Abstract
Vascular smooth muscle cells (SMCs) play an important role in vascular remodeling. Heterogeneity and phenotypic changes in SMCs are usually accompanied by a morphological difference, i.e., elongated/spindle-like versus spread-out or epithelioid/rhomboid cell shapes. However, it is not known whether the cell shape directly regulates SMC proliferation, and what the underlying mechanisms are. In this study, microgrooves and micropatterned matrix islands were used to engineer the cell shape and investigate the associated biophysical and biological mechanisms. Compared to spread-out SMCs on nonpatterned surfaces, SMCs on micropatterned surfaces demonstrated elongated morphology, significantly lower cell and nucleus shape indexes, less spreading, a lower proliferation rate, and a similar response (but to a lesser extent) to platelet-derived growth factor, transforming growth factor-β, and mechanical stretching. DNA microarray profiling revealed a lower expression of neuron-derived orphan receptor-1 (NOR-1) in elongated SMCs. Knocking down NOR-1 suppressed DNA synthesis in SMCs, suggesting that NOR-1 is a mediator of cell elongation effects. Regulation of DNA synthesis in SMCs by the cell shape alone and a decrease in DNA synthesis in the case of small cell spreading area were achieved by micropatterning SMCs on matrix islands of different shapes and spreading areas. Changes in the cell shape also affected the nucleus shape, whereas variations in the cell spreading area modulated the nucleus volume, indicating a possible link between nucleus morphology (both shape and volume) and DNA synthesis. The findings of this investigation provide insight into cell shape effects on cell structure and proliferation, and have direct implications for vascular pathophysiology.
Introduction
It is well known that vascular smooth muscle cells (SMCs) play an important role in vascular remodeling and disease development. In atherosclerotic lesions, SMCs migrate from the vascular wall into the lumen, demonstrating a phenotypic change from contractile to proliferative. It has been reported that in vitro isolation of SMCs from three-dimensional (3D) extracellular matrix (ECM), where the cells exhibit elongated, spindle-shape morphology, and subculture in culture dishes promotes a spread-out morphology and proliferative phenotype (1–3). Distinct SMC populations with different cell shapes (spindle or epithelioid/rhomboid) and phenotypes (differentiated or proliferative) have also been isolated from arteries (4). However, it is not known whether the cell shape directly affects SMC proliferation. In this study, in vitro culture systems were used to examine the effects of SMC morphology on SMC proliferation.
Micropatterning technology offers powerful tools to manipulate the microenvironment for cell growth, migration, and differentiation (5,6). Recent studies have shown that micropatterned matrix strips and microgrooves restrict SMC spreading in one direction, resulting in a more elongated SMC morphology and lower proliferation rate (7–9). However, the underlying mechanisms require further investigation. Although the micropatterning methods used in these studies provide an effective means of manipulating the SMC morphology for biochemical and biological analyses, changes in the SMC morphology are usually accompanied by changes in the spreading area. The cell spreading area has been observed to regulate cell proliferation in many cell types (10–13). To distinguish the effects of the cell shape and spreading area, micropatterned islands of well-defined shape and area were used in this study to evaluate cell shape effects on SMC proliferation.
Soluble chemical factors in the vascular system are important modulators of SMC functions. Platelet-derived growth factor (PDGF) and transforming growth factor-β (TGF-β) are two important growth factors that regulate SMC proliferation and differentiation. PDGF is a known mitogen of SMCs that yields the synthetic or proliferative phenotype, whereas TGF-β induces the differentiated phenotype of SMCs and upregulates the SMC markers (1,14). In addition to soluble factors, SMCs in artery walls are constantly subjected to cyclic mechanical stretching due to the pulsatile nature of the blood flow. Accumulating evidence indicates that mechanical strain regulates the vascular SMC phenotype, functions, and matrix remodeling (15). However, the effects of changes in the SMC morphology on the cell response to these growth factors and mechanical stretching are not known.
In this investigation, micropatterning techniques were used to engineer the shape of SMCs to study the effects of changes in the cell shape on DNA synthesis in SMCs in the absence and presence of growth factors and mechanical strain. Gene expression and gene knockdown analyses revealed that the neuron-derived orphan receptor-1 (NOR-1) is a mediator of the cell elongation effect on SMC proliferation. Through precise control of the cell shape and spreading area, the decrease in the cell shape index (CSI) alone was found to correlate with the decrease in SMC proliferation. A relationship was obtained between the cell shape and spreading area and the nucleus shape and volume. The results illustrate significant cell shape effects on SMC structure and proliferation, and have important implications for vascular pathophysiology.
Materials and Methods
Cell micropatterning by microgrooves and microstamps
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO), Polysciences (Warrington, PA), and Pierce Biotechnology (Rockford, IL) unless otherwise specified. Micropatterned poly(dimethylsiloxane) (PDMS) membranes and microstamps were fabricated using soft-lithography techniques with minor modifications (7). To fabricate molds with microgrooves (see Fig. S1 in the Supporting Material), an ∼2.8-μm-thick layer of I-Line positive photoresist (PR; OCG OiR 897-10i) was deposited on primed single-crystal Si wafers by spin coating. Microgrooves were patterned on the PR-coated wafers by exposure to UV light through a photomask with 10-μm-wide strips. The PR was developed to form a mold with a micropattern. To construct elastic membranes with microgrooves, PDMS was poured onto a Si wafer with the micropatterned mold, which was then spun to form a 250-μm-thick PDMS membrane. After curing, the micropatterned PDMS membrane was peeled off from the mold. Nonpatterned PDMS membranes were fabricated by the same method, except that the Si molds were blank Si wafers. Before the PDMS membranes were coated with 2% bovine gelatin (Sigma-Aldrich) under exposure to UV light, the surface hydrophilicity was increased by means of an oxygen plasma treatment. Subsequently, the cells were seeded in culture media on both micropatterned and nonpatterned surfaces.
A 2.8-μm-thick layer of PR and a patterned array of holes with a designed shape were used to fabricate microstamps (Fig. S2). This microstamping process was used to fabricate PDMS stamps with posts of the desired shape.
To obtain nonfouling surfaces for microstamping of matrix proteins, an ultrathin poly(acrylic acid-co-ethylene glycol) p(AA-co-EG) coating was formed on aminosilane slides, followed by covalent immobilization of di-amino-polyethylene glycol (PEG) molecules. PEG 1000 monomethyl ether (0.02 g/mL; Polysciences), N,N-methylene bisacrylamide (0.01 g/mL; (Polysciences), photoinitiator [3-(3,4-dimethyl-9-oxo-9H-thioxanthen-2-yloxy)-2-hydroxypropyl]trimethylammonium chloride (0.01 g/mL; Sigma-Aldrich) and acrylic acid (0.1 mL/mL; Polysciences) were dissolved in a 1:1 mixture of isopropyl alcohol (Sigma-Aldrich) and distilled, deionized water (ddH2O). Aminosilane slides (Sigma-Aldrich) were coated with the solution (200 μL/slide), incubated in the dark for 6 min, and then UV-treated for 2.5 min using a UVB40 UV transilluminator (Ultra-Lum, Claremont, CA). The slides were then thoroughly washed to remove unreacted monomers. The p(AA-co-EG) slides were then incubated with di-amino-PEG (0.15 g/mL; Sigma-Aldrich), [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride] (0.06 g/mL; Pierce), and N-hydroxysulfosuccinimide (0.02 g/mL; Pierce) dissolved in 0.5 M 2-morpholinoethanesulfonic acid (MES) buffer (pH 7.2; Pierce) for 1 h (0.3 mL/slide).
PDMS stamps were inked with fibronectin (0.33 mg/mL ddH2O) for 30 min while the di-amino-PEG slides were simultaneously incubated in the dark with a heterobifunctional cross-linker N-sulfosuccinimidyl-6-[4′-azido-2′-nitrophenylamino] hexanoate) (sulfo-SANPAH) (1 mg/mL in HEPES buffer, pH 7.5, 0.5 mL per slide). The stamps and slides were washed with ddH2O, dried, and placed in contact with each other for 30 min. The PDMS stamps were then peeled off from the slides. Fibronectin was cross-linked to the surface by UV activation of the sulfo-SANPAH nitrophenyl azide groups and the incubation in the phosphate-buffered saline (PBS) for 15 min. Finally, the unreacted sulfo-SANPAH molecules were quenched by immersing the slides in 10% glycine in PBS solution. An alternative method for single-cell micropatterning was achieved by combining the grafting of nonfouling PEG-like films and surface chemical patterning with a shadow mask. Details of this surface chemical micropatterning process can be found elsewhere (16).
Cell culture and treatment
Human aortic SMCs (Cascade, Portland, OR) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS), all obtained from Gibco-BRL (Grand Island, NY). Cell cultures were maintained in a humidified 95% air/5% CO2 incubator at 37°C. SMCs between passages 6 and 20 were used in the experiments. These cells expressed low levels of SMC contractile markers and had a proliferative phenotype. SMCs were seeded at ∼60% confluency (unless otherwise specified) and cultured on micropatterned and nonpatterned surfaces in DMEM supplemented with 1% FBS and 1% PS. For growth factor treatment, 10 ng/mL PDGF (Cell Signaling, Beverly, MA) or 10 ng/mL TGF-β (Sigma-Aldrich) was added to the culture medium. The concentration of the growth factors was selected based on information obtained from earlier studies. The mechanical device used to subject the PDMS membranes to uniaxial cyclic stretching was described previously (17). SMCs were seeded on PDMS membranes (with or without pattern) for 24 h and either kept as static controls or subjected to cyclic uniaxial mechanical stretching (1 Hz, 5% strain) for 24 h before performing a proliferation analysis.
Immunostaining, microscopy, and morphological analysis
Immunostaining and microscopy were performed as described previously (7). Briefly, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 1% bovine serum albumin (BSA). For cytoskeletal staining, samples were incubated in fluorescein isothiocyanate (FITC) or rhodamine-conjugated phalloidin for 30 min to stain all filamentous actin (F-actin). Nucleus staining was achieved with ToPro or DAPI dye (Molecular Probes, Eugene, OR). Fluorescence images were collected by a Leica TCL SL (Leica, Bannockburn, IL) confocal microscope system (40× objective) or a Zeiss LSM (Carl-Zeiss Microimaging, Thornwood, NY) confocal microscope (100× objective). Multiple Z-section images (∼0.3–0.4 μm thick sections) were obtained for each specimen. All of the images of a given group were collected using the same hardware and software settings. For 3D analysis, serial pictures of the nucleus were reconstructed into a 3D structure using the software Imaris (Bitplane, St. Paul, MN), which was then used to calculate the nucleus volume. For two-dimensional (2D) analysis, the optical sections were projected onto a single plane to construct an overall image of the specimen. The CSI and the nucleus shape index (NSI) were calculated from the 2D images of the cells. The cell boundaries were outlined with IMAGE software (Scion, Fredrick, MD). The spreading area (i.e., cell projected area) and the perimeter of the cell were measured, and the CSI was calculated from the relationship CSI = 4π · area/(perimeter)2. The CSI assumes values between one (circular shape) and zero (elongated, linear morphology). A similar relationship was used to calculate the NSI in terms of the measured nucleus projection area and nucleus perimeter.
Cell proliferation analysis
SMCs seeded on different surfaces and cultured for 24 h were incubated for 2 h with 10 μM 5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich). Then the samples were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated for 30 min in 2N HCl at 37°C. Subsequently, the samples were washed and incubated for 30 min in a buffer containing PBS, 0.05% Tween 20, and 1 mg/mL BSA to minimize background adsorption of antibodies. The samples were incubated overnight at 4°C with the primary antibodies against BrdU (PharMingen, San Diego, CA) and an FITC-anti-mouse antibody. After washing with PBS, the stained samples were counterstained with Hoechst nuclear dye (Sigma-Aldrich) for 5 min, mounted in VectaShield (Vector Laboratories, Burlingame, CA), and observed with a confocal microscope. The percentage of SMCs that incorporated BrdU (i.e., the cells with DNA synthesis) was correlated to the proliferation rate of SMCs.
Immunoblotting analysis
Immunoblotting analysis was performed with antibodies NOR-1, α-actin, and tubulin as described previously (17,18). The NOR-1 and tubulin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and the α-actin was obtained from Sigma-Aldrich. Briefly, cells were lysed in a lysis buffer containing 25 mM Tris, pH 7.4, 0.5 M NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 1 mM phenylmethylsulphonyl fluoride (PMSF), 10 μg/mL leupeptin, and 1 mM Na3VO4. Protein lysates were centrifuged, and the supernatant was removed and quantified using DC Protein Assay (Bio-Rad Laboratories, Hercules, CA). Protein samples were run in SDS polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were blocked with 3% nonfat milk and incubated with the primary antibody diluted in TBS-T buffer containing 25 mM Tris-HCl, pH 7.4, 60 mM NaCl, and 0.05% Tween 20. This was followed by incubation with either HRP-conjugated anti-mouse or anti-rabbit IgG secondary antibody (Santa Cruz Biotechnologies) as appropriate. Protein bands were visualized using the ECL detection system (Amersham Biosciences, Piscataway, NJ).
RNA isolation, DNA microarray analysis, and quantitative reverse transcription-polymerase chain reaction
Gene expression analysis was performed as described previously (17,19). Cells were lysed with RNA STAT-60 reagent (Tel-Test, Friendswood, TX). RNA was extracted with the use of chloroform and phenol extractions, and precipitated with isopropanol. The resulting RNA pellet was washed with 75% ethanol. For microarray analysis, the RNA pellet was resuspended in 12 μL diethyl pyrocarbonate-treated H2O and analyzed with a UV spectrophotometer to confirm a purity of A260/A280 ≥ 1.80. The RNA concentration was determined using the RiboGreen quantification assay (Molecular Probes, Eugene, OR). An amount of 3 μg of RNA from each sample was used in the DNA microarray experiments performed with a high-throughput array (HTA) system (Affymetrix, Santa Clara, CA). Human U133A2.0 chips with 22,944 probe-sets were used in this study. Data management and statistical analysis were performed with GeneTraffic Microarray Data Management and Analysis software (Stratagene, La Jolla, CA). The ratios of gene expression between micropatterned and nonpatterned control samples were calculated.
For quantitative reverse transcription-polymerase chain reaction (qRT-PCR), cDNA was synthesized by a two-step reverse-transcription with the ThermoScript RT-PCR system (Invitrogen, San Diego, CA), followed by qRT-PCR using SYBR-green reagent and the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Primers for the genes of interest were all designed using the ABI Prism Primer Express software v.2.0 (Applied Biosystems). The gene expression levels were normalized to 18 S levels from the same sample.
Knocking down NOR-1 gene expression
Short interference RNAs (siRNAs) were transfected into SMCs as described elsewhere (18). The siRNAs for NOR-1 or negative control were obtained from Dharmacon (Chicago, IL). Briefly, siRNAs (100 pmol/10 cm2 culture area) were transfected into ∼50–60% confluent SMCs using Lipofectamine 2000 reagent (Invitrogen) as recommended by the manufacturer. After 5-h incubation with the mixture of siRNA and Lipofectamine 2000, complete medium was added to the cells. The cells were cultured for 1–2 days before use in the experiments. FITC-conjugated control siRNAs (Cell Signaling Technology, Danvers, MA) were transfected into SMCs as siRNA transfection controls, which showed that siRNA transfection had >90% efficiency in SMCs. The effects of siRNA knockdown were verified by qRT-PCR.
Statistical analysis
Mean and standard deviation (SD) values were calculated for each group of data. Analysis of variance (ANOVA) was performed to detect whether a significant difference existed between groups of different treatments, and a multiple comparison procedure, the Holm t-test, was used to identify any differences. Student's t-test was used to analyze experimental groups with two samples.
Results
Proliferation rates of elongated cells
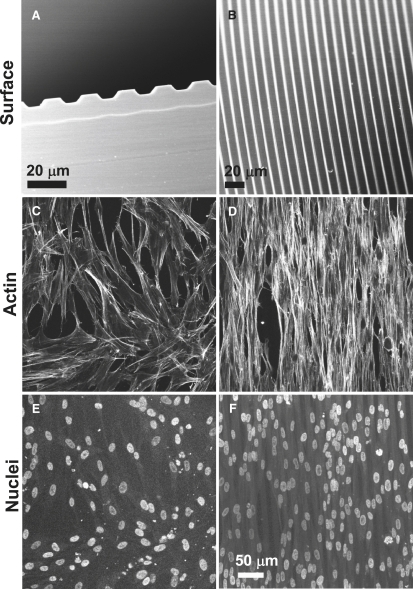
To manipulate the cell shape, soft lithography (Fig. S1) was used to fabricate parallel microgrooves (10 μm in width and 2.8 μm in depth) on elastic PDMS membranes, as shown in Fig. 1, A and B. Pilot studies suggested that a microgroove width less than the SMC size (<20 μm) and a microgroove depth of >2 μm were needed to effectively restrict cell spreading in the direction perpendicular to the microgrooves. SMCs on nonpatterned surfaces spread out, exhibiting fibroblastic morphology (Fig. 1 C). In contrast, SMCs on microgrooves demonstrated an elongated morphology (Fig. 1 D). The cells and actin filaments aligned uniformly in the direction of the grooves, mimicking the morphology and organization of SMCs in arteries. Of interest, the elongated cell morphology was accompanied by an elongated nucleus shape (Fig. 1 F).
Figure 1.
SEM images showing the effect of topographic micropatterning on SMCs on micropatterned and nonpatterned PDMS membranes (24-h culture). (A) Cross-sectional view of 10-μm-wide microgrooves. (B) Top view of microgrooves. (C and D) Phalloidin staining of actin stress fibers on (C) nonpatterned and (D) micropatterned surfaces (80–90% confluency). (E and F) ToPro nuclear staining on (E) nonpatterned and (F) micropatterned surfaces. (C–F, scale bar = 50 μm.)
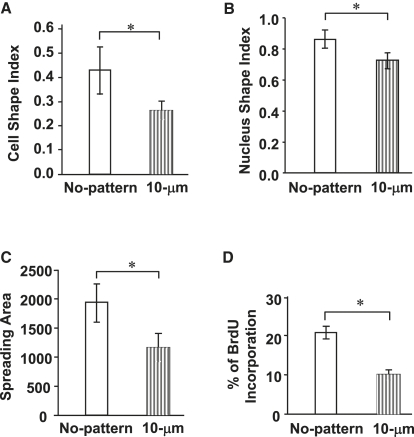
To further compare the morphological differences of SMCs cultured on nonpatterned and micropatterned surfaces, the cell shape, nucleus shape, and cell spreading area were evaluated in a 60–70% confluent culture of SMCs (Fig. 2). The 10-μm-wide microgrooves resulted in significantly lower CSI values (∼0.25) compared to the nonpatterned surface (∼0.45) (Fig. 2 A). Forcing the cells to spread unidirectionally also decreased the NSI from 0.87 (nonpatterned surface) to 0.75 (micropatterned surface) (Fig. 2 B), suggesting that the change in the cell shape also affected significantly the nucleus shape. In addition to the effects of micropatterning on the cell and nucleus shapes, the SMC spreading area decreased by ∼40%, from 1950 μm2 (nonpatterned surface) to 1150 μm2 (micropatterned surface) (Fig. 2 C). Therefore, in this culture system, the elongated SMC morphology was characterized by changes in both the shape and spreading area of the cells, and in the nucleus shape.
Figure 2.
Micropatterning effects on cell shape, nucleus shape, cell spreading area, and cell proliferation rate. SMCs were cultured on nonpatterned and micropatterned (10-μm-wide grooves) surfaces (24-h culture, unless otherwise specified). (A) CSI determined from fluorescent images of SMCs (50 cells per group; Fig. S3). (B) NSI determined from nucleus fluorescent staining images of SMCs (50 cells per group). (C) Spreading area of SMCs (50 cells per group). (D) SMC proliferation. After a 48-h culture, SMCs were incubated with BrdU for 2 h and then fixed and double-stained for BrdU and nuclei. The percentage of BrdU-positive cells (cells in S phase) was calculated using >500 cells per group. Bars represent mean ± SD. Data were extracted from at least three experiments. The asterisk indicates the statistically significant difference (p < 0.05) between specified groups.
To determine the effect of cell elongation on SMC proliferation, SMCs were cultured on micropatterned (10-μm-wide grooves) and nonpatterned surfaces for 48 h in 1% serum and then incubated with BrdU. The proliferation rate of SMCs (indicated by the percentage of SMCs with BrdU incorporation) grown on the micropatterned PDMS surfaces was found to be ∼50% less than that of the SMCs cultured on the nonpatterned PDMS surfaces (Fig. 2 D), suggesting that cell elongation modulated the SMC proliferation rate.
Effects of growth factors and mechanical stretching on cell proliferation
Since PDGF and TGF-β are known to have opposite effects on SMC proliferation, SMCs cultured on nonpatterned and micropatterned surfaces were treated with PDGF or TGF-β for 24 h to examine the effects of these growth factors on SMC proliferation. SMCs on micropatterned surfaces generally exhibited lower proliferation rates (Fig. S4 a). PDGF significantly enhanced cell proliferation in both spread-out and elongated SMCs, but the proliferation rate of the elongated cells was still much lower in the presence of PDGF, suggesting that elongated SMCs still responded to PDGF but to a less extent. In contrast to PDGF, TGF-β significantly suppressed cell proliferation in spread-out SMCs but had no significant effect on the elongated SMCs, possibly because of the already low proliferation rate of the SMCs cultured on the micropatterned surface.
SMCs in native blood vessel walls possess an elongated morphology; align in a circumferential direction, exhibiting a well-organized structure (20–22); and experience cyclic mechanical stretching mainly in the circumferential direction. Therefore, a cyclic (1 Hz) uniaxial strain of 5% was applied to SMCs in the direction of their alignment on the micropatterned and nonpatterned surfaces. Consistent with previous studies (17,23), SMCs grown on the nonpatterned surfaces aligned perpendicular to the direction of mechanical stretching and were more elongated, whereas SMCs grown on the micropatterned surfaces remained aligned in the stretching direction parallel to the microgrooves (data not shown here). Mechanical stretching suppressed SMC proliferation on both nonpatterned and micropatterned surfaces (Fig. S4 b). However, under the stretching condition, differences in SMC proliferation between nonpatterned and micropatterned surfaces were statistically insignificant.
Regulation of gene expression by cell elongation
A DNA microarray analysis was performed to investigate the effects of cell elongation on gene expression in SMCs. The focus was on genes that experienced more than a twofold change in expression after a 1-day culture of SMCs on micropatterned and nonpatterned surfaces. Only a small group of genes demonstrated more than twofold changes (five increased and two decreased, average of two experiments; Table S1). Most of the genes, including SMC contractile markers (α-actin, SM-22, calponin, and smooth muscle myosin heavy chain) and related transcriptional factors (serum response factor (SRF) and Kruppel-like factor 4 (KLF4)) did not exhibit more than a twofold change. Of interest, the expression of NOR-1 (NR4A3) (24) and the nuclear receptor subfamily 4 group A member 2 (NR4A2) in the SMCs cultured on the micropatterned (10-μm-wide grooves) surfaces showed more than a twofold decrease. The decrease in the gene and protein expression of NOR-1 in the elongated SMCs is supported by the fact that NOR-1 is a transcription factor that modulates SMC proliferation and plays a role in the acceleration of atherosclerosis (25).
NOR-1 as a mediator of the cell elongation effect on SMC proliferation
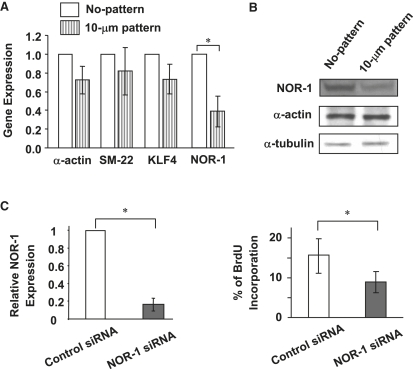
To verify the microarray data, qRT-PCR (Fig. 3 A) and immunoblotting (Fig. 3 B) were performed to examine the expression of NOR-1 and some SMC-related genes. The experiments revealed lower mRNA and protein levels of NOR-1 in elongated SMCs. However, the gene expression of the SMC contractile markers (e.g., α-actin and SM-22) and transcriptional factor KLF4 did not change significantly. These data are consistent with the DNA microarray results.
Figure 3.
Micropatterning effects on gene expression and role of NOR-1 in SMC proliferation. (A) Gene expression quantified by qRT-PCR experiments (n = 3, 24-h culture). (B) Immunoblotting analysis of NOR-1, α-actin, and tubulin expression (48-h culture). (C) NOR-1 knock-down effect on NOR-1 expression and SMC proliferation (>300 cells per group, three independent experiments, 48-h culture). Bars show mean ± SD. The asterisk indicates the statistically significant difference (p < 0.05) between specified groups.
A NOR-1 knockdown experiment with siRNA was performed to determine whether the decrease in NOR-1 expression was sufficient to suppress SMC proliferation (Fig. 3 C). It was observed that NOR-1 siRNA knocked down NOR-1 gene expression by 80% after 1 day. Two days after the silencing of the NOR-1 gene, there was significantly less SMC proliferation than in the negative control. These results suggest that NOR-1 can mediate the cell elongation effect on SMC proliferation.
Regulation of SMC proliferation by the cell shape and spreading area
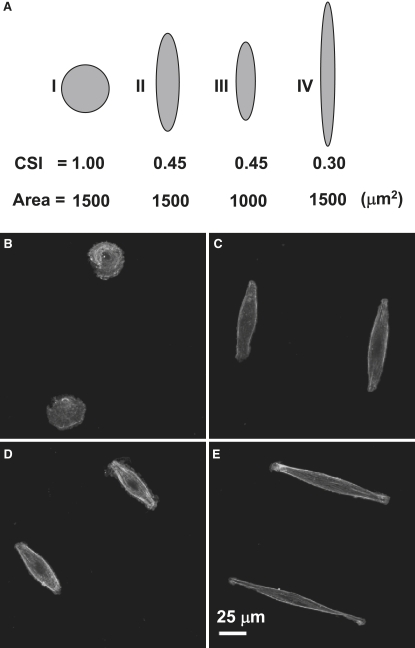
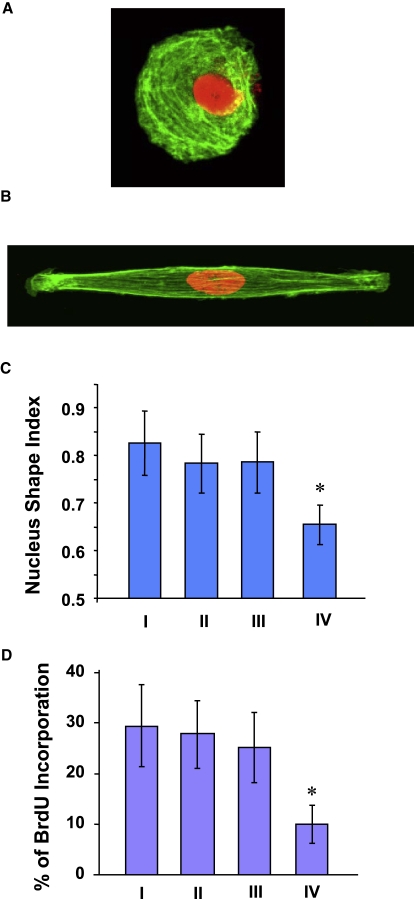
In view of the decrease of the CSI and the cell spreading area on the micropatterned surfaces (Fig. 2), experiments were carried out to further distinguish the effects of both the cell shape and the cell spreading area on SMC proliferation. Arrays of adhesion islands with different CSI values and spreading areas were created by a microstamping method (Fig. S2). Fig. 4 A shows a pictorial representation of CSI and spreading area combinations designed based on the cell morphological parameters shown in Fig. 2. The median spreading area (∼1500 μm2) of spread-out and elongated cells (Fig. 2 C) was selected in these experiments. To examine the cell shape effect, the spreading area was fixed at 1500 μm2 and three circular or oval shapes were fabricated with CSI equal to 1.0, 0.45, and 0.30 (cases I, II, and IV, respectively; Fig. 5 A). To study the effect of the cell spreading area, the CSI was set at 0.45 and the spreading area was decreased to 1000 μm2 (case III; Fig. 5 A). Actin staining showed that SMCs attached and conformed to microstamped adhesion islands (Fig. 4, B–E). All of the SMCs on the oval islands demonstrated elongated and bipolar shapes, except for the cells on the circular adhesion islands.
Figure 4.
Micropatterning effects on SMC shape. (A) Schematic representation of CSI versus spreading area combinations used in the experiments of this study. (B–E) Phalloidin staining of actin stress fibers for cases I–IV, respectively (24-h culture).
Figure 5.
Cell shape effects on SMC proliferation. (A) Actin (green) and nuclear (red) staining in an SMC micropatterned in a circular shape (24-h culture). (B) Actin (green) and nuclear (red) staining in an SMC micropatterned in a spindle-like shape (24-h culture). (C) NSI determined from nuclear fluorescent staining images of SMCs cultured on nonpatterned and micropatterned surfaces (>15 cells per group, 24-h culture). (D) BrdU incorporation for cases I–IV (24-h culture; Fig. 4A). Bars represent mean ± SD. The asterisk indicates the statistically significant difference (p < 0.05) compared to all other groups (>200 cells per group, three independent experiments).
Exemplified cells of circular and elongated shapes are shown in Fig. 5, A and B. The actin filaments in the circular SMCs preferentially lined up in the circumferential direction. In contrast, actin filaments aligned in the direction of the long axis in elongated SMCs. Similar to the effect of microgroove patterning, changes in the nucleus shape were observed in the elongated SMCs. A marked decrease in NSI was obtained with the decrease of the CSI to 0.30 (Fig. 5 C), an indication of a significant effect on the nucleus shape for this low CSI value.
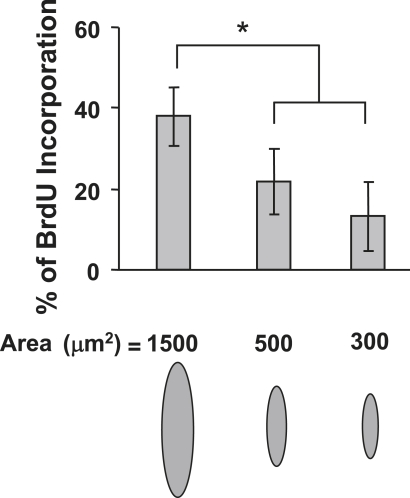
The effect of decreasing the CSI from 1.0 to 0.45 on the SMC proliferation was found to be secondary (Fig. 5 D). However, decreasing the CSI from 0.45 to 0.30 yielded a decrease in SMC proliferation by ∼60%, similar to that produced in the presence of microgrooves. Of interest, decreasing the cell spreading area from 1500 to 1000 μm2 while keeping the CSI fixed at ∼0.45 (cases II and III) did not produce an effect on the SMC proliferation rate. These results clearly demonstrate that the cell shape was responsible for the lower SMC proliferation rate when the spreading area was maintained constant (or within a range in which cell spreading was sufficient) and that SMC proliferation was sensitive to the decrease of the CSI below 0.45. Although the change in the spreading area from 1500 to 1000 μm2 did not produce a significant effect on SMC proliferation, restricting cell spreading over a much smaller area (e.g., 500 or 300 μm2) resulted in a profound decrease in SMC proliferation (Fig. 6). These results reveal the existence of a threshold for cell shape and spreading area effects on SMC proliferation.
Figure 6.
Cell spreading area effects on SMC proliferation (24-h culture). SMCs were cultured on micropatterned matrix islands of the same shape (CSI ≈ 0.45) and different spreading area (i.e., 1500, 500, and 300 μm2), and BrdU incorporation was subsequently analyzed (∼50 cells per group, three independent experiments). Bars represent mean ± SD. The asterisk indicates the statistically significant difference (p < 0.05) between specified groups.
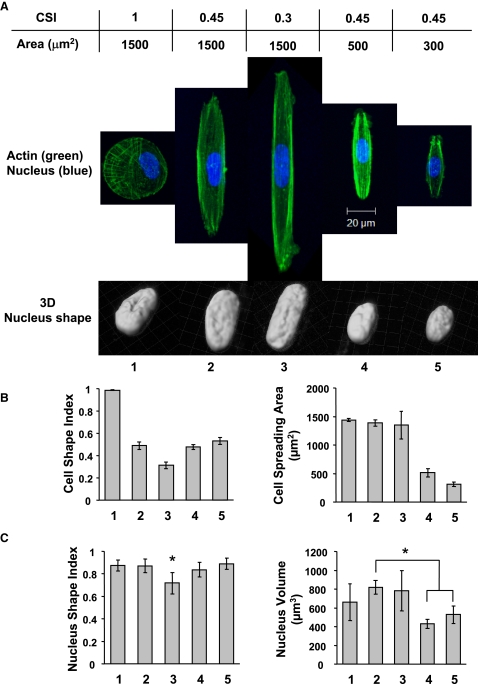
Regulation of the nucleus shape and volume by the cell shape and spreading area
It was postulated that the cell shape and spreading area modulate the nucleus morphology, affecting DNA synthesis and SMC proliferation. Three cases of cell spreading area equal to 1500 μm2 and different micropattern shapes, and three cases of CSI ≈ 0.45 and different spreading areas were examined to elucidate the dependence of the nucleus morphology on the cell shape and spreading area and SMC proliferation (Fig. 7 A). As evidenced from Fig. 7, A and B, the cells conformed to the designed shape of the spreading area with small deviations. Elongated cells (CSI ≈ 0.30) exhibited a more elongated nucleus with insignificant volume change; however, the decrease in the spreading area did not significantly affect the nucleus shape (Fig. 7 C), implying a greater sensitivity of the change in nucleus shape on the cell shape change. In contrast, the decrease in the cell spreading area produced a significant decrease in the nucleus volume, suggesting that the change of the nucleus volume is mostly regulated by the cell spreading area. Since both the cell shape and the cell spreading area regulate SMC proliferation, it is likely that both the shape and the volume of the nucleus affect DNA synthesis and, in turn, cell proliferation.
Figure 7.
Cell shape and spreading area effects on nucleus morphology (24-h culture). SMCs of specific shape and spreading area were subjected to fluorescence staining for actin filaments (green) and nucleus (blue). (A) Confocal microscopy images of actin filaments and 3D reconstruction of the nucleus shape. (B) Calculated CSI and cell spreading area (3–10 cells per group). (C) Calculated NSI and nucleus volume (3–10 cells per group). The asterisk indicates the statistically significant difference (p < 0.05) between specified groups or compared to all other groups.
Discussion
Microgrooves and micropatterned matrix islands were used to engineer the cell shape and examine how the cell shape and the cell spreading area regulate SMC proliferation. SMCs on microgrooved surfaces showed elongated morphologies and lower proliferation rates, and responded to PDGF to a lesser extent relative to spread-out SMCs on nonpatterned surfaces. These results have significant implications for vascular biology and pathophysiology. For example, they provide an explanation, at least in part, for the higher in vitro proliferation rates of spread-out SMCs compared to in vivo proliferation rates of elongated, spindle-shaped SMCs, and also shed some light on why spindle SMCs exhibit a differentiated phenotype whereas epithelioid/rhomboid SMCs are more proliferative (4). It is also worth noting that cell elongation does not contribute significantly to other SMC phenotypic changes, such as contractile marker expression and matrix synthesis.
DNA microarray and gene knocking-down analyses suggested that NOR-1 is a mediator of cell elongation, affecting SMC proliferation. NOR-1 has been identified as an early-response gene in SMCs and has a wide range of effects on SMCs, including advancing atherogenesis and modulating SMC proliferation (25–27). Several factors implicated in atherogenesis, such as PDGF, have been shown to upregulate NOR-1 in SMCs (25), which could explain why PDGF can partially recover the proliferation of elongated SMCs. However, it is not clear how SMC elongation regulates NOR-1 expression. A recent study has shown that intracellular signaling molecules, such as protein kinase C (PKC) and extracellular-regulated protein kinase (ERK), can modulate NOR-1 expression (27). The study presented here shows that phosphorylation of PKC but not ERK decreased in elongated SMCs, suggesting the involvement of a PKC pathway in the NOR-1 expression. Since microgrooves resulted in the decrease of both CSI and cell spreading area, the decrease in the NOR-1 expression cannot be attributed only to the effect of the cell shape change. Furthermore, the gene expression changes in the elongated SMCs were not extensive, implying that other mechanisms, such as posttranslational modification of proteins, could be regulated by changes in the cell shape and spreading area. These issues await further studies in the near future.
One of the most important findings of this investigation is that the cell shape directly regulates DNA synthesis and, in turn, SMC proliferation. This was achieved by micropatterning SMCs on matrix islands with different CSI values and the same cell spreading area, hence establishing an effective means of obtaining what to our knowledge is the first direct evidence of cell shape effects on cell proliferation. Since DNA synthesis decreased in the SMCs characterized by a CSI of ∼0.30 but not for ∼0.45, a threshold of the cell shape effect on proliferation may exist for CSI between 0.45 and 0.30. The results also revealed the existence of a threshold of the cell spreading effect on proliferation. Although a dependence of cell proliferation on the cell spreading area (varied in the range of 0–3000 μm2) has been reported for endothelial cells (12), the study presented here shows that SMC proliferation is independent of the variation of the cell spreading area in the range of 1000–1500 μm2, and decreases when cell spreading is confined within much smaller areas (e.g., 300 and 500 μm2). A plausible explanation for this behavior is that SMC proliferation is not affected when there is sufficient cell spreading (e.g., >1000 μm2) and decreases only when spreading is restricted to significantly smaller areas (e.g., 300–500 μm2). These differences in the spreading area dependence between endothelial cells and SMCs may be related to cell type differences.
Important insight into the threshold of shape and spreading effects on SMC proliferation was obtained by analyzing the nucleus morphology (shape and volume). The change in the nucleus shape was found to be sensitive to the CSI decrease to a low value (e.g., 0.30) and to changes in the cell shape, but not to changes in the cell spreading area. The change in the nucleus shape showed a good correlation with the decrease in DNA synthesis for CSI ≈ 0.30. This finding suggests that the change in the nucleus shape mediates the cell shape effect on DNA synthesis, and provides evidence for the existence of a CSI threshold for the nucleus shape effect. On the other hand, the change in the nucleus volume was sensitive to changes in the cell spreading area but not the cell shape. A significant decrease in the nucleus volume was encountered by confining cell spreading over small areas (e.g., 300 and 500 μm2), indicating a good correlation with the decrease in DNA synthesis in SMCs. These results provide strong evidence that changes in the nucleus morphology (either shape or volume) can modulate DNA synthesis and, in turn, SMC proliferation. It was recently shown that DNA synthesis in endothelial cells is affected by the nucleus volume but not the cell shape (28). It is possible that the change in cell shape in that study did not reach the threshold that would trigger changes in nucleus shape and DNA synthesis. Alternatively, this may be a cell-type-dependent property. It is evident that more in-depth studies must be performed to determine the relationship between changes in nucleus morphology and DNA synthesis, and to fully elucidate the underlying mechanisms.
Supporting Material
Four figures and a table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00488-3.
Supporting Material
Acknowledgments
This research was supported by the National Institutes of Health under grant HL 078534, and the National Science Foundation under grant CMS-0528506.
References
- 1.Owens G.K. Regulation of differentiation of vascular smooth muscle cells. Physiol. Rev. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 2.Thyberg J. Phenotypic modulation of smooth muscle cells during formation of neointimal thickenings following vascular injury. Histol. Histopathol. 1998;13:871–891. doi: 10.14670/HH-13.871. [DOI] [PubMed] [Google Scholar]
- 3.Dilley R.J., McGeachie J.K., Prendergast F.J. A review of the proliferative behaviour, morphology and phenotypes of vascular smooth muscle. Atherosclerosis. 1987;63:99–107. doi: 10.1016/0021-9150(87)90109-2. [DOI] [PubMed] [Google Scholar]
- 4.Hao H., Ropraz P., Verin V., Camenzind E., Geinoz A. Heterogeneity of smooth muscle cell populations cultured from pig coronary artery. Arterioscler. Thromb. Vasc. Biol. 2002;22:1093–1099. doi: 10.1161/01.atv.0000022407.91111.e4. [DOI] [PubMed] [Google Scholar]
- 5.Whitesides G.M., Ostuni E., Takayama S., Jiang X., Ingber D.E. Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 6.Tsang V.L., Bhatia S.N. Three-dimensional tissue fabrication. Adv. Drug Deliv. Rev. 2004;56:1635–1647. doi: 10.1016/j.addr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Thakar R.G., Ho F., Huang N.F., Liepmann D., Li S. Regulation of vascular smooth muscle cells by micropatterning. Biochem. Biophys. Res. Commun. 2003;307:883–890. doi: 10.1016/s0006-291x(03)01285-3. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar S., Dadhania M., Rourke P., Desai T.A., Wong J.Y. Vascular tissue engineering: microtextured scaffold templates to control organization of vascular smooth muscle cells and extracellular matrix. Acta Biomater. 2005;1:93–100. doi: 10.1016/j.actbio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Yim E.K., Leong K.W. Significance of synthetic nanostructures in dictating cellular response. Nanomedicine. 2005;1:10–21. doi: 10.1016/j.nano.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Folkman J., Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 11.Singhvi R., Kumar A., Lopez G.P., Stephanopoulos G.N., Wang D.I. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 12.Chen C.S., Mrksich M., Huang S., Whitesides G.M., Ingber D.E. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 13.Bhadriraju K., Hansen L.K. Extracellular matrix- and cytoskeleton-dependent changes in cell shape and stiffness. Exp. Cell Res. 2002;278:92–100. doi: 10.1006/excr.2002.5557. [DOI] [PubMed] [Google Scholar]
- 14.Thyberg J. Differentiated properties and proliferation of arterial smooth muscle cells in culture. Int. Rev. Cytol. 1996;169:183–265. doi: 10.1016/s0074-7696(08)61987-7. [DOI] [PubMed] [Google Scholar]
- 15.Kurpinski K., Park J., Thakar R.G., Li S. Regulation of vascular smooth muscle cells and mesenchymal stem cells by mechanical strain. Mol. Cell. Biomech. 2006;3:21–34. [PubMed] [Google Scholar]
- 16.Cheng Q., Komvopoulos K. Surface chemical patterning for controlled cell adhesion. Proc. Int. Joint Tribology Conf. 2009 Abstract IJTC2009–15134. [Google Scholar]
- 17.Kurpinski K., Chu J., Hashi C., Li S. Anisotropic mechanosensing by mesenchymal stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:16095–16100. doi: 10.1073/pnas.0604182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D., Park J.S., Chu J.S., Krakowski A., Luo K. Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor β1 stimulation. J. Biol. Chem. 2004;279:43725–43734. doi: 10.1074/jbc.M407368200. [DOI] [PubMed] [Google Scholar]
- 19.Park J.S., Chu J.S., Cheng C., Chen F., Chen D. Differential effects of equiaxial and uniaxial strain on mesenchymal stem cells. Biotechnol. Bioeng. 2004;88:359–368. doi: 10.1002/bit.20250. [DOI] [PubMed] [Google Scholar]
- 20.Canham P.B., Mullin K. Orientation of medial smooth muscle in the wall of systemic muscular arteries. J. Microsc. 1978;114:307–318. doi: 10.1111/j.1365-2818.1978.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 21.Walmsley J.G., Campling M.R., Chertkow H.M. Interrelationships among wall structure, smooth muscle orientation, and contraction in human major cerebral arteries. Stroke. 1983;14:781–790. doi: 10.1161/01.str.14.5.781. [DOI] [PubMed] [Google Scholar]
- 22.Peters M.W., Canham P.B., Finlay H.M. Circumferential alignment of muscle cells in the tunica media of the human brain artery. Blood Vessels. 1983;20:221–233. doi: 10.1159/000158475. [DOI] [PubMed] [Google Scholar]
- 23.Qu M.J., Liu B., Wang H.Q., Yan Z.Q., Shen B.R. Frequency-dependent phenotype modulation of vascular smooth muscle cells under cyclic mechanical strain. J. Vasc. Res. 2007;44:345–353. doi: 10.1159/000102278. [DOI] [PubMed] [Google Scholar]
- 24.Ohkura N., Ito M., Tsukada T., Sasaki K., Yamaguchi K. Structure, mapping and expression of a human NOR-1 gene, the third member of the Nur77/NGFI-B family. Biochim. Biophys. Acta. 1996;1308:205–214. doi: 10.1016/0167-4781(96)00101-7. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Gonzalez J., Rius J., Castello A., Cases-Langhoff C., Badimon L. Neuron-derived orphan receptor-1 (NOR-1) modulates vascular smooth muscle cell proliferation. Circ. Res. 2003;92:96–103. doi: 10.1161/01.es.0000050921.53008.47. [DOI] [PubMed] [Google Scholar]
- 26.Maxwell M.A., Muscat G.E. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rius J., Martinez-Gonzalez J., Crespo J., Badimon L. Involvement of neuron-derived orphan receptor-1 (NOR-1) in LDL-induced mitogenic stimulus in vascular smooth muscle cells: role of CREB. Arterioscler. Thromb. Vasc. Biol. 2004;24:697–702. doi: 10.1161/01.ATV.0000121570.00515.dc. [DOI] [PubMed] [Google Scholar]
- 28.Roca-Cusachs P., Alcaraz J., Sunyer R., Samitier J., Farre R. Micropatterning of single endothelial cell shape reveals a tight coupling between nuclear volume in G1 and proliferation. Biophys. J. 2008;94:4984–4995. doi: 10.1529/biophysj.107.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.