16V)•GDP, sw1 was partially ordered and was found to be directed towards the periphery of the G domain of EF-G (Hansson et al, 2005b). This unusual sw1 orientation was puzzling and attributed to intermolecular crystal contacts. Interestingly, if one superpositions EF-G(G16V)•GDP onto T. thermophilus ribosome-bound EF-G•GDPNP, the location of residue 58 is consistent with the flipped-out sw1 conformation, which we have characterized. Thus, these comparisons suggest that, similar to Ras, sw1 adopts dissimilar conformations in EF-G•GDP and EF-Tu•GDP.
Further insights into sw1 dynamics can be gleaned by comparing our findings to several recent cryo-EM studies. We have made extensive use of the work of Connell et al (2007) who, examining T. thermophilus EF-G, localized its sw1 to a pocket below its G domain and between rRNA helices 14 and 95 of the 30S and 50S subunits, respectively. Their work focused exclusively on the GDPNP complex and relied on the unusually well ordered sw1 in their accompanying crystal structure of T. thermophilus EF-G-2. An independent study by Taylor et al (2007) examined S. cerevisiae eEF2, which was ADP-ribosylated and trapped on the 80S ribosome in GDPNP and GDP/sordarin states. They observed electron density, attributed to sw1, which was present in the former state and disappeared in the latter. These results suggested that sw1 had become disordered after GTP hydrolysis. In a follow-up study, Sengupta et al (2008) examined ribosome-bound eEF2•GDP•AlF4−, a complex designed to mimic the transition state of the hydrolysis reaction as Pi is being severed from GTP. They found electron density, unique to this complex, near the shoulder of the small ribosomal subunit, which was assigned to a relocated sw1 helix.
Two very recent cryo-EM studies examined E. coli EF-Tu stalled on the 70S ribosome in the GDP/kirromycin state (Schuette et al, 2009; Villa et al, 2009). At their higher resolutions (6–7 Å), α-helices of EF-Tu could be discerned, whereas sw1 remained fragmented (partially disordered) and seen making contact with 16S rRNA nucleotides at the junction of its helices 8 and 14. The orientation of sw1 appears distinct from either EF-Tu•GDPNP or EF-Tu•GDP crystal structures. Interestingly, the same nucleotides were cleaved by FeBABE attached to sw1 of EF-G in the GDP/fus state, as well as other nucleotides more exterior to the ribosome cavity (Figure 4B). These comparisons suggest that the sw1 elements of EF-Tu and EF-G may relax into similarly disordered, flipped-out conformations after GTP hydrolysis on the ribosome. However, genetic swapping experiments indicated that their sw1 elements are not functionally interchangeable (Kolesnikov and Gudkov, 2002).
Functional roles of sw1 in driving the EF-G cycle
EF-Tu displays many typical properties of GTPases: EF-Tu•GTP binds to the ribosome (its effector and GTPase activating particle); GTP hydrolysis comes after its regulatory function (codon–anticodon recognition) on the ribosome; EF-Tu•GDP dissociates from the ribosome, and it requires EF-Ts (its GDP/GTP exchange factor; or GEF) off the ribosome. EF-G is similar to EF-Tu in some respects, but unusual in others: its ribosome-independent GTP hydrolysis is unusually high; its GTP hydrolysis comes before (and is independent of) its regulatory function (translocation); and it does not require a GEF.
The collective properties of EF-G have inspired a long history of ideas on the roles of its GTP hydrolysis and, by implication, conformational changes in its switches. Two (nonexclusive) mechanisms have generally been considered: (i) GTP hydrolysis reduces EF-G affinity for the ribosome (Kaziro, 1978; Spirin, 1985, 2009) through altered contacts between its switches and ribosomal components; (ii) GTP hydrolysis mechanically drives translocation (Lipmann, 1969; Rodnina et al, 1999) through rotation of EF-G supradomains, induced by the switches at their interface (Frank et al, 2007). As sw1 flips out from the ribosome cavity, our results argue against sw1 playing either of these roles. Whether sw2 fulfils either role remain open questions.
Rather, we have shown that the flipped-out conformation of sw1 correlates with a greatly accelerated dissociation of GDP, relative to GDPNP, from ribosome-bound EF-G. This correlation suggests that sw1 regulates GTP binding to, and GDP release from, EF-G during its catalytic cycle. Our data agree with results of related experiments indicating that GDPNP is released slowly and inhibits EF-G turnover from the ribosome (Kaziro, 1978; Katunin et al, 2002; Wilden et al, 2006).
We propose the following scenario of events (Figure 6). On EF-G•GTP binding to a pretranslocational ribosome, its sw1 becomes tucked inside the ribosome cavity, through spring-loaded interactions of its conserved Thr62 with the γ-phosphate of GTP (Supplementary Figure S6), which locks the GTP substrate between the G domain and rRNA helix 95. GTP hydrolysis leads to Pi release and sw1 flips away from the G domain and out from the ribosome cavity. In its flipped-out conformation, sw1 provides an exit pathway for the GDP product.
Figure 6.
Model of sw1 conformational changes, driving the EF-G catalytic cycle. (A) Sw1 in ribosome-bound EF-G locks GTP in the ribosome. Structure is based on T. thermophilis EF-G•GDPNP•ribosome (Connell et al, 2007). Its electron density (mesh, contoured at 3σ) and underlying molecular model (ribbon) are shown in white. Highlighted elements in this model include sw1 (red), GDPNP (cyan), G domain of EF-G (yellow), and helix 95 of 23S rRNA (pink). (B) Flipped-out sw1 permits the escape of GDP and Pi after GTP hydrolysis. Represented here is ribosome-bound EF-G, before and after GTP hydrolysis. The view is orthogonal to that in panel A, and only domain I of EF-G is shown for clarity. Two structures are superpositioned onto ribosome-bound EF-G: Left structure (before GTP hydrolysis) is T. thermophilus EF-G-2-GDPNP (Connell et al, 2007); right structure (after hydrolysis) is E. coli EF-G(G16V)•GDP (Hansson et al, 2005b). Colour scheme is described in panel A, except that GDP here is dark blue and Pi is cyan. (C) Proposed mechanism for sw1 conformational changes occurring within the EF-G cycle. The general format here follows Figure 1A. Proceeding left to right, the diagram starts with EF-G bound to the ribosome, after subunit rotation and before GTP hydrolysis. The squiggly line represents the nascent polypeptide chain attached to tRNA in the A/P hybrid state. Sw1 is represented by the red flap. Sw1 flips out from the ribosome after GTP hydrolysis (centre diagram). The diagram ends with release of EF-G from the ribosome, and the cycle is completed with a fresh GTP substrate binding to EF-G off the ribosome.
We further suggest that GDP release from ribosome-bound EF-G may facilitate rapid release of EF-G from the ribosome. Our reasoning stems from several observations. First, we have observed that fus suppresses release of both mant-GDP and EF-G(OG) from the ribosome. These effects could arise if fus inhibits release of binary EF-G•GDP complex, or stepwise slow release of GDP followed rapidly by EF-G. Second, we have measured a large acceleration in EF-G(OG) release, comparing complexes with GDPNP or GDP (860-fold difference). In contrast, release rates of EF-G(OG) and mant-GDP were similar (6-fold difference). Third, kinetic experiments under saturating nucleotide conditions indicated that EF-G•GTP and EF-G•GDP bind to the ribosome with similar and relatively high affinities (KM=0.22 and 0.15 μM, respectively; Baca et al, 1976). EF-G turnover from the ribosome is inhibited by both GDPNP and GDP (Katunin et al, 2002). Fourth, we have observed that EF-G(OG) lacking nucleotide does not bind to the ribosome, in agreement with an early study indicating that EF-G alone has a much lower affinity for the ribosome (kd≈50 μM; Lin and Bodley, 1976). An EF-G•ribosome complex forms at higher concentrations (>10 μM), but represents a dead-end that inhibits turnover in GTP hydrolysis (Rohrback and Bodley, 1976; Nechifor et al, 2007).
Alternatively, EF-G•GDP may dissociate from the ribosome, as a consequence of broken interactions between the ribosome and sw1. This possibility seems less likely, because we observed that fus does not prevent sw1 from flipping out of the ribosome, whereas fus strongly stabilizes EF-G•GDP on the ribosome and suppresses GDP release. Moreover, we found that fus can efficiently exert its stabilizing effects, even when it is present in solution and rapidly mixed with EF-G•GDP•ribosome complexes. Although fusR mutations in EF-G imply that fus binds to an interdomain crevice in EF-G (Laurberg et al, 2000), fus does not bind to free EF-G but binds specifically to ribosome-bound EF-G after GTP hydrolysis (Willie et al, 1975). Thus, our results suggest that fus can rapidly diffuse into the ribosome cavity and access its binding site in EF-G, through the opening created by the flipped-out sw1.
To complete the cycle in Figure 6, we propose that soon after EF-G comes off the ribosome, it quickly binds a fresh GTP from the cellular pool (since GTP predominates over GDP in healthy cells), which re-closes the sw1 flap, and facilitates EF-G•GTP binding to another pretranslocational ribosome. This phase of the cycle is supported by our trypsin probing experiments showing that sw1 is cleaved more rapidly in its GDP(free) and (free) states, relative to its GTP(free) state. Finally, our results suggest that sw1 retains its dynamic character, on and off the ribosome, according to both trypsin and FeBABE probing experiments. In particular, sw1 in the GTP(free) and GDPNP states was weakly cleaved by trypsin, suggesting that sw1 exits mainly in its flipped-in conformation but flips out occasionally. Conversely, experiments with 58C-FeBABE showed that sw1 in the GDP/fus state exists mainly in its flipped-out conformation but flips into the ribosome cavity occasionally.
Coupling of sw1 movements to ribosomal translocation
Apart from the spring-loaded mechanism that is intrinsic to GTPases, a conformational ‘feedback' exchange may occur between EF-G and the ribosome. Conformational changes in EF-G triggered by GTP hydrolysis may be propagated into the ribosome to unlock the translocation process (Spirin, 1985, 2009). In turn, conformational changes in the ribosome may feedback to control when EF-G is released from the ribosome. In particular, the flipped-out conformation of sw1 may be triggered not only by GTP hydrolysis, but also may be sterically prevented by the ribosome until both steps of translocation are completed. This feedback mechanism would ensure an efficient coupling between the events of GTP hydrolysis on the G domain of EF-G, and the events of translocation on the ribosome.
Our FeBABE probing experiments provide support for such a coupling mechanism by showing that sw1 remains tucked into the ribosome cavity in the GDPNP state (before GTP hydrolysis and partial 30S translocation) or in the GDP/vio state (after GTP hydrolysis and before 30S translocation). The flipped-out sw1 was only observed after both GTP hydrolysis and both steps of translocation (GDP/fus state). On the other hand, our trypsin probing experiments seem to contradict the coupling mechanism. The flipped-out sw1 was still observed in GDP/fus complexes in which 50S translocation was artificially impeded by N-acetyl-Phe-tRNAPhe in the P site. Although this experiment was designed presuming a strict requirement of deacylated P-site tRNA for ribosomal subunit rotation (Valle et al, 2003), recent studies suggest that 50S translocation and subunit rotation can be decoupled (Cornish et al, 2008; Marshall et al, 2008). After the subunits have rotated, sw1 appears unobstructed by the ribosome and could sample its flipped-out conformation (Figure 6A). EF-G stabilizes 50S translocation and subunit rotation, independent of GTP hydrolysis (Valle et al, 2003; Spiegel et al, 2007). These discrepancies suggest that sw1 movements may be controlled not only by GTP hydrolysis and subunit rotation, but also by some other event linked to 30S translocation.
Conclusions
This study defines the movements of sw1, a central regulatory element in EF-G and other GTPases, in the context of the unidirectional EF-G cycle. Our observations rest on six functionally distinct states of EF-G, trapped on and off the ribosome, which presumably approximate the transient intermediate species in the uninhibited cycle. The approaches we have used for tracking sw1 movements complement earlier structural investigations of free and ribosome-bound EF-G. By functionally characterizing the trapped complexes, and structurally probing sw1 conformation in parallel, we have been able to correlate movements of sw1 with respect to specific events in the EF-G cycle.
We have combined the new structural and functional information into a model, which highlights the central role of sw1 in driving the EF-G cycle. In this model, we envision a two-state conformational oscillation in sw1 that accelerates several steps in the cycle: binding of EF-G•GTP to the ribosome, GTP hydrolysis, release of GDP and EF-G from the ribosome, and nucleotide exchange by free EF-G. Our model raises new questions, which motivate future experimentation to elucidate the full functional significance of sw1 at each step. The spotlight we have focused onto sw1 of EF-G in this study should, of course, be viewed also in the broader perspective of the multilayered, dynamic, and highly coordinated process of cellular protein synthesis.
Materials and methods
Materials
Materials are listed in Supplementary data. Assays described below were conducted in aqueous buffer A (80 mM HEPES-KOH pH 7.7, 50 mM NH4Cl, 10 mM MgCl2, 1 mM DTT).
Enzymatic probing of EF-G
The free states of EF-G (20 μl) were formed by mixing wild-type E. coli EF-G (2.0 μM), without or with nucleotide (GTP or GDP; 0.5 mM), followed by incubation (20°C, 10 min). Trypsin (4.5 μg/ml) was added, and further incubated (20°C). Samples (2 μl) were removed at different time intervals (see Figure 2), denatured in sodium dodecyl sulfate (SDS, 1%; 90°C; 5 min), and resolved by SDS polyacrylamide gel electrophoresis (SDS–PAGE). Protein bands in the gels were stained with Coomassie.
The ribosome-bound states of EF-G were formed with a pretranslocational ribosome (60 μl), assembled as follows. Uncharged E. coli tRNAfMet (2.4 μM) was bound to the 30S P site of the E. coli 70S ribosome (2.0 μM), containing mRNA (5′…AUG UUU…3′; 2.4 μM) derived from phage T4 gene 32. After incubation (37°C, 30 min), uncharged E. coli tRNAPhe (2.4 μM) was bound to the 30S A site, and further incubated (37°C, 30 min). The stepwise assembly of this complex was monitored by toeprinting (Wilson and Nechifor, 2004).
The pretranslocational ribosome was divided into three equal volumes. The GDP/vio state was formed by adding vio (250 μM; 20°C, 10 m), followed by EF-G•GTP (1.7 μM; 0.5 mM). The GDPNP state was formed with EF-G•GDPNP (1.7 μM; 0.5 mM). The GDP/fus state was formed with EF-G•GTP (1.7 μM; 0.5 mM) and fus (0.5 mM). After EF-G addition, all reactions were incubated (37°C, 15 m; 0°C, 5 m). Free EF-G was removed from ribosome-bound EF-G by centrifugal filtration (Millipore Microcon YM-100): reactions were diluted in 500 μl buffer A (supplemented, as appropriate, with 20 μM fus or 100 μM vio) at 0°C, and re-concentrated to ∼10 μl. The ribosome-bound EF-G complexes were probed with trypsin and analysed by SDS–PAGE (as above).
Chemical probing of the ribosome
Modification of the ribosome with dimethyl sulfate and kethoxal, and primer extension analysis of the extracted rRNAs, were performed as described (Stern et al, 1988).
Hydroxyl radical probing experiments were conducted as described (Wilson and Noller, 1998), with minor modifications. EF-G(FeBABE) (2 μM; Supplementary data) was bound to a pretranslocational ribosome (0.5 μM) in three states (defined above). Hydroxyl radicals were generated from the attached FeBABE by adding H2O2 (0.05%) and ascorbic acid (5 mM). Reactions were incubated (0°C; 10 min), quenched with thiourea (100 mM), and ethanol precipitated. rRNAs were extracted and analysed by primer extension. Specific experiments are described in Figures 3 and 4.
Time-resolved fluorescence
Experiments were conducted by using a QM-6 fluorimeter (Photon Technology International), equipped with a stopped-flow device (MiniMixer, KinTek) connected in-line to a cuvette in the flourimeter's sample chamber.
To measure nucleotide release, ribosome-bound EF-G complexes (1.6 ml) were formed with mant-GDPNP or mant-GDP (2 μM), wild-type EF-G (2 μM), vacant ribosome (3 μM), and antibiotic (fus or vio; 100 μM). Complexes were transferred to syringe A of the stopped-flow device and into syringe B was loaded chase nucleotide (2.4 mM unlabelled GDPNP or GDP; with 100 μM fus in some experiments). Samples (∼200 μl) from each syringe were rapidly mixed together (deadtime: ∼3.5 msec) and injected into the fluorimeter's cuvette. Fluorescence (excitation: 362 nm; emission: 444 nm) was monitored over 50–1800 sec, with measurements taken every 10–1000 msec, depending on the reaction rate. Data were fitted to a single exponential decay equation: Ft=F0+A × exp(−kd × t), where F0 and Ft are the fluorescence intensities at time 0 and t, A is the amplitude of the fluorescent change, and kd is the dissociation rate constant.
To measure EF-G release, EF-G mutant 426C was conjugated with OG fluorescent probe (see Supplementary data). This EF-G(OG) protein (2 μM) was bound to the vacant ribosome (3 μM) with nucleotides (GDPNP or GDP; 200 μM) and antibiotic (fus or vio; 100 μM). Complexes were transferred to syringe A and into syringe B was loaded EF-G (20 μM unlabelled E. coli wild-type protein, with 100 μM fus in some experiments). After mixing, fluorescence (excitation: 492 nm; emission: 518 nm) was monitored (as above). Data were fitted to a single exponential rise equation: Ft=F0+A × [1–exp(−kd × t)].
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
This work was funded by a grant from Canadian Institutes of Health Research. We thank Julian Davies for viomycin, and Léa Brakier-Gingras and Richard Fahlman for discussions and comments on the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abel K, Yoder MD, Hilgenfeld R, Jurnak F (1996) An alpha to beta conformational switch in EF-Tu. Structure 4: 1153–1159 [DOI] [PubMed] [Google Scholar]
- Ævarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J 13: 3669–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J (2008) Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell 32: 190–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca OG, Rohrbach MS, Bodley JW (1976) Equilibrium measurements of the interactions of guanine nucleotides with Escherichia coli elongation factor G and the ribosome. Biochemistry 15: 4570–4574 [DOI] [PubMed] [Google Scholar]
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365: 126–132 [DOI] [PubMed] [Google Scholar]
- Connell SR, Takemoto C, Wilson DN, Wang H, Murayama K, Terada T, Shirouzu M, Rost M, Schuler M, Giesebrecht J, Dabrowski M, Mielke T, Fucini P, Yokoyama S, Spahn CM (2007) Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell 25: 751–764 [DOI] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T (2008) Spontaneous intersubunit rotation in single ribosomes. Mol Cell 30: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czworkowski J, Moore PB (1997) The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry 36: 10327–10334 [DOI] [PubMed] [Google Scholar]
- Czworkowski J, Wang J, Steitz TA, Moore PB (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J 13: 3661–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Spiegel PC, Majumdar ZK, Hickerson RP, Clegg RM, Noller HF (2007) The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol 14: 493–497 [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Frank J, Gao H, Sengupta J, Gao N, Taylor DJ (2007) The process of mRNA–tRNA translocation. Proc Natl Acad Sci USA 104: 19671–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Wieden HJ, Rodnina MV (2002) Kinetic mechanism of elongation factor Ts-catalyzed nucleotide exchange in elongation factor Tu. Biochemistry 41: 162–169 [DOI] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT (2005a) Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett 579: 4492–4497 [DOI] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT (2005b) Structural insights into fusidic acid resistance and sensitivity in EF-G. J Mol Biol 348: 939–949 [DOI] [PubMed] [Google Scholar]
- Johansen SK, Maus CE, Plikaytis BB, Douthwaite S (2006) Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol Cell 23: 173–182 [DOI] [PubMed] [Google Scholar]
- Jørgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 10: 379–385 [DOI] [PubMed] [Google Scholar]
- Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W (2002) Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry 41: 12806–12812 [DOI] [PubMed] [Google Scholar]
- Kaziro Y (1978) The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta 505: 95–127 [DOI] [PubMed] [Google Scholar]
- Kolesnikov A, Gudkov AT (2002) Elongation factor G with effector loop from elongation factor Tu is inactive in translocation. FEBS Lett 514: 67–69 [DOI] [PubMed] [Google Scholar]
- Kothe U, Wieden HJ, Mohr D, Rodnina MV (2004) Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol 336: 1011–1021 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Kristensen O, Martemyanov K, Gudkov AT, Nagaev I, Hughes D, Liljas A (2000) Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol 303: 593–603 [DOI] [PubMed] [Google Scholar]
- Lin L, Bodley JW (1976) Binding interactions between radiolabeled Escherichia coli elongation factor G and the ribosome. J Biol Chem 251: 1795–1798 [PubMed] [Google Scholar]
- Lipmann F (1969) Polypeptide chain elongation in protein biosynthesis. Science 164: 1024–1031 [DOI] [PubMed] [Google Scholar]
- Marshall RA, Dorywalska M, Puglisi JD (2008) Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA 105: 15364–15369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meares CF, Datwyler SA, Schmidt BD, Owens J, Ishihama A (2003) Principles and methods of affinity cleavage in studying transcription. Methods Enzymol 371: 82–106 [DOI] [PubMed] [Google Scholar]
- Mesters JR, Potapov AP, de Graaf JM, Kraal B (1994) Synergism between the GTPase activities of EF-Tu•GTP and EF-G•GTP on empty ribosomes. J Mol Biol 242: 644–654 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF (1988) Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334: 362–364 [DOI] [PubMed] [Google Scholar]
- Mohr D, Wintermeyer W, Rodnina MV (2002) GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry 41: 12520–12528 [DOI] [PubMed] [Google Scholar]
- Nechifor R, Murataliev M, Wilson KS (2007) Functional interactions between the G' subdomain of bacterial translation factor EF-G and ribosomal protein L7/L12. J Biol Chem 282: 36998–37005 [DOI] [PubMed] [Google Scholar]
- Nechifor R, Wilson KS (2007) Crosslinking of translation factor EF-G to proteins of the bacterial ribosome before and after translocation. J Mol Biol 368: 1412–1425 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Nygard O (1991) Altered sensitivity of eukaryotic elongation factor 2 for trypsin after phosphorylation and ribosomal binding. J Biol Chem 266: 10578–10582 [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Ann Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Pan D, Kirillov SV, Cooperman BS (2007) Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell 25: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Wintermeyer W (1999) Dynamics of translation on the ribosome: molecular mechanics of translocation. FEMS Microbiol Rev 23: 317–333 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Ann Rev Biochem 70: 415–435 [DOI] [PubMed] [Google Scholar]
- Rohrback MS, Bodley JW (1976) Steady state kinetic analysis of the mechanism of guanosine triphosphate hydrolysis catalyzed by Escherichia coli elongation factor G and the ribosome. Biochemistry 15: 4565–4569 [DOI] [PubMed] [Google Scholar]
- Samaha RR, Green R, Noller HF (1995) A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377: 309–314 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W (2003) An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 11: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Kjeldgaard M, Nissen P, Frank J (2008) Visualization of the eEF2-80S ribosome transition-state complex by cryo-electron microscopy. J Mol Biol 382: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K (2009) Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol 4: 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skar DC, Rohrbach MS, Bodley JW (1975) Limited trypsinolysis of native Escherichia coli elongation factor G. Biochemistry 14: 3922–3926 [DOI] [PubMed] [Google Scholar]
- Sørensen MA, Pedersen S (1991) Absolute in vivo translation rates of individual codons in Escherichia coli. J Mol Biol 222: 265–280 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jørgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF (2007) Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA 13: 1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS (1985) Ribosomal translocation: facts and models. Prog Nucleic Acid Res Mol Biol 32: 75–114 [DOI] [PubMed] [Google Scholar]
- Spirin AS (2009) The ribosome as a conveying thermal ratchet machine. J Biol Chem 284 (in press; doi:10.1074/jbc.X109.001552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S, Moazed D, Noller HF (1988) Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol 164: 481–489 [DOI] [PubMed] [Google Scholar]
- Studer SM, Feinberg JS, Joseph S (2003) Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol 327: 369–381 [DOI] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J (2007) Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J 26: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, Frank J (2009) Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA 106: 1063–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W (2006) Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA 103: 13670–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willie GR, Richman N, Godtfredsen WP, Bodley JW (1975) Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome-elongation factor G complexes. Biochemistry 14: 1713–1718 [DOI] [PubMed] [Google Scholar]
- Wilson KS, Nechifor R (2004) Interactions of translational factor EF-G with the bacterial ribosome before and after mRNA translocation. J Mol Biol 337: 15–30 [DOI] [PubMed] [Google Scholar]
- Wilson KS, Noller HF (1998) Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell 92: 131–139 [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Frank R, Leberman R (1980) Composition and properties of trypsin-cleaved elongation factor Tu. Eur J Biochem 108: 423–431 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M (2003) Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell 114: 113–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File