Abstract
Pig kidney Na+,K+-ATPase was studied by means of reaction-induced infrared difference spectroscopy. The reaction from E1Na3+ to an E2P state was initiated by photolysis of P3-1-(2-nitrophenyl)ethyl ATP (NPE caged ATP) in samples that contained 3 mM free Mg2+ and 130 mM NaCl at pH 7.5. Release of ATP from caged ATP produced highly detailed infrared difference spectra indicating structural changes of the Na+,K+-ATPase. The observed transient state of the enzyme accumulated within seconds after ATP release and decayed on a timescale of minutes at 15°C. Several controls ensured that the observed difference signals were due to structural changes of the Na+,K+-ATPase. Samples that additionally contained 20 mM KCl showed similar spectra but less intense difference bands. The absorbance changes observed in the amide I region, reflecting conformational changes of the protein backbone, corresponded to only 0.3% of the maximum absorbance. Thus the net change of secondary structure was concluded to be very small, which is in line with movement of rigid protein segments during the catalytic cycle. Despite their small amplitude, the amide I signals unambiguously reveal the involvement of several secondary structure elements in the conformational change. Similarities and dissimilarities to corresponding spectra of the Ca2+-ATPase and H+,K+-ATPase are discussed, and suggest characteristic bands for the E1 and E2 conformations at 1641 and 1661 cm−1, respectively, for αβ heterodimeric ATPases. The spectra further indicate the participation of protonated carboxyl groups or lipid carbonyl groups in the reaction from E1Na3+ to an E2P state. A negative band at 1730 cm−1 is in line with the presence of a protonated Asp or Glu residue that coordinates Na+ in E1Na3+. Infrared signals were also detected in the absorption regions of ionized carboxyl groups.
Introduction
The Na+,K+-ATPase (EC 3.6.3.9) (1) actively transports Na+ and K+ in opposite directions across the plasma membrane of animal cells, thereby maintaining ion concentration gradients that are essential for nerve conduction and secondary transport processes. The transport of three Na+ out of the cell and two K+ into it is coupled to the hydrolysis of one ATP molecule, and this process accounts for 20–30% of the total ATP consumption in resting mammalian cells (2). The Na+,K+-ATPase belongs to the family of P-type ATPases (3), which are characterized by an aspartylphosphate phosphoenzyme that transiently forms in the transport process.
The Na+,K+-ATPase consists of three subunits denoted as α, β, and γ, with molecular masses of 113, 35, and 7 kDa, respectively. The α-subunit contains the catalytic machinery (nucleotide-binding site, phosphorylation site, and ion-binding sites) and is homologous to single subunit P-type ATPases such as the Ca2+-ATPase (4). The glycosylated β-subunit is important for correct insertion of the α-subunit into the membrane (5). The γ-subunit belongs to the FXYD protein family and regulates the activity of the ATPase (6,7).
The reaction cycle of the Na+,K+-ATPase is commonly described according to the Albers-Post model (8,9) and has been reviewed extensively (10–12). The E2 form of the enzyme occludes two K+ (E2[K2+]). Low-affinity ATP binding promotes the transition to the E1 form and the release of K+ to the cytoplasm (E2[K2+] + ATP → E1ATP + 2 K+). E1 binds three Na+ from the cytosol (E1ATP → E1ATP Na3+). ATP phosphorylates Asp-369 of the α-subunit, which occludes the three sodium ions in the ADP-sensitive phosphoenzyme intermediate E1P [Na3+]. Isomerization to the ADP-insensitive phosphoenzyme E2P leads to dissociation of Na+ to the extracellular space. The E2P phosphoenzyme binds two K+ with high affinity, which induces its rapid dephosphorylation and the occlusion of K+ (E2[K2+]).
The atomic structure of the Na+,K+-ATPase in an E2 state with two rubidium ions bound was recently solved by x-ray crystallography (13). The structural organization of the α-subunit is similar to that of the Ca2+-ATPase of the sarcoplasmic reticulum membrane (SERCA1a) (14), with 10 transmembrane helices and three cytoplasmic domains (the actuator, nucleotide-binding, and phosphorylation domains). However, there are distinct differences: the Na+,K+-ATPase has a smaller N domain, a protrusion in the P domain, a kink in transmembrane helix M7, and a strongly positive C-terminus that may convey sensitivity to the membrane potential.
The β-subunit consists of a small cytosolic N-terminal segment, one transmembrane helix that is in contact with transmembrane helices M7 and M10 of the α-subunit, and an extracellular domain. The γ-subunit consists of a transmembrane helix close to M9 of the α-subunit.
To characterize structural changes of the active Na+,K+-ATPase, we used infrared spectroscopy, which has yielded inconsistent results in previous studies. Studies that analyzed absorption spectra of the Na+ and K+ forms detected no significant change (15) or changes of 3% in α-helix content that were observed only in H2O (and not in D2O) and were close to the error margin (16). Similarly, secondary structure changes involving <3% of the residues—again close to experimental error—were concluded from the analysis of a larger number of states (17). On the other hand, this approach of analyzing absorption spectra revealed clear changes upon binding of metal-ATP analogs to E1Na3 (18). Analysis of the infrared dichroism indicated a reorientation of NH bonds and carboxylate groups between the Na+, K+, and Tris forms, and it was estimated that 13% of the residues undergo a conformational change from the Tris form to the K+ form (19). Unexpectedly large absorbance changes (on the order of 10% of the maximum absorbance) were recently observed with the use of reaction-induced difference spectroscopy (20).
To clarify the situation, we present our results obtained with reaction-induced difference spectroscopy. We use the photolytic release of ATP from caged ATP to trigger the catalytic reactions of Na+,K+-ATPase as was done in previous work by our group (21) and others (20) on Na+,K+-ATPase, as well as on Ca2+-ATPase (22,23) and gastric H+,K+-ATPase (24). The absorbance spectrum before release of ATP is then subtracted from the absorbance spectrum after ATP release. Only groups that change their absorption upon ATP release show in the difference spectrum, i.e., only those protein groups are detected that actively participate in the induced reactions. The absorption of passive groups, inactive protein, or impurities cancels. Therefore, this approach is sensitive enough to detect single bonds in large proteins (25,26). We find only small changes in infrared absorption, which, however, provide detailed and informative difference spectra. Several control experiments ensured that the absorption changes were due to the catalytic activity of the Na+,K+-ATPase.
Materials and Methods
Sample preparation
Na,K-ATPase was isolated according to the slightly modified procedure of Jørgensen (27,28) and Grell et al. (29) with tissues obtained from pig kidney, rectal salt gland of Squalus acanthias, and nasal salt gland of duck. All activities were higher than 28 μmol mg−1 min−1 in 30 mM histidine/HCl, 130 mM NaCl, 20 mM KCl, 3 mM MgCl2, and 3 mM ATP, at pH 7.5 and 37°C. Details of the analytical characterizations (29) and the preparation of P3-1-(2-nitrophenyl)ethyl ATP (NPE caged ATP) and its [15N]nitro-isotopomer (30,31) were described previously.
For a typical experiment, 150 μg (∼23 μL) of Na,K-ATPase in 5 mM 4-(2-hydroxyethyl)-piperazine-1-ethane-sulfonic acid (HEPES)/Tris pH 7.5 and 1 μL each of other solutions were pipetted onto a CaF2 window with a trough of 5 μm depth and vacuum-concentrated within 15 min to a volume of ∼1 μL at 10°C. Approximate final concentrations were 1 mM Na+,K+-ATPase, 150 mM HEPES/Tris pH 7.5, 20% glycerol, 5 mM dithiothreitol I (DTT), 5 mM NPE caged ATP, 2 mM EDTA, 8 mM MgCl2 (3 mM free Mg2+), and 130 mM NaCl in H2O solution. Some samples additionally contained 20 mM KCl. The infrared cuvette was closed with a second CaF2 window and placed in the purged spectrometer, where it was thermostated at 5°C or 15°C. The time between sample preparation and infrared measurement varied typically between 30 and 90 min.
Infrared experiments
Infrared transmission measurements were performed with a modified Bruker IFS 66 spectrometer at 4 cm−1 spectral resolution. After the measured absorption became stable, a reference spectrum of 300 interferometer scans was recorded. Then, NPE caged ATP was photolyzed with one or two subsequent light flashes from a Radiant Dyes (Wermelskirchen, Germany) excimer laser RD-EXC-200, which released ∼3 mM ATP under the chosen conditions. After the flash, rapid scan spectra were recorded with an initial time resolution of 65 ms for the first 10 spectra after the flash. The interferograms were zero-filled using a factor of 2 and apodized with a Blackman-Harris-4-term function. All spectra were normalized to an equal protein concentration as previously described (32). They are therefore directly comparable with our spectra of the Ca2+-ATPase (23,33,34).
Infrared sample changer with temperature control
To significantly improve the efficiency of the infrared measurements, a sample changer was developed. It consisted of a round aluminum disc and was designed to hold up to five infrared cuvettes. The disc-shaped cuvette holder was equipped with a platinum resistance thermometer PT 100 for temperature control and connected with a RE 204 thermostat (Lauda, Lauda-Königshofen, Germany), containing a water/ethylene glycol mixture. The complete unit was mounted on a mechanical XZ positioner to position one of the cells into the focus of the infrared beam. For selection and variation of the cuvette positions, the disc was rotated with a freely programmable step motor driven by a separate microprocessor control unit employing the program system BASIC-Tiger (Wilke, Aachen, Germany). For electronic control of the cuvette positions, an angle encoder was used in combination with two inductive sensors, mounted on the XZ positioner, to mark the start/end positions of disc rotation.
Subtraction of photolysis bands
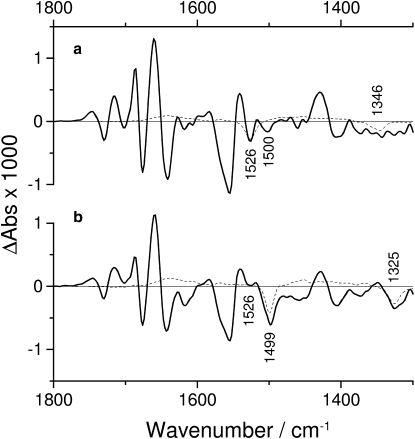
The photolysis of NPE caged ATP produced changes in infrared absorption that were subtracted from the difference spectra of the Na+,K+-ATPase samples. For this a difference spectrum of NPE caged ATP photolysis in the same medium but without Na+,K+-ATPase, recorded at the same temperature and in the same time interval, was used. The correct subtraction factor was determined in the spectral regions of the photolysis bands at 1526 and 1346 cm−1 by comparison with a spectrum obtained with [15N]NPE caged ATP, for which these regions are clear of overlap by photolysis bands (35,36) as shown in Fig. 1. Unless mentioned otherwise, the spectra shown have been corrected for photolysis bands.
Figure 1.
Comparison of uncorrected Na+,K+-ATPase spectra (solid lines) and normalized photolysis spectra (dashed lines) in the presence of 130 mM NaCl and 3 mM free Mg2+, recorded 0.66–3.24 s after the photolysis flash at 15°C. (a) Unlabeled caged ATP. (b) [15N] Caged ATP. The spectra are an average of three experiments performed with different samples.
Activity estimation in infrared samples
Hydrolysis of ATP leads to changes in the infrared absorption that can be used to measure the hydrolytic activity of ATPases directly in infrared samples (22,37). The protein concentrations used in these studies ranged from 7.5 to 150 mg/mL. In accordance with a previous study (37), we evaluated the negative ATP band at 1246 cm−1, which was integrated between 1260 and 1224 cm−1 with respect to a baseline drawn between 1296 and 1168 cm−1 using integration method C of the program OPUS (Bruker Optics). The concentration of released ATP was 2.8 mM, corresponding to a photolysis yield of 56%. This was determined using a sample without protein but with otherwise identical composition. This sample was flashed 20 times to photolyze the caged ATP completely. The photolysis yield is the signal after the first flash divided by the total signal after 20 flashes.
The evaluation of ATPase activity assumed that all ATP was hydrolyzed at the end of the time-resolved experiment, i.e., that the difference between the initial and final band areas of the 1246 cm−1 band corresponded to 2.8 mM. With this knowledge, ATPase activity can be calculated from the initial velocity of the reaction (37). This was calculated from the integrated band area of the 1246 cm−1 band of two early spectra: the first spectrum was averaged in the first 660 ms (at 15°C) or 2 s (at 5°C) after the flash, and the second spectrum was averaged 0.66–3.24 s (15°C) or 2–5 s (5°C) after the flash.
Calculation of the COBSI index
The change of backbone structure and interaction (COBSI) index was calculated as described previously (38). In short, the band area of positive and negative bands between 1700 and 1610 cm−1 in the difference spectrum were added and then divided by two and by the band area of the absorption spectrum in the same spectral region. The absorption spectrum was recorded for dry samples containing 150 μg protein to avoid inaccuracies due to subtraction of a water spectrum. Thirteen samples from three different preparations were averaged.
Results
Subtraction of photolysis bands
Fig. 1 shows difference spectra obtained 0.66–3.24 s after the photolysis flash and illustrates the subtraction of photolysis bands. Negative and positive bands are characteristic of the sample before and after ATP release, respectively. The full lines show spectra of Na+,K+-ATPase samples, whereas the dashed lines are the respective spectra in the absence of protein (termed “photolysis spectra”). It is evident that caged ATP photolysis contributes little to the spectra, particularly above 1530 cm−1, where protein conformational changes and several side chains contribute to the spectra.
The spectra in Fig. 1 a were obtained with unlabeled caged ATP, and those in Fig. 1 b were obtained with [15N]caged ATP. Labeling of the nitro group shifts the photolysis bands at 1526 and 1346 cm−1 to 1499 and 1325 cm−1. These bands have been assigned to the antisymmetric and symmetric stretching vibrations of the nitro group, respectively (22). The photolysis spectrum of unlabeled caged ATP was normalized such that its subtraction from the protein spectrum produced a corrected spectrum that was similar to the uncorrected spectrum obtained with [15N]caged ATP (Fig. 1 b) in the region of the nitro bands. The corrected spectrum is shown in Fig. 2 a. Comparison with the [15N]caged ATP protein spectrum is important for correct subtraction of the photolysis spectrum. It reveals, for example, that there is a small negative protein band at 1526 cm−1 that overlaps with the larger photolysis band of unlabeled caged ATP (Fig. 1 a) but is free from overlap in Fig. 1 b. This band should not vanish in the subtraction.
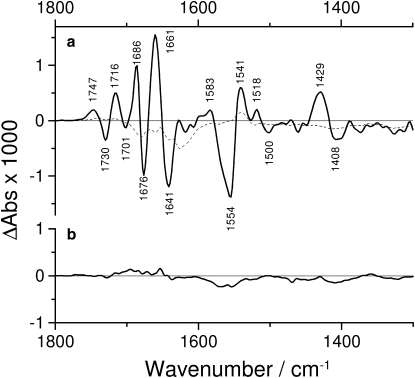
Figure 2.
Photolysis-corrected spectra of Na+,K+-ATPase in the presence of 130 mM NaCl and 3 mM free Mg2+, recorded at 15°C. (a) The full-line and dashed-line spectra were recorded 0.66–3.24 s and 400 s after the photolysis flash, respectively. (b) Control spectrum with FITC-labeled ATP recorded 0.66–3.24 s after the photolysis flash.
The photolysis spectrum of [15N]caged ATP was normalized accordingly in Fig. 1 b. In addition, a small protein band at 1500 cm−1 overlaps with the photolysis band at 1499 cm−1 and is revealed without overlap in Fig. 1 a.
Na+,K+-ATPase spectra and control spectra
Fig. 2 shows photolysis-corrected Na+,K+-ATPase spectra in Fig. 2 a and a control spectrum in Fig. 2 b. The full line spectrum in Fig. 2 a was obtained 0.66–3.24 s after the photolysis flash under the same conditions as those shown in Fig. 1 a. Different sets of three independent experiments were averaged in the two figures. The full line spectra in Figs. 1, a and b, and 2 a are virtually identical, apart from the isotope shifts. This demonstrates the excellent reproducibility of our experiments.
The dashed line shows a later spectrum, recorded 400 s after the flash. This spectrum exhibits comparably small absorbance changes indicating that the enzyme state after 400 s is similar to that before photolysis. The full line spectrum therefore characterizes the formation of a transient enzyme state within 2 s that returns to the initial state at longer times, when the released ATP has been hydrolyzed.
Spectra similar to that shown in Fig. 2 a with reduced band amplitude in the amide I region were obtained at 20 mM Na+ (21). Similar spectra were also obtained for the shark rectal gland and the duck salt gland enzymes at 130 mM Na+ (data not shown). The similarity holds for band positions and overall band amplitudes. The main difference between the shark and duck enzyme spectra on the one hand and the pig enzyme spectrum on the other was a relatively enhanced amplitude of the 1661 cm−1 band.
Fig. 2 b shows a spectrum obtained with fluorescein isothiocyanate (FITC)-labeled ATPase in the same time interval as those in panel a and in Fig. 1. Since FITC blocks the ATP-binding site (39), this modification abolished the infrared signals.
Fig. 2 demonstrates several important findings. The kinetic behavior of the difference signals is as expected, i.e., a transient state is adopted shortly after ATP release and decays at later times due to the hydrolytic activity of the Na+,K+-ATPase. The spectrum recorded at 400 s indicates that possible baseline instabilities are much smaller than the signals obtained after 2 s. Indeed, the independent baseline control runs conducted before the flash photolysis experiments revealed absorbance changes of <2 × 10−4 after 2 s (data not shown), an order of magnitude smaller than the signals observed in Fig. 2 a. The FITC sample demonstrates that the infrared signals are only observed when the ATP-binding site is accessible. The remaining small signals with a blocked ATP site are likely due to baseline instability.
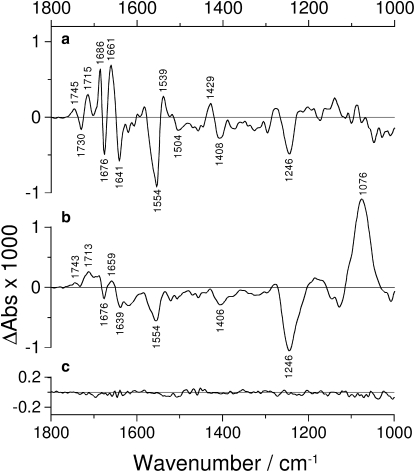
Fig. 3 presents further controls. The spectra were recorded in the additional presence of 20 mM KCl at 5°C under otherwise identical conditions. Fig. 3 a shows photolysis-corrected spectra obtained 0.66–3.24 s after ATP release that are nearly identical in shape to the full line spectrum in Fig. 2 a. However, the band amplitudes are considerably smaller. Fig. 3 b shows a spectrum after 320 s. The protein bands above 1530 cm−1 have largely decayed in this spectrum. In contrast, relatively large bands at 1246 and 1076 cm−1 appear. They can be attributed to the hydrolysis of ATP (22,37) and demonstrate therefore that the Na+,K+-ATPase in our infrared samples is catalytically active. A control with 4 mM vanadate did not produce difference bands as shown in Fig. 3 c for a spectrum obtained 16 s after the photolysis flash. Spectra acquired at earlier times were similar but had more noise because of the shorter acquisition time. Hydrolysis bands have also been observed for shark and duck enzymes (40) (although only within the first 5 s for the shark enzyme). This is in line with our previous observation that concentration of the shark enzyme leads to inactivation (41). On the other hand, phosphate production of the shark enzyme has been found to be linear at least up to a concentration of 1 mg/mL, indicating full activity at this concentration (Mikael Esmann, University of Aarhus, personal communication, 2009).
Figure 3.
Photolysis-corrected spectra of Na+,K+-ATPase in the presence of 130 mM NaCl, 3 mM free Mg2+, and 20 mM KCl recorded at 5°C, (a) 0.66–3.24 s and (b) 320 s after the photolysis flash. (c) Control sample containing 4 mM vanadate. Spectrum recorded 16 s after the photolysis flash. The spectra are an average over three experiments performed with different samples.
Taken together, Figs. 2 and 3 demonstrate that the full line spectrum in Fig. 2 a must be due to structural changes of the active ATPase induced by ATP release. Further controls recorded in the absence of Na+ and Mg2+ or in the presence of 1200 mM NaCl and 20 mM EDTA showed no infrared bands (40).
Hydrolytic activity
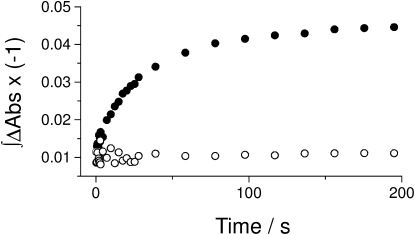
Fig. 4 shows a measurement of the hydrolytic activity in infrared samples, with the 1246 cm−1 band used to monitor the progress of the reaction. We obtained specific activity values of ∼50 nmol mg−1min−1 at 5°C and ∼170 nmol mg−1min−1 at 15°C. For comparison, the activity in the diluted suspension was measured by determining the produced phosphate (42) as a function of time. The conditions were 40 nM ATPase in 150 mM HEPES/Tris pH 7.5, 1 mM DTT, 20% glycerol, 20 mM KCl, 3 mM MgCl2, and 130 mM NaCl. The specific activities were 0.3 and 2 μmol mg−1min−1 at 5°C and 15°C, respectively. Thus the ATPase in concentrated infrared samples and in the presence of unphotolyzed caged ATP retained 9–17% of the specific activity of control samples in diluted suspensions. We previously found a retained activity of 23% at 37°C (41). We conclude that the activity in our infrared samples is reduced to 10–20% of the reference value in diluted suspension and attribute this to inhibition by caged ATP (43–45), to the high viscosity of the concentrated infrared samples (40), and to their different hydration state as compared with diluted suspensions (46). Of importance, however, our sample preparation does not lead to irreversible inhibition of the Na+,K+-ATPase (41).
Figure 4.
Measurement of hydrolytic activity in infrared samples at 5°C. The integrated 1246 cm−1 band is plotted against time. Selected spectra of this experiment are shown in Fig. 3. Full circles: Na+,K+-ATPase in the presence of 130 mM NaCl, 3 mM free Mg2+, and 20 mM KCl. Open circles: Control sample containing 4 mM vanadate.
Discussion
Nature of the initial and transient states
NPE caged ATP binds to the Na+,K+-ATPase with an affinity in the 10–50 μM range (43–45). Thus, the E1Na3+ complex with NPE caged ATP is the initial state in our samples in the presence of Na+ and the absence of K+. Upon ATP release in a low Na+ and no K+ medium, the ADP-insensitive phosphoenzyme E2P accumulates (44,47–49). A spectrum similar to the full line spectrum in Fig. 2 a but with reduced band amplitude has also been obtained in the presence of only 20 mM Na+ (21). We conclude that an E2P-type state accumulates in our experiments at 130 mM Na+ in the absence of K+. Thus we infer that the full line spectrum in Fig. 2 a reflects the reaction from E1Na3+ to an E2P state.
We note that the spectrum in the presence of K+ (Fig. 3 a) is virtually the same as in its absence (Fig. 2 a), apart from smaller band amplitudes, and conclude that the same or very similar states accumulate transiently under both conditions. The state that is expected to accumulate in the presence of K+ is E2[K2+] (50). There are two possible explanations for the observation of similar spectra in different expected states: 1), E2[K2+] and the E2P state observed here may have very similar backbone structures and side-chain interactions, which seems plausible only if our E2P state retains some of the Na+ ions. If not, one would expect differences in cation coordination that reflect in the spectral regions of protonated carboxyl groups (1800–1700 cm−1) and carboxylate groups (1600–1520 cm−1 and ∼1400 cm−1). 2), The state accumulating in the presence of K+ may not be exclusively E2[K2+] but may be at least partially E2P under our conditions. It is possible that the rate-limiting step of the catalytic cycle in our high-viscosity samples is different from that in the low-viscosity samples used in most other studies. In this case, E2[K2+] could have a similar absorption as our initial state E1Na3+, which would explain the reduced signal amplitude in the presence of K+. However, we consider a similar absorption of E2[K2+] and E1Na3+ as unlikely since marker bands for the E1-E2 transition have also been observed for the H+,K+-ATPase in states that resemble the E2[K2+] and E1Na3+ states of the Na+,K+-ATPase, as discussed below. Rather, we think that the reduced signal amplitude in the presence of K+ is due to the partial accumulation of E1 states under steady-state conditions. A K+ impurity in the experiment shown in Fig. 2 a can be excluded because experiments with the additional presence of 5 mM K+ chelator [2,2,2]cryptand gave the same difference spectra.
Extent of conformational change
The absorbance changes we observed are very small. The amplitude of the largest band in the amide I region in Fig. 2 a has only 0.3% of the maximum protein absorbance. This region reflects the conformation of the protein backbone and can be used to estimate the net secondary structure change that is associated with a protein reaction. From the spectrum shown in Fig. 2 a, we estimated the net secondary structure change in the reaction E1Na3+ → E2P. We used the COBSI index (38), which relates the band area in the difference spectrum to the band area of the absorption spectrum. The index gives the relative band area that is redistributed in the reaction. For a uniform secondary structure change that involves all residues, a COBSI index of ∼0.4 is expected. The COBSI index for the full line spectrum in Fig. 2 a is 6.3 × 10−4. The COBSI index may become larger when the observed lower hydrolytic activity in infrared samples is taken into account. This inhibition may or may not decrease the amide I signals, depending on its cause. For example, if the high viscosity slows down partial reactions of the pump cycle in infrared samples without affecting the conformations of the intermediate states, no reduction in the extent of the observed conformational change is expected. If, however, binding of caged ATP blocks access of released ATP to the active site, the number of active ATPases will be reduced and the observed signals will be smaller. In the following, we will discuss a worst-case scenario by assuming that the signals in the amide I region are reduced to ∼15%, the average value of our activity measurements in infrared samples. Hydrolytic activity was measured in the presence of KCl, and the respective spectra are shown in Fig. 3. The omission of KCl increases the infrared signals in the amide I region by a factor of ∼2 (compare Figs. 2 and 3), implying that the signal amplitudes in Fig. 2 are at least 30% of those theoretically expected in the case of full activity of the infrared samples. The same band amplitudes were observed at 5°C (40). Thus the expected COBSI index for fully active samples in this worst-case scenario is 3.3 times the observed value, i.e., 2 × 10−3. This is 200-fold smaller than it would be for a 100% secondary structure change, indicating that <1% of the amino acids contribute to a net change in secondary structure. The actual number of amino acids experiencing a change in secondary structure may be larger if some changes are compensated for by others.
Similar COBSI indices have been obtained for the Ca2+-ATPase (38), which undergoes a rearrangement of its domains under catalysis. Most of these changes can be described as rigid-body movements of domains or of secondary structure elements that are connected by small, flexible hinges. The COBSI index is not sensitive to the movement of rigid domains itself. Thus the small COBSI indices are not in contrast to the current structural models and give realistic values, as discussed previously (51–53). We conclude that our data demonstrate a largely preserved secondary structure composition in the transition from E1Na3+ with bound caged ATP to an E2P state. This is consistent with the assumption of rigid-body movements of domains and protein segments in the catalytic cycle of the Na+,K+-ATPase.
Our finding of absorbance changes of 1% of the maximum protein absorbance (allowing for the reduced activity in our infrared samples) are in line with several studies that analyzed the infrared absorption spectrum and found only small secondary structure changes (15–17). However, they seem to be in contrast to a recent work by Pratap et al. (20), who used the same experimental approach. They, found absorbance changes corresponding to 10% of the maximum absorbance with the enzyme from the salt glands of ducks upon photolysis of dimethoxybenzoin (DMB) caged ATP in 2H2O and in the presence of ∼80 mM NaCl and ∼2.5 mM EDTA. The amide I′ absorbance changes in the study presented here increased slowly to a value of 0.1 on a timescale of 2 h. The end point of the reaction was not reached after 2 h. The maximum absorbance change observed here was 0.0015, or 0.005 after correction for the reduced activity according to the worst-case scenario. The difference spectrum in the previous study was attributed to ATP binding and showed a negative band at 1659 cm−1 and a positive band at 1620 cm−1, consistent with a considerable conversion of α-helices to β-sheets. The authors concluded that their absorbance changes were at least one order of magnitude larger than those observed for other P-type ATPases (38,54), possibly due to the use of duck enzyme or DMB caged ATP, which has a low affinity for the Na+,K+-ATPase (55).
Our study is not directly comparable to that of Pratap et al. (20) because we investigated the reaction from E1Na3+ with bound NPE caged ATP to E2P, whereas the previous study followed the reaction from E1Na3+ to the E1Na3+ complex with ATP (E1ATP Na3+). However, it seems unlikely that ATP binding would produce secondary structure changes that are more than 10 times larger than the transition from the E1 form of the enzyme to the E2P form. Nor has this been observed for the Ca2+-ATPase, where the extent of net secondary structure change upon ATP binding was reported to be similar to that of phosphoenzyme conversion from Ca2E1P to E2P (38). The enzyme source is not an explanation for the apparent discrepancy between the two studies, since our experiments with the duck enzyme gave spectra that were similar in band position and band amplitude to those of the pig kidney enzyme shown here.
A further explanation could be that NPE caged ATP (used here) induces large structural changes when it binds to the Na+,K+-ATPase in the E1Na3+ state, making our initial state similar to our transient state E2P. Photolysis of caged ATP would then lead to only relatively small changes upon formation of E2P. In that case, our initial state likely would resemble that of E1ATP Na3+, again implying much larger conformational changes upon nucleotide binding than upon phosphorylation and phosphoenzyme conversion to E2P.
NPE caged ATP binding to the Ca2+-ATPase does not explain the discrepancy between our previous finding of small net secondary structure changes for the Ca2+-ATPase and that of Pratap et al. (20) of large changes on the Na+,K+-ATPase. This is because caged ATP does not bind to the Ca2+-ATPase under our conditions, as described for the close ATP analog β,γ-imidoadenosine 5′-triphosphate (AMPPNP) (56), because of the lower affinity of NPE caged ATP for the Ca2+-ATPase (45,57–59), which is ∼100–150 μM in diluted suspensions (57).
At present, the discrepancy between our study and that by Pratap et al. (20) remains unexplained in molecular terms. However, we would like to point out that our infrared signals show the expected kinetic behavior of a transient change, and that our controls prove that the transient absorbance changes are due to a structural change of the active ATPase.
Molecular interpretation
Carbonyl groups of acids, lipids, and the aspartyl phosphate absorb between 1800 and 1700 cm−1. In this region, three bands are observed: positive bands at 1745 and 1715 cm−1, and a negative band at 1730 cm−1. The band position of carbonyl groups is sensitive to hydrogen bonding and indicates no or weak hydrogen bonding for the 1745 cm−1 band, intermediate hydrogen bonding for the 1730 cm−1 band, and hydrogen bonding as strong as found in aqueous solution for the 1716 cm−1 band (60,61). These bands may originate from the following causes: An environmental change around carbonyl groups of lipids or protonated carboxyl groups may lead to a band shift in this spectral region, giving rise to a positive and a negative band. The negative and positive bands are characteristic of the initial state and the state that accumulates after ATP release, respectively. Alternatively, the bands may appear because carbonyl groups are formed or disappear in the observed reaction, for example, by protonation/deprotonation of carboxylate groups or phosphorylation of Asp-369. Formation of carbonyl groups causes positive bands, whereas their disappearance results in negative bands.
The carbonyl group of the aspartyl phosphate is expected to contribute to one of the positive bands. The model compound acetyl phosphate absorbs at 1718 cm−1 in aqueous solution (32). At a similar position, a band has been tentatively assigned to the aspartyl phosphate of the Ca2+-ATPase (33). This assignment is supported by the sensitivity of this band to the divalent cation that binds at the catalytic site (33,62). In analogy, we tentatively assign the 1716 cm−1 band or part of it to the carbonyl group of the aspartyl phosphate of Na+,K+-ATPase. The remaining absorbance changes in the carbonyl region can be due either to changes in hydrogen bonding or to protonation (positive bands) and deprotonation (negative bands) of carboxyl groups. At present, it is not possible to distinguish between these possibilities. However, the involvement of carboxyl groups in the reaction seems to be evident since signals are also observed in the spectral regions of the antisymmetric (1600–1520 cm−1) and symmetric (∼1400 cm−1) stretching vibrations of the carboxylate group. Again, they can be due to protonation (negative bands), deprotonation (positive bands), or an environmental change (e.g., a change in cation coordination).
The amide I region (1700–1610 cm−1) is adjacent to the carbonyl region. It is usually dominated by the absorption of the amide I vibration, which is predominantly a vibration of the amide carbonyl group. The amide I vibration is sensitive to the three-dimensional structure of the protein backbone and depends in particular on the secondary structure. One can tentatively assign the 1686 cm−1 band to turns or β-sheets, the 1676 cm−1 band to turns, the 1661 cm−1 band to turns or α-helices, and the 1641 cm−1 band to α-helices, irregular structure, or β-sheets. It is tempting to speculate that the 1661 and 1641 cm−1 bands originate from a band shift from a lower to a higher wave number. In this case, the band pair could reflect straightening (63,64), shortening (65), or less solvent exposure (66–68) of an α-helix. We note that side chains such as Asn, Gln, and Arg may also contribute in the amide I region.
Comparison with other P-type ATPases
A spectrum very similar to the one shown in Fig. 2 a was previously obtained for the E1-E2 transition of the gastric H+,K+-ATPase (54). The E1 and E2 forms of this enzyme were stabilized by Na+ and K+, respectively, and rise to a negative E1 band at 1640 cm−1 and a positive E2 band at 1654 cm−1. The respective bands for the E1Na3+ to E2P transition of the Na+,K+-ATPase are found at 1661 cm−1 and 1641 cm−1. As the Na+,K+-ATPase, the H+,K+-ATPase contains a β-subunit. The similarity between the spectra indicates that the main bands at 1661 cm−1 and 1641 cm−1 are characteristic of the E2 and E1 conformations, respectively.
Spectra of the corresponding reaction of the Ca2+-ATPase (Ca2E1 → E2P) have been published (33,34,38). They show some similarities in the carbonyl region of the spectrum (1800–1700 cm−1). The Ca2+-ATPase exhibits two broad positive bands with maxima at 1758 and 1710 cm−1 that are composed of at least four different bands (34). At similar positions, two positive bands at 1747 and 1716 cm−1 were observed for the Na+,K+-ATPase in this study. In contrast to the observation of a clear negative band at 1730 cm−1 for the Na+,K+-ATPase, the Ca2+-ATPase spectrum does not show negative bands in this region. This characteristic feature of the Na+,K+-ATPase has also been observed for duck and shark enzymes (40). This indicates that a lipid carbonyl or a Glu or Asp residue that is protonated in E1Na3+, is involved in the reaction from E1Na3+ to E2P. It is tempting to speculate that one or more of the Na+ coordinating acidic residues of sites I and II (pig kidney sequence according to the crystal structure: Glu-327, Glu-779, Asp-804, Asp-808) (13,69,70) is protonated in the Na+,K+-ATPase because the two positive charges bound to these sites in the Na+,K+-ATPase require less charge compensation than the four positive charges bound to the Ca2+-ATPase.
The amide I spectral range of the Na+,K+-ATPase spectrum is dissimilar to that of the Ca2+-ATPase spectrum. The only band common to both enzymes is the 1641 cm−1 band found at 1643 cm−1 for the Ca2+-ATPase. Above 1641 cm−1, both spectra are mirror images of each other. Where the Na+,K+-ATPase has positive bands at 1686 and 1661 cm−1, the Ca2+-ATPase has negative bands at 1687 and 1662 cm−1. Where the Na+,K+-ATPase has a negative band at 1676 cm−1, the Ca2+-ATPase has a positive band at 1671 cm−1. There are several possible explanations for these spectral differences:
-
1.
Opposite structural changes could occur in both enzymes. However, this explanation seems to be unlikely since a common reaction mechanism is usually assumed.
-
2.
Corresponding structural elements could absorb at different wavenumbers for the two enzymes because of different local structures or different strengths in hydrogen bonding to the amide groups. In line with this assumption, a relatively broad positive band is found at 1661 cm−1 and 1671 cm−1 for the Na+,K+-ATPase and Ca2+-ATPase, respectively. The similar bandwidths might indicate that the two bands are caused by corresponding structural elements.
-
3.
The β-and γ-subunits could contribute to the Na+,K+-ATPase spectrum. These subunits are not present in the Ca2+-ATPase.
Notable is the absence of a positive band near 1630 cm−1 for the Na+,K+-ATPase. This band appears prominently upon ATP binding to the Ca2+-ATPase and has tentatively been assigned (52) to a conformational change of the β-sheet in the P domain (71) that persists until dephosphorylation of E2P (72). The absence of the 1630 cm−1 band may indicate that the β-sheet is less perturbed upon phosphorylation of the Na+,K+-ATPase. An alternative explanation is that binding of NPE caged ATP to E1Na3+ already causes a structural change in this β-sheet. It would then be altered in the initial state of our Na+,K+-ATPase samples but not in the Ca2+-ATPase samples because NPE caged ATP does not bind to the Ca2+-ATPase under our conditions, as found for the close ATP analog AMPPNP (56).
Conclusions
We obtained infrared difference spectra of the Na+,K+-ATPase for the reaction from E1Na3+ with bound NPE caged ATP to E2P. The spectra show highly detailed absorbance changes that can be attributed to structural changes of the enzyme based on a number of control experiments. The reactions studied here affect protonated carboxyl groups or lipid carbonyl groups. The data do not support a previous finding that the conformational changes of the Na+,K+-ATPase are exceptionally large (20).
Acknowledgments
We thank A. Schacht for enzyme preparation, and J. Reichert for construction of the sample changer.
References
- 1.Skou J.C. The influence of some cations on an adenosine triphosphatase from pheripheral nerves. Biochim. Biophys. Acta. 1957;23:394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- 2.Rolfe D.F.S., Brown G.C. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 3.Axelsen K.B., Palmgren M.G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 4.Hasselbach W., Makinose M. Die Calciumpumpe der “Erschlaffungsgrana” des Muskels und ihre Abhängigkeit von der ATP-Spaltung [The calcium pump of the relaxing granules of muscle and its dependence on ATP splitting] Biochem. Z. 1961;333:518–528. [PubMed] [Google Scholar]
- 5.Geering K. The functional role of β subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- 6.Therien A.G., Blostein R. Mechanism of sodium pump regulation. Am. J. Physiol. Cell Physiol. 2000;279:C541–C566. doi: 10.1152/ajpcell.2000.279.3.C541. [DOI] [PubMed] [Google Scholar]
- 7.Garty H., Karlish S.J.D. Role of FXYD proteins in ion transport. Annu. Rev. Physiol. 2006;68:431–459. doi: 10.1146/annurev.physiol.68.040104.131852. [DOI] [PubMed] [Google Scholar]
- 8.Albers R.W. Biochemical aspects of active transport. Annu. Rev. Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 9.Post R.L., Kume S., Tobin T., Orcutt B., Sen A.K. Flexibility of an active center in sodium-plus-potassium adenosine triphosphatase. J. Gen. Physiol. 1969;54:306–326. doi: 10.1085/jgp.54.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen P.L., Pedersen P.A. Structure-function relationships of Na+, K+, ATP, or Mg2+ binding and energy transduction in Na,K-ATPase. Biochim. Biophys. Acta. 2001;1505:57–74. doi: 10.1016/s0005-2728(00)00277-2. [DOI] [PubMed] [Google Scholar]
- 11.Skou J.C., Esmann M. The Na,K-ATPase. J. Bioenerg. Biomembr. 1992;24:249–261. doi: 10.1007/BF00768846. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen P.L., Håkansson K.O., Karlish S.J.D. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- 13.Morth J.P., Pedersen B.P., Toustrup-Jensen M.S., Sørensen T.L.-M., Petersen J. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 14.Toyoshima C., Nakasako M., Nomura H., Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 15.Chetverin A.B., Brazhnikov E.V. Do sodium and potassium forms of Na,K-ATPase differ in their secondary structure? J. Biol. Chem. 1985;260:7817–7819. [PubMed] [Google Scholar]
- 16.Neault J.F., Malonga H., Diamantoglou S., Carpentier R., Stepp R.L. Secondary structural analysis of the Na+,K+-ATPase and its Na+ (E1) and K+ (E2) complexes by FTIR spectroscopy. J. Biomol. Struct. Dyn. 2002;20:173–178. doi: 10.1080/07391102.2002.10506833. [DOI] [PubMed] [Google Scholar]
- 17.Chetverin A.B., Venyaminov S.Y., Emelyanenko V.I., Burstein E.A. Lack of gross protein structure changes in the working cycle of (Na+,K+)-dependent adenosinetriphosphatase. Eur. J. Biochem. 1980;108:149–156. doi: 10.1111/j.1432-1033.1980.tb04706.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanfani F., Linnertz H., Obsil T., Krumscheid R., Urbanova P. Effects of fluorescent pseudo-ATP and ATP-metal analogs on secondayr structure of Na+/K+-ATPase. Biochim. Biophys. Acta. 2000;1457:94–102. doi: 10.1016/s0005-2728(00)00055-4. [DOI] [PubMed] [Google Scholar]
- 19.Fringeli U.P., Apell H.-J., Fringeli M., Läuger P. Polarized infrared absorption of Na+/K+-ATPase studied by attenuated total reflection spectroscopy. Biochim. Biophys. Acta. 1989;984:301–312. doi: 10.1016/0005-2736(89)90297-6. [DOI] [PubMed] [Google Scholar]
- 20.Pratap P.R., Dediu O., Nienhaus G.U. FTIR study of ATP-induced changes in Na+/K+-ATPase from duck supraorbital glands. Biophys. J. 2003;85:3707–3717. doi: 10.1016/S0006-3495(03)74787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolz M., Lewitzki E., Mäntele W., Barth A., Grell E. Inhibition and partial reactions of Na,K-ATPase studied by Fourier transform infrared difference spectroscopy. Biopolymers. 2005;82:368–372. doi: 10.1002/bip.20427. [DOI] [PubMed] [Google Scholar]
- 22.Barth A., Mäntele W., Kreutz W. Molecular changes in the sarcoplasmic reticulum Ca2+ ATPase during catalytic activity. A Fourier transform infrared (FTIR) study using photolysis of caged ATP to trigger the reaction cycle. FEBS Lett. 1990;277:147–150. doi: 10.1016/0014-5793(90)80830-c. [DOI] [PubMed] [Google Scholar]
- 23.Barth A. Structural dynamics of the Ca2+-ATPase studied by time-resolved infrared spectroscopy. Spectroscopy. 2008;22:63–82. [Google Scholar]
- 24.Scheirlinckx F., Buchet R., Ruysschaert J.-M., Goormaghtigh E. Monitoring of secondary and tertiary structure changes in the gastric H+/K+-ATPase by infrared spectroscopy. Eur. J. Biochem. 2001;268:3644–3653. doi: 10.1046/j.1432-1327.2001.02266.x. [DOI] [PubMed] [Google Scholar]
- 25.Barth A. Selective monitoring of 3 out of 50,000 protein vibrations. Biopolymers. 2002;67:237–241. doi: 10.1002/bip.10085. [DOI] [PubMed] [Google Scholar]
- 26.Barth A., Bezlyepkina N. P-O bond destabilization accelerates phosphoenzyme hydrolysis of sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 2004;279:51888–51896. doi: 10.1074/jbc.M410867200. [DOI] [PubMed] [Google Scholar]
- 27.Jørgensen P.L. Purification and characterisation of (Na+ + K+)-ATPase. 3. Purification from outer medulla of mammalian kidney after selective removal of membrane components by sodium dodecylsulfate. Biochim. Biophys. Acta. 1974;356:36–52. doi: 10.1016/0005-2736(74)90292-2. [DOI] [PubMed] [Google Scholar]
- 28.Jørgensen P.L. Purification and characterisation of (Na+ + K+)-ATPase. 4. Estimation of purity and of molecular-weight and polypeptide content per enzyme unit in preparations from outer medulla of rabbit kidney. Biochim. Biophys. Acta. 1974;356:53–67. doi: 10.1016/0005-2736(74)90293-4. [DOI] [PubMed] [Google Scholar]
- 29.Grell E., Schick E., Lewitzki E. Membrane receptor calorimetry: cardiac glycoside interaction with Na,K-ATPase. Thermochim Acta. 2001;380:245–254. [Google Scholar]
- 30.Walker J.W., Reid G.P., McCray J.A., Trentham D.R. Photolabile 1-(2-nitrophenyl)ethyl phosphate esters of adenine nucleotide analogues. Synthesis and mechanism of photolysis. J. Am. Chem. Soc. 1988;110:7170–7177. [Google Scholar]
- 31.Corrie J.E.T. Synthesis of [15N] and [side-chain 1-13C] isotopomers of 1-(2-nitrophenyl)ethyl phosphates. J. Labelled Comp. Radiopharm. 1996;36:289–300. [Google Scholar]
- 32.Barth A., Mäntele W. ATP-induced phosphorylation of the sarcoplasmic reticulum Ca2+ ATPase: molecular interpretation of infrared difference spectra. Biophys. J. 1998;75:538–544. doi: 10.1016/S0006-3495(98)77543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barth A., Kreutz W., Mäntele W. Changes of protein structure, nucleotide microenvironment, and Ca2+ binding states in the catalytic cycle of sarcoplasmic reticulum Ca2+ ATPase: investigation of nucleotide binding, phosphorylation and phosphoenzyme conversion by FTIR difference spectroscopy. Biochim. Biophys. Acta. 1994;1194:75–91. doi: 10.1016/0005-2736(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 34.Andersson J., Hauser K., Karjalainen E.-L., Barth A. Protonation and hydrogen bonding of Ca2+ site residues in the E2P phosphoenzyme intermediate of sarcoplasmic reticulum Ca2+-ATPase studied by a combination of infrared spectroscopy and electrostatic calculations. Biophys. J. 2008;94:600–611. doi: 10.1529/biophysj.107.114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth A., Corrie J.E.T., Gradwell M.J., Maeda Y., Mäntele W. Time-resolved infrared spectroscopy of intermediates and products from photolysis of 1-(2-nitrophenyl)ethyl phosphates: reaction of the 2-nitrosoacetophenone byproduct with thiols. J. Am. Chem. Soc. 1997;119:4149–4159. [Google Scholar]
- 36.Von Germar F., Barth A., Mäntele W. Structural changes of the sarcoplasmic reticulum Ca2+-ATPase upon nucleotide binding studied by Fourier transform infrared spectroscopy. Biophys. J. 2000;78:1531–1540. doi: 10.1016/S0006-3495(00)76705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thoenges D., Barth A. Direct measurement of enzyme activity with infrared spectroscopy. J. Biomol. Screen. 2002;7:353–357. doi: 10.1177/108705710200700407. [DOI] [PubMed] [Google Scholar]
- 38.Barth A., von Germar F., Kreutz W., Mäntele W. Time-resolved infrared spectroscopy of the Ca2+-ATPase. The enzyme at work. J. Biol. Chem. 1996;271:30637–30646. doi: 10.1074/jbc.271.48.30637. [DOI] [PubMed] [Google Scholar]
- 39.Karlish S.J.D. Characterisation of conformational changes in (Na,K) ATPase labeled with fluorescein at the active side. J. Bioenerg. Biomembr. 1980;12:111–136. doi: 10.1007/BF00744678. [DOI] [PubMed] [Google Scholar]
- 40.Stolz, M. The pump cycle of the Na,K-ATPase: a characterization of changes in secondary structure and protein micro-environment with reaction-induced and time-resolved FTIR-difference spectrosocopy. PhD thesis. Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany. 2006.
- 41.Thoenges D., Zscherp C., Grell E., Barth A. Preparation of active enzyme samples for IR studies of Na+/K+-ATPase. Biopolymers. 2002;67:271–274. doi: 10.1002/bip.10108. [DOI] [PubMed] [Google Scholar]
- 42.Fiske C.H., Subbarow Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 43.Forbush B. Na+ movement in a single turnover of the Na pump. Proc. Natl. Acad. Sci. USA. 1984;81:5310–5314. doi: 10.1073/pnas.81.17.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke R.J., Kane D.J., Apell H.J., Roudna M., Bamberg E. Kinetics of Na+-dependent conformational changes of rabbit kidney Na+,K+-ATPase. Biophys. J. 1998;75:1340–1353. doi: 10.1016/S0006-3495(98)74052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel G., Fendler K., Grell E., Bamberg E. Na+ currents generated by the purified (Na+ + K+)-ATPase on planar lipid membranes. Biochim. Biophys. Acta. 1987;901:239–249. doi: 10.1016/0005-2736(87)90120-9. [DOI] [PubMed] [Google Scholar]
- 46.Grell E., Geoffroy A., Stolz M., Lewitzki E., von Raumer M. Membrane proteins in thin films. A DSC study. J. Therm. Anal. Calorim. 2007;89:723–727. [Google Scholar]
- 47.Kuriki Y., Racker E. Inhibition of (Na+,K+)adenosine triphosphatase and its partial reactions by quercetin. Biochemistry. 1976;15:4951–4956. doi: 10.1021/bi00668a001. [DOI] [PubMed] [Google Scholar]
- 48.Nørby J.G., Klodos I., Christiansen N.O. Kinetics of Na-ATPase activity by the Na, K Pump. Interactions of the phosphorylated intermediates with Na+, Tris+, and K+ J. Gen. Physiol. 1983;82:725–759. doi: 10.1085/jgp.82.6.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornelius F., Fedosova N.U., Klodos I. E2P phosphoforms of Na,K-ATPase. II. Interaction of substrate and cation-binding sites in π phosphorylation of Na,K-ATPase. Biochemistry. 1998;37:16686–16696. doi: 10.1021/bi981571v. [DOI] [PubMed] [Google Scholar]
- 50.Lüpfert C., Grell E., Pintschovius V., Apell H.J., Cornelius F. Rate limitation of Na+,K+-ATPase pump cycle. Biophys. J. 2001;81:2069–2081. doi: 10.1016/S0006-3495(01)75856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barth A. Infrared spectroscopy. In: Uversky V.N., Permyakov E.A., editors. Methods in Protein Structure and Stability Analysis: Vibrational Spectroscopy. Nova Science Publishers; New York: 2007. pp. 69–151. [Google Scholar]
- 52.Liu M., Barth A. Phosphorylation of the sarcoplasmic reticulum Ca2+-ATPase from ATP and ATP analogs studied by infrared spectroscopy. J. Biol. Chem. 2004;279:49902–49909. doi: 10.1074/jbc.M408062200. [DOI] [PubMed] [Google Scholar]
- 53.Barth A., Zscherp C. What vibrations tell us about proteins. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 54.Scheirlinckx F., Raussens V., Ruysschaert J.-M., Goormaghtigh E. Conformational changes in gastric H+/K+-ATPase monitored by difference Fourier-transform infrared spectroscopy and hydrogen/deuterium exchange. Biochem. J. 2004;382:121–129. doi: 10.1042/BJ20040277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokolov V.S., Apell H.J., Corrie J.E., Trentham D.R. Fast transient currents in Na,K-ATPase induced by ATP concentration jumps from the P3-[1-(3′,5′-dimethoxyphenyl)-2-phenyl-2-oxo]ethyl ester of ATP. Biophys. J. 1998;74:2285–2298. doi: 10.1016/S0006-3495(98)77938-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M., Barth A. TNP-AMP binding to the sarcoplasmic reticulum Ca2+-ATPase studied by infrared spectroscopy. Biophys. J. 2003;85:3262–3270. doi: 10.1016/S0006-3495(03)74744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartung K., Froehlich J.P., Fendler K. Time-resolved charge translocation by the Ca-ATPase from sarcoplasmic reticulum after an ATP concentration jump. Biophys. J. 1997;72:2503–2514. doi: 10.1016/S0006-3495(97)78895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartung K., Grell E., Hasselbach W., Bamberg W. Electrical pump currents generated by the Ca2+-ATPase of sarcoplasmic reticulum vesicles absorbed on black lipid membranes. Biochim. Biophys. Acta. 1987;900:209–220. doi: 10.1016/0005-2736(87)90335-x. [DOI] [PubMed] [Google Scholar]
- 59.McCray J.A., Herbette L., Kihara T., Trentham D.R. A new approach to time-resolved studies of ATP-requiring biological systems: laser flash photolysis of caged ATP. Proc. Natl. Acad. Sci. USA. 1980;77:7237–7241. doi: 10.1073/pnas.77.12.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dioumaev A.K., Braiman M.S. Modeling vibrational spectra of amino acid side chains in proteins: the carbonyl stretch frequency of buried carboxylic residues. J. Am. Chem. Soc. 1995;117:10572–10574. [Google Scholar]
- 61.Chirgadze Y.N., Fedorov O.V., Trushina N.P. Estimation of amino acid residue side chain absorption in the infrared spectra of protein solutions in heavy water. Biopolymers. 1975;14:679–694. doi: 10.1002/bip.1975.360140402. [DOI] [PubMed] [Google Scholar]
- 62.Andersson J., Barth A. FTIR studies on the bond properties of the aspartyl phosphate moiety of the Ca2+-ATPase. Biopolymers. 2006;82:353–357. doi: 10.1002/bip.20436. [DOI] [PubMed] [Google Scholar]
- 63.Heimburg T., Schuenemann J., Weber K., Geisler N. Specific recognition of coiled coils by infrared spectroscopy: analysis of the three structural domains of type III intermediate filament proteins. Biochemistry. 1996;35:1375–1382. doi: 10.1021/bi9515883. [DOI] [PubMed] [Google Scholar]
- 64.Reisdorf W.C., Krimm S. Infrared amide I′ band of the coiled coil. Biochemistry. 1996;35:1383–1386. doi: 10.1021/bi951589v. [DOI] [PubMed] [Google Scholar]
- 65.Nevskaya N.A., Chirgadze Y.N. Infrared spectra and resonance interactions of amide-I and II vibrations of α-helix. Biopolymers. 1976;15:637–648. doi: 10.1002/bip.1976.360150404. [DOI] [PubMed] [Google Scholar]
- 66.Mukherjee S., Chowdhury P., Gai F. Infrared study of the effect of hydration on the amide I band and aggregation properties of helical peptides. J. Phys. Chem. B. 2007;111:4596–4602. doi: 10.1021/jp0689060. [DOI] [PubMed] [Google Scholar]
- 67.Manas E.S., Getahun Z., Wright W.W., DeGrado W.F., Vanderkooi J.M. Infrared spectra of amide groups in α-helical proteins: evidence for hydrogen bonding between helices and water. J. Am. Chem. Soc. 2000;122:9883–9890. [Google Scholar]
- 68.Walsh S.T.R., Cheng R.P., Wright W.W., Alonso D.O.V., Daggett V. The hydration of amides in helices; a comprehensive picture from molecular dynamics, IR, and NMR. Protein Sci. 2003;12:520–531. doi: 10.1110/ps.0223003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogawa H., Toyoshima C. Homology modeling of the cation binding sites of Na+K+-ATPase. Proc. Natl. Acad. Sci. USA. 2002;99:15977–15982. doi: 10.1073/pnas.202622299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li C., Capendeguy O., Geering K., Horisberger J.-D. A third Na+-binding site in the sodium pump. Proc. Natl. Acad. Sci. USA. 2005;102:12706–12711. doi: 10.1073/pnas.0505980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sørensen T.L.-M., Møller J.V., Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 72.Toyoshima C., Nomura H., Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]