Abstract
Mechanisms of bacterial pathogenesis have become an increasingly important subject as pathogens have become increasingly resistant to current antibiotics. The adhesion of microorganisms to the surface of host tissue is often a first step in pathogenesis and is a plausible target for new antiinfective agents. Examination of bacterial adhesion has been difficult both because it is polyvalent and because bacterial adhesins often recognize more than one type of cell-surface molecule. This paper describes an experimental procedure that measures the forces of adhesion resulting from the interaction of uropathogenic Escherichia coli to molecularly well defined models of cellular surfaces. This procedure uses self-assembled monolayers (SAMs) to model the surface of epithelial cells and optical tweezers to manipulate the bacteria. Optical tweezers orient the bacteria relative to the surface and, thus, limit the number of points of attachment (that is, the valency of attachment). Using this combination, it was possible to quantify the force required to break a single interaction between pilus and mannose groups linked to the SAM. These results demonstrate the deconvolution and characterization of complicated events in microbial adhesion in terms of specific molecular interactions. They also suggest that the combination of optical tweezers and appropriately functionalized SAMs is a uniquely synergistic system with which to study polyvalent adhesion of bacteria to biologically relevant surfaces and with which to screen for inhibitors of this adhesion.
Keywords: polyvalency, optical tweezers, self-assembled monolayers
Adhesion of microorganisms to biological surfaces often correlates with pathogenicity (1). This observation suggests blocking adhesion as a strategy for limiting pathogenicity. Polyvalency is a tactic often used by microorganisms to remain attached to host cells when exposed to shear forces from flowing liquid (2). For example, Escherichia coli [the primary causative agent of urinary tract infections (3, 4)] use the FimH adhesin located on the tips of its type 1 pili to bind to mannose groups in oligosaccharides present on the surface of bladder epithelial cells (5–7). The small number of techniques that can be used to study adhesion of complex, polyvalent systems in biologically relevant circumstances has limited understanding of microbial adhesion and has slowed the development of new medicinal agents whose mechanism involves blocking adhesion.
We have quantified successfully the forces of adhesion of uropathogenic E. coli to mannose-presenting surfaces by using gradient force optical traps—better known as optical tweezers—in conjunction with self-assembled monolayers (SAMs). Several characteristics of optical tweezers make them particularly well suited for studies of polyvalent adhesion involving bacteria: (i) they generate an optical trap (a focal region ≈0.1–0.3 μm in size) that has dimensions similar to those of a typical bacterial cell (≈0.5–1.0 μm) (8, 9); (ii) they can be used in biological media (so long as these media are transparent at the frequency of the tweezers) and are compatible with studies of both bacterial and mammalian cells in culture; and (iii) they can be used to orient nonspherical microorganisms relative to surfaces of SAMs or cells.
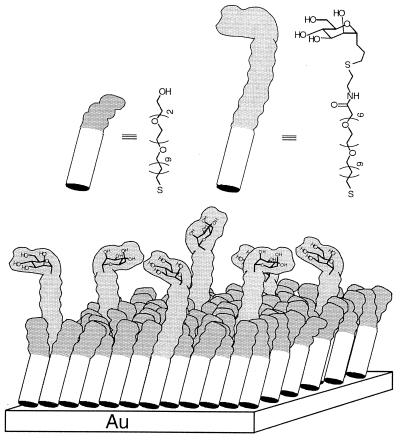
SAMs of alkanethiolates on gold were used as surfaces. By working with model surfaces that had only a single ligand recognized by E. coli (Fig. 1), mannose, we simplified the problem of understanding the mechanisms substantially relative to that presented by the heterogeneous mixture of ligands present on the surface of a cell. By using SAMs as model surfaces, we were also able to exploit a number of their attractive properties: (i) their preparation and biophysically relevant properties are well developed; (ii) using available synthetic techniques, we could both control the density of ligands in them and eliminate nonspecific adsorption to them (10–12); and (iii) they are mechanically rigid, and their deformation contributes less to the force required to detach the bacteria than would a more compliant surface. The SAMs used in these studies were composed of two components: one presented α-C-mannoside units, and the second presented triethylene glycol units (Fig. 1); these latter groups prevented nonspecific adsorption of proteins or adhesion of bacteria to the SAM (13). We examined SAMs that had four mole fractions of mannose (χMan): 0, 10−6, 10−3, or 10−1.
Figure 1.
Schematic representation of a mixed SAM in which an (EG)6 group terminating in a mannose ligand is presented above a background of (EG)3 groups. In this illustration, a SAM with χMan = 10−1 is shown. The triethylene glycol thiol extends 22 Å from the gold surface, whereas the extended mannose thiol, with three extra ethylene glycol units, extends 30 Å from the gold surface (18). The (EG)n groups prevent nonspecific adhesion of proteins or bacteria to the surface of the SAM (18).
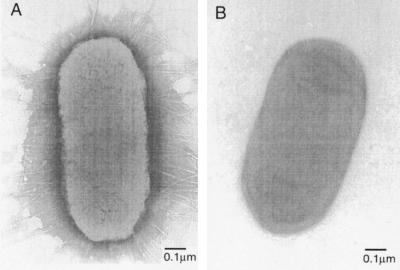
The two strains of E. coli used in these studies initially were isolated from patients with acute pyelonephritis by Shaw Warren (Massachusetts General Hospital, Boston). Transmission electron micrographs (TEMs) of strain RB128 showed pili evenly distributed across the bacterial surfaces (a peritrichous distribution) (Fig. 2A); strain RB124 was nonpiliated (Fig. 2B). Strain RB124 did not adhere to surfaces presenting mannose and served as a control for our experiments.
Figure 2.
(A) Electron micrograph of a representative RB128 cell. (B) Electron micrograph of RB124, an E. coli strain that is mannose-insensitive in a hemagglutination assay and does not express pili.
Materials and Methods
Preparation of SAMs.
The formation of thiol-based SAMs on gold has been described previously (10, 13). The SAMs presenting mannose units were prepared by using two different methods. In one, the shorter triethyleneglycol-terminated thiol [HS(CH2)11(OCH2CH2)3OH] was mixed with the thiol HS(CH2)11(OCH2CH2)6OCH2CON(CH2)2S(CH2)3-α-C-Man in varying concentrations to produce SAMs having χMan = 10−1, 10−3, or 10−6. We assume that the relative mole fractions of these two thiols in solution and on the surfaces are the same. Alternatively, the triethyleneglycol-terminated thiol was mixed with the carboxylic acid-terminated thiol [HS(CH2)11(OCH2CH2)6OCH2CO2H]. The compound NH2(CH2)2S(CH2)3-α-C-mannose then was coupled to the surface carboxylic acid groups, using N-hydroxysuccinimide (NHS) and 1-ethyl-3-dimethylaminopropylcarbodiimide to form the NHS ester, followed by displacement of the NHS ester with the amino group of the ligand (12). SAMs formed from both methods yielded indistinguishable results. The experiments were performed with thicknesses of the gold film of 12 nm, 25 nm, 38 nm, and 100 nm. Indistinguishable results were obtained in all cases.
Characterization of SAMs.
For long-chain alkyl disulfides and thiols chemisorbed on Au (111), the sulfur atoms are spaced by a distance of 4.99 Å (14). Assuming this spacing, SAMs with χMan = 10−6 presented, on average, 4 mannose μm−2, whereas SAMs with χMan = 10−3 presented 4 × 103 mannose μm−2, and SAMs with χMan = 10−1 presented 4 × 105 mannose μm−2. These values also assume that the two components in the mixed monolayers do not phase-segregate into macroscopic islands (greater than a few tens of angstroms across). Several studies (15–17) have examined the phase behavior of two-component SAMs and have concluded that, for a two-component system of alkanethiolates of gold in equilibrium with alkanethiols in solution, a single phase is preferred over a phase-separated system. The closest analog to our system was one in which a mixed monolayer was prepared with HS(CH2)11(OCH2CH2)3OH and HS(CH2)11(OCH2CH2)6OH (18). In this mixed SAM, the midpoint of the change in thickness of the SAM (corresponding to a SAM containing equal numbers of molecules of the two thiolates) occurred roughly where the mole fractions of each thiolate in solution were equal.
Bacterial Growth Conditions.
RB128 were grown from frozen cultures in trypticase soy broth (TSB) as well as defined M9 medium (42 mM sodium phosphate/22 mM potassium phosphate/18.7 mM ammonium chloride/8.5 mM sodium chloride/1 mM magnesium sulfate/11.1 mM glucose, pH 7.4). Bacteria were grown at 37°C under static conditions and also with shaking at 150 rpm, until reaching an OD650 of 0.8 (≈2 × 109 cells/ml). Expression of type 1 pili was verified by TEM and by mannose-sensitive agglutination of guinea pig erythrocytes. The cultures either were used directly in the experiments, diluted 100-fold into fresh medium (TSB or M9), or harvested by centrifugation (5,000 × g for 5 min), followed by resuspension in 10 ml of PBS (pH 7.4). Experiments were conducted with bacteria grown under each of these conditions, and indistinguishable results were obtained in all cases.
TEMs.
From a solution of 107 RB128 cells/ml M9 medium, a 5-μl aliquot was placed onto a carbon-coated grid that had been made hydrophilic by a 30-sec exposure to a glow discharge in an Edwards Auto 306 vacuum evaporator. After 5 min, excess liquid was removed by blotting with filter paper (no. 1; Whatman), and bacteria that had adsorbed onto the carbon grid were stained with 1% uranyl acetate for 1 min. The grids were examined by using a JEOL 1200EX transmission electron microscope. The TEM of a representative RB128 cell (Fig. 2A) illustrates that each bacterium displays roughly 100 pili evenly distributed across its surface. If we treat the bacterium as a three-dimensional cylinder with length of 1 μm and diameter of 0.5 μm, there are approximately 60 pili per μm2 of E. coli. End-on orientation of the bacterium on the SAM allows pili on only one end to attach, and only pili within some arbitrary angle θ are in the correct geometry to bind. If we set θ = 45°, this angle encompasses a surface of 0.025 μm2. At a density of 60 pili μm−2, an area of 0.025 μm2 corresponds to 1 or 2 pili available for attachment.
Optical Tweezers Apparatus.
All experiments used a single-beam optical tweezers apparatus with a cesium diode laser of wavelength 854 nm (19). We calibrated the curve of force vs. laser power for our optical tweezers by measuring the maximum velocity at which tweezers with a given laser power could drag a 1.0 μm diameter polystyrene sphere through water (19). We converted this maximum velocity to a maximum force by equating the Stokes drag at that velocity with the maximum force from optical tweezers. Every 4.1 mW of laser power corresponded to 1.0 pN of force exerted on the object. Similar Stokes drag measurements on RB128 bacteria suggested that the force on an E. coli with a length of 1.0 μm is one-third to one-fourth that of the force on a 1.0 μm diameter polystyrene sphere. Although the absolute magnitude of our reported forces may vary based on the method of calibration, the force to detach E. coli in an end-on orientation from a SAM with χMan = 10−1 will always be twice the value of the force of detachment from a SAM with χMan = 10−6.
Results and Discussion
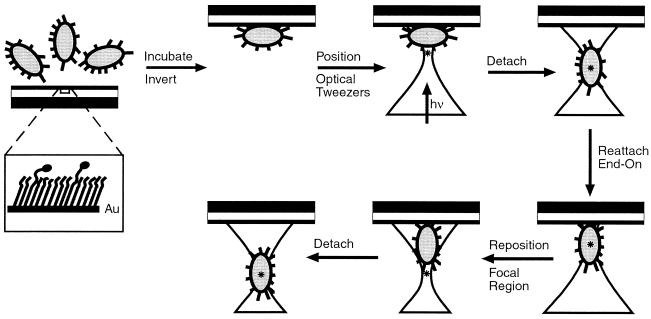
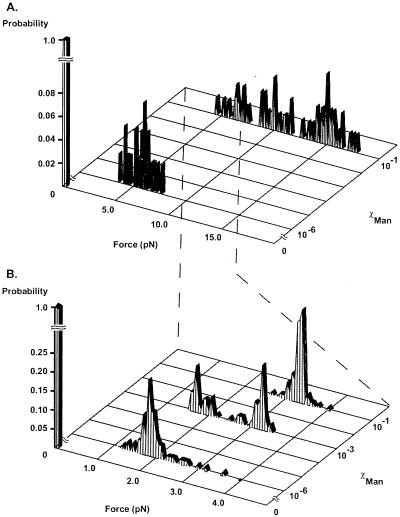
In initial experiments, SAMs with χMan = 10−1 or 10−6 were incubated with a suspension of RB128. After incubation for 30 min, bacteria adhered to both surfaces. All bacteria attached with their long axis parallel to the surface; we assumed that this orientation allowed the maximal number of adhesive contacts with the surface. We used the optical tweezers to apply force to the microorganisms and to detach them from the surface of the SAM. Optical tweezers exert a force that tends to move the material with the highest index of refraction into the region of highest light intensity. To maximize the trapping force, we placed the focal spot at the edge of the bacterium (≈0.5–1.0 μm away from the surface); in this configuration, the gradients in both light intensity and index of refraction are highest. We increased the power of the laser approximately linearly over an interval of 20 sec, until the bacterium detached from the surface (Fig. 3). The moment at which detachment occurred was observed easily because the bacterium changed its orientation from an alignment parallel to the surface to vertical to the surface and “fell” into the optical trap. Fig. 4A shows the levels of laser power required for initial detachment of 50 bacteria from each surface with different values of χMan. Power readings then were converted to forces, using a calibration based on calculating the Stokes drag on a 1 μm polystyrene bead.
Figure 3.
Schematic illustration of the procedure for using optical tweezers to detach E. coli from SAMs presenting α-C-mannoside ligands. Bacteria were allowed to settle by gravity onto SAMs with χMan = 10−1, 10−3, or 10−6 and left for 30 minutes to adhere without control over their orientation. The substrates were inverted, and the focal region of the optical tweezers was brought to the edge of a bacterium. The bacterium was detached from the SAM and allowed to reattach end-on. The focal region of the optical trap again was placed at the edge of the bacterium, and the tweezers were used immediately to detach microorganisms from this end-on orientation. The location of the focal region of the optical trap is indicated by an asterisk.
Figure 4.
(A) Histogram of forces for initial detachment of RB128 E. coli attached to SAMs presenting α-C-mannoside groups at χMan = 10−1, χMan = 10−6, and χMan = 0. The data at each value of χMan were obtained by using 50 different bacteria, chosen at random among the bacteria that had attached to the surface. (B) Histogram of detachment forces for RB128 attached in an end-on orientation to SAMs presenting α-C-mannoside groups at χMan = 10−1, χMan = 10−3, χMan = 10−6, and χMan = 0. Data from each surface were a result of analyzing 50 individual bacteria with 5–10 force measurements per bacterium. The probability indicates the percentage of bacteria that would detach at a given force value; for each surface, the sum of the data set equals a probability of 1.0.
Bacteria that attached to SAMs with χMan = 10−1 required forces ranging from 3.5 to ≥18 pN for detachment (Fig. 4A). These SAMs displayed essentially a complete monolayer of α-C-mannoside ligand. With the assumption that a discrete value of force is required to rupture the complex between the FimH receptor and the mannoside ligand, this distribution suggested that each bacterium was attached by a different number of bonds; that is, that the attachment was polyvalent (2). The maximum force observed—18 pN—for initial detachment corresponded to the highest laser power (75 mW) available in our apparatus. When this magnitude of power was not enough to detach a bacterium immediately, we were still ultimately able to detach it by a procedure in which we swept the position of the optical trap across the entire length of the bacterium; the detachment seemed to occur in a “Velcro-like” process. We believe that sweeping caused sequential dissociation of individual interactions between α-C-mannoside ligands and FimH receptors, by concentrating force on different points of the microorganism. This sweeping motion transformed the bacterium from a state in which it was attached by multiple FimH–mannose interactions to one in which only a few attachments remained. At this point, the bacterium would detach completely when subjected to the force exerted at the highest laser power.
Bacteria that attached to a SAM presenting α-C-mannoside at χMan = 10−6 also had a configuration with their long axis parallel to the plane of the surface. Detachment required much smaller forces (3.8–7.8 pN) than those required for χMan = 10−1 (Fig. 4A). These values still suggested multipoint attachment of bacteria to the surface. On average, however, this SAM presented only four α-C-mannoside groups per μm−2, so each bacterium could attach to the surface by using only a few pili. That a force of 7.8 pN was the highest value required to remove any bacterium suggested that polyvalency was less important in adhesion on this surface than it was on a SAM with χMan = 10−1.
From these data, we inferred that side-on attachment of the bacteria led to multivalent attachment mediated by multiple pili. To reduce the number of points of attachment, we oriented the bacteria so that they attached in an end-on orientation. A key advantage of optical tweezers in studying adhesion of bacteria was the ability of this technique to present oblate biological objects to the surface in a particular orientation. In our system, detached E. coli spontaneously aligned along the axis of the optical trap; that is, perpendicular to the plane of the surface (Fig. 3). When bacteria were reattached to the surface in an end-on orientation, we estimated the number of pili able to find an accessible mannose ligand on the surface to be only one or two, even on a SAM with χMan = 10−1 (see Materials and Methods).
Bacteria initially were allowed to adhere to the SAMs without control over their orientation. They then were detached from the SAMs, oriented vertically in the optical trap, brought back to the surface, and allowed to reattach. From the end-on orientation (Fig. 3), the bacteria were immediately detached with the optical tweezers. We examined 50 different bacteria on each surface and sampled the forces required to detach bacteria from the end-on attachment 5–10 times per bacterium.
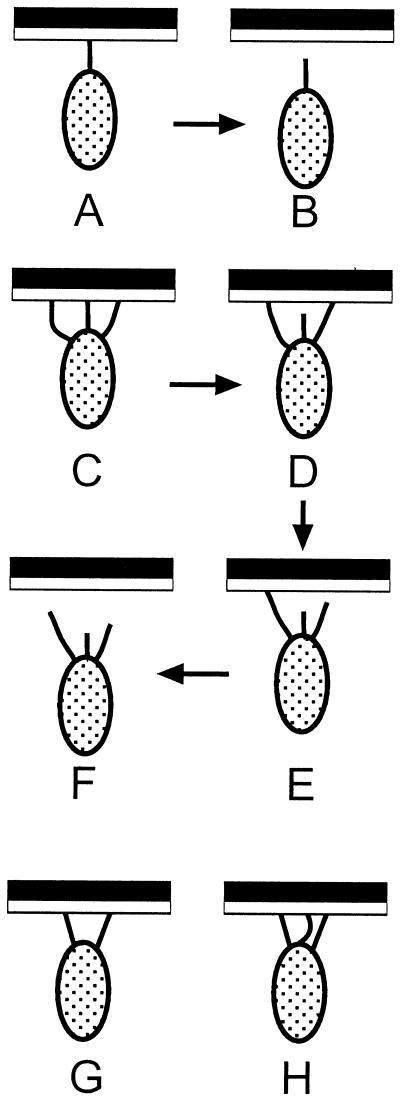
The distribution of forces needed to detach bacteria oriented end-on was much different from that for detachment of bacteria oriented side-on. The forces were lower and were characterized by a sharp unimodal or bimodal distribution (Fig. 4B). The observation of two, well defined peaks in Fig. 4B, with the one twice the value of the other, suggests that the lowest value of force—1.7 pN—has fundamental significance. We identify it with the force required to break one interaction between pilus and surface-bound α-mannoside, whether by dissociation of a FimH-α-C-mannoside complex or by some other process (e.g., rupture of the pilus). To require twice this value of force would require that two pili come under equal tension at the same time. A hypothetical set of events such as C → D → E → F (Fig. 5), in which three pili detach, independently but over a distance of movement of the bacterium sufficiently small that the force exerted by the tweezers does not change significantly, would be indistinguishable in this type of experiment from simple detachment of one pilus (A → B). Similarly, it would not be possible, under conditions of these experiments, to distinguish the type of detachment shown in G (with two, equally stressed pili) from that suggested by H (in which detachment would first dissociate two stressed pili and only then disconnect a third). Types of information that would be required to resolve these types of ambiguities include the following: (i) an understanding of the ability of the bacterium to equalize stress among pili by moving laterally in the optical trap or by twisting its long axis away from that of the direction of force, (ii) an understanding of the rates of dissociation of the FimH-α-C-mannoside complex with the pilus under tension, (iii) information concerning the elasticity of the pili, and (iv) a description of the potential function for dissociation and of the contributions to it from dissociation of ligand from receptor and from deformation of the receptor, the pilus, the membrane of the bacterium, and the SAM and the (EG)6 group.
Figure 5.
Simplified schematic of hypothetical detachment mechanisms for bacteria bound to SAMs presenting α-C-mannoside groups. The sequence A → B represents the simplest case in which one pilus is attached. The sequence C → D → E → F represents a case in which three pili bind to the surface but only one pili comes under tension at a time. In this case, the pili can dissociate sequentially, and the force required to remove the bacterium would be indistinguishable from the force required to remove a bacterium attached through a single pilus. Similarly, the force required to remove the bacterium in G might be indistinguishable from that required to remove the bacterium in H.
After a bacterium was subjected to multiple rounds of end-on presentation and detachment, it was allowed to adhere in a side-on orientation. The forces required to remove bacteria that had reattached in this orientation were similar in magnitude and distribution to the forces required for its initial detachment. This experiment suggests that neither exposure to the photon flux in the optical trap nor repeated cycles of attachment and detachment alter the bacterium in a way that makes the values of the forces obtained in the experiments dependent on their history. It is also compatible with (but does not demand) the hypothesis that attachment and release are reversible, as might be expected of attachment mediated by noncovalent association/dissociation of FimH and mannoside ligand.
Three separate experiments confirmed that the interactions between E. coli and mannose-presenting SAMs were mediated by the interactions between receptors present on the pili and α-C-mannoside ligands presented by the SAMs. First, RB124, a strain of E. coli devoid of type 1 pili, failed to bind any of the mannose-presenting SAMs. Second, strain RB128 did not bind to SAMs presenting only (EG)3OH groups and no mannose ligands. Third, the addition of 1 mM mannose, which was shown in an earlier study to be the concentration required to inhibit adhesion of E. coli to yeast mannan by 50% (IC50) (20), completely blocked binding of RB128 to the surfaces presenting α-C-mannoside ligand.
In summary, this study demonstrates that optical tweezers, used synergistically in conjunction with SAMs, are a uniquely powerful tool for studying adhesion of bacteria to surfaces. Tweezers provide the ability to orient the bacteria relative to the surface, to measure the force required to detach a microorganism from the surface, and to make a statistically significant number of measurements, all without having to prepare and examine multiple samples. We have measured what we interpret to be the force of detachment for a single, monovalent, α-C-mannoside–pilus interaction (1.7 pN). We cannot interpret quantitatively the polyvalent adhesion of the microorganism to the surface, but the best evidence for polyvalent adhesion comes from the observation that the bacteria detached in a “Velcro-like” process from SAMs with high values of χMan. The forces to detach bacteria in a side-on (maximal number of adhesive contacts) vs. an end-on orientation (one or two adhesive contacts) demonstrates the potency of polyvalency in attaining an overall strong attachment to the surface. Although in this report we only studied the ability of α-d-mannopyranoside to antagonize the adhesion of E. coli to mannose-bearing surfaces, polyvalent inhibitors also can be easily assayed in this experimental setup for enhanced inhibition of adhesion.
The magnitude of force for detachment in our system is of the same order of magnitude as the unbinding force and force of mechanical interactions between an actin filament and a single motor molecule of myosin, as well as isometric forces generated by translocation of the mechanochemical enzyme kinesin along microtubules (21–23), but it is several orders of magnitude smaller than the force necessary to break an actin–actin bond (24). The studies described in this report were conducted by using a SAM as a model for the surface of a cell. We believe, however, that similar types of experiments can be conducted, with appropriate modification, on systems involving other types of interactions, including cell–bacterium, cell–virus, and cell–cell. This technique also provides an assay to screen for compounds that block adhesion and, in principle, for those that catalyze detachment.
Acknowledgments
We thank H. S. Warren (Massachusetts General Hospital, Boston) for providing us with the strains of E. coli. We are also grateful to M. Ericsson and the Harvard Medical School Electron Microscope Facility (Boston) for assistance in obtaining the TEM images. The work was funded by Defense Advanced Research Projects Agency/Office of Naval Research/Space and Naval Warfare Systems Center, the National Science Foundation (PHY-9312572 and PHY-9732449), and Materials Research Science and Engineering Centers (DMR-9809363). S.J.M. thanks the National Institutes of Health (Grant AI10013) for a postdoctoral fellowship.
Abbreviations
- SAM
self-assembled monolayer
- TEM
transmission electron micrograph
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230451697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230451697
References
- 1.Finlay B B, Cossart P. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 2.Mammen M, Choi S-k, Whitesides G M. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinker J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, et al. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 4.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 5.Abraham S N, Sun D X, Dale J B, Beachey E H. Nature (London) 1988;336:682–684. doi: 10.1038/336682a0. [DOI] [PubMed] [Google Scholar]
- 6.Jones C H, Pinkner J S, Roth R, Heuser J, Nicholes A V, Abraham S N, Hultgren S J. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogfelt K A, Bergmans H, Klemm P. Infect Immun. 1990;58:1995–1998. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashkin A. Proc Natl Acad USA. 1997;94:4853–4860. doi: 10.1073/pnas.94.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashkin A. Biophys J. 1992;61:569–582. doi: 10.1016/S0006-3495(92)81860-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mrksich M, Whitesides G M. Annu Rev Biophys Biomol Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- 11.Whitesides G M, Gorman C B. Self-Assembled Monolayers: Models for Organic Surface Chemistry. Boca Raton, FL: CRC; 1995. [Google Scholar]
- 12.Lahiri J, Isaacs L, Tien J, Whitesides G M. Anal Chem. 1999;71:777–790. doi: 10.1021/ac980959t. [DOI] [PubMed] [Google Scholar]
- 13.Prime K L, Whitesides G M. J Am Chem Soc. 1993;115:10714–10721. [Google Scholar]
- 14.Bain C D, Whitesides G M. Angew Chem, Intl Ed Eng. 1989;28:506–512. [Google Scholar]
- 15.Folkers J P, Laibinis P E, Whitesides G M, Deutch J. J Phys Chem. 1994;98:563–571. [Google Scholar]
- 16.Laibinis P E, Nuzzo R G, Whitesides G M. J Am Chem Soc. 1992;96:5097–5105. [Google Scholar]
- 17.Bain C D, Whitesides G M. J Am Chem Soc. 1989;111:7164–7175. [Google Scholar]
- 18.Pale-Grosdemange C, Simon E S, Prime K L, Whitesides G M. J Am Chem Soc. 1991;113:12–20. [Google Scholar]
- 19.Smith S P, Bhalotra S R, Brody A L, Brown B L, Boyda E K, Prentiss M. Am J Phys. 1999;67:26–35. [Google Scholar]
- 20.Sokurenko E V, Chesnokova V, Doyle R J, Hasty D L. J Biol Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- 21.Kuo S C, Sheetz M P. Science. 1993;260:232–234. doi: 10.1126/science.8469975. [DOI] [PubMed] [Google Scholar]
- 22.Molloy J E, Burns J E, Sparrow J C, Tregear R T, Kendrick-Jones J, White D C. Biophys J. 1995;68:298S–303S. [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizaka T, Miyata H, Yoshikawa H, Ishiwata S, Kinosita K., Jr Nature (London) 1995;377:251–254. doi: 10.1038/377251a0. [DOI] [PubMed] [Google Scholar]
- 24.Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Proc Natl Acad Sci USA. 1996;93:12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]