Abstract
Cullin (Cul)-based E3 ubiquitin ligases are activated through the attachment of Nedd8 to the Cul protein. In yeast, Dcn1 (defective in Cul neddylation 1 protein) functions as a scaffold-like Nedd8 E3-ligase by interacting with its Cul substrates and the Nedd8 E2 Ubc12. Human cells express 5 Dcn1-like (DCNL) proteins each containing a C-terminal potentiating neddylation domain but distinct amino-terminal extensions. Although the UBA-containing DCNL1 and DCNL2 are likely functional homologues of yeast Dcn1, DCNL3 also interacts with human Culs and is able to complement the neddylation defect of yeast dcn1Δ cells. DCNL3 down-regulation by RNAi decreases Cul neddylation, and overexpression of a Cul3 mutant deficient in DCNL3 binding interferes with Cul3 function in vivo. Interestingly, DCNL3 accumulates at the plasma membrane through a conserved, lipid-modified motif at the N terminus. Membrane-bound DCNL3 is able to recruit Cul3 to membranes and is functionally important for Cul3 neddylation in vivo. We conclude that DCNL proteins function as nonredundant Cul Nedd8-E3 ligases. Moreover, the diversification of the N termini in mammalian Dcn1 homologues may contribute to substrate specificity by regulating their subcellular localization.
Keywords: Cullin, DCUN1D, Nedd8, ubiquitin, squamous cell carcinoma-related oncogene
Ubiquitination is a major protein modification that targets specific proteins for degradation by 26S proteasomes (1). Substrate specificity is achieved mainly through the recognition of target proteins by ubiquitin E3 ligases. Cullin (Cul)-Ring-Ligases (CRLs) comprise a large class of E3 ligases (2), which are assembled around a Cul scaffold that interacts through conserved regions with substrate adaptors and the ring-finger protein Rbx1.
The assembly and activity of Cul-based ligases is regulated through reversible conjugation of Nedd8, a ubiquitin-like protein, which is covalently attached to a conserved lysine residue in the Cul backbone (3). In mammals, this modification, termed neddylation, is essential for CRL activity by at least two mechanisms. First, it promotes assembly of the Cul complex by dissociating the assembly inhibitor CAND1 (4). Second, Cul neddylation enhances the recruitment of the activated E2 ubiquitin-conjugating enzyme to the Cul ligase (5) by inducing a conformational change in its carboxyl-terminal domain (6, 7).
Cullins are not the only targets that are regulated by Nedd8 modification (3). For example, neddylation has been reported to inhibit the transcriptional activation of the tumor suppressor p53 and the breast cancer-associated protein 3 (BCA3) (8, 9). Like ubiquitination, neddylation of substrates is achieved by an enzymatic cascade involving the Nedd8-activating enzyme APP-BP1(ULA1)/UBA3 and the Nedd8-conjugating enzyme encoded by Ubc12 (3). Although the RING-finger protein Rbx1 promotes Cul neddylation (10–12), recent data suggest that Dcn1 (defective in Cul neddylation 1) functions as an E3 ligase for Cul neddylation in yeast and Caenorhabditis elegans (13). Indeed, yeast Dcn1 directly binds to the Cul and the Nedd8 E2 enzyme and promotes Nedd8 conjugation through formation of this complex (14, 15). Human cells harbor 5 Dcn1-like proteins termed DCNL1–DCNL5 (also named DCUN1D 1–5 for defective in Cul neddylation 1 domain-containing protein 1–5) (Fig. S1). These DCNLs have distinct amino-terminal domains, but share a conserved C-terminal potentiating neddylation (PONY) domain, which in yeast Dcn1 is necessary and sufficient for Cul neddylation in vivo and in vitro (14). The Cul interaction surface at the C terminus of the PONY domain, the DAD patch (D226, A253, D259 in scDcn1), is conserved in all human DCNLs. Like yeast Dcn1, DCNL1 and DCNL2 harbor a predicted amino-terminal UBA domain, which directly binds ubiquitin. DCNL1 is highly amplified in various tumors including squamous cell carcinomas and based on this finding is also referred to as SCCRO (squamous cell carcinoma-related oncogene) (16). In contrast, DCNL3 levels appear to decrease in liver, bladder, and renal tumors (17), suggesting that modulation of DCNLs may be important for cancer development. Although DCNL1 was shown to neddylate Culs (14, 18), the functional significance and targets of other DCNLs remain to be investigated.
In this study, we functionally characterized the human DCNL proteins with respect to Cul neddylation in vivo and in vitro. We found that DCNL1, DCNL2, and DCNL3 interact with Culs and modulate Cul neddylation in a nonredundant manner. Unlike DCNL1 and DCNL2, DCNL3 localizes to the plasma membrane through a conserved amino-terminal membrane motif. Our results suggest that this membrane localization is essential for DCNL3-mediated Cul3 neddylation in vivo, implying that Cul3 may be activated at least in part at membranes.
Results
DCNL3 Promotes Cul Neddylation in Vivo in a Nonredundant Manner.
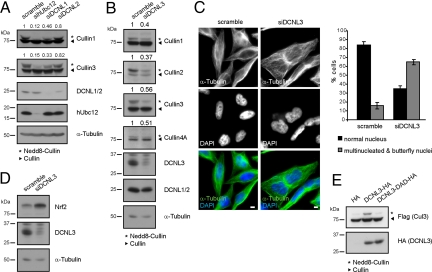
Like yeast Dcn1, human DCNL1 and DCNL2 possess an amino-terminal UBA domain and a conserved Cul interaction domain (DAD patch), which was required for its interaction with multiple Culs [Figs. S1A and S2 A and B (14, 18)]. Moreover, both DCNL1 and DCNL2 coimmunoprecipitated hUbc12 (Fig. S2C), and their down-regulation by RNAi decreased the neddylation status of Cul1 and Cul3 (Fig. 1A and Fig. S3 A and C for statistics), implying that DCNL1 and to some extent DCNL2 promote Cul neddylation in vivo.
Fig. 1.
Human DCNL proteins function in a nonredundant manner for Cul neddylation in vivo. (A) HeLa cells were transfected with control siRNA (scramble) or siRNA duplexes that specifically down-regulate hUbc12, DCNL1, or DCNL2. After 72 h, the neddylation state of Cul1 and Cul3 was analyzed by immunoblotting, and the ratio between neddylated and unneddylated Cul protein was quantified by LI-COR and normalized to scramble siRNA controls. The mean ratio from 4 independent experiments is indicated (see Fig. S3C for statistics). DCNL1 and DCNL2 comigrate and were detected with an antibody that recognizes both proteins (see Fig. S4C); mRNA levels were quantified by quantitative RT-PCR (Fig. S3A). (B) HeLa cells were transfected with scramble control siRNA or DCNL3 siRNA duplexes, and the neddylation state of Cul1–4 was analyzed after 72 h as described in A. The corresponding statistical analysis and mRNA quantifications are listed in Fig. S3 B and C. (C) HeLa cells transfected with scramble (Left) or DCNL3-specific siRNA duplexes (Right) were analyzed after 5 days by indirect immunofluorescence and DAPI staining. Multinucleated and butterfly nuclei were quantified from 3 independent experiments (bar graph). (Bars: 5 μm.) (D) Extracts from cells depleted for DCNL3 or treated with scramble RNAi controls (from B) were analyzed by immunoblotting for the Cul3 substrate Nrf2. (E) HA-tagged DCNL3 or the DCNL3 DAD-patch mutant were overexpressed in HeLa cells together with Flag-tagged Cul3. Total cell extracts were analyzed by immunoblotting with specific antibodies.
In addition to DCNL1 and DCNL2, human cells express other DCNL-members, which all share a conserved PONY domain but lack an amino-terminal UBA motif (Fig. S1A). Like DCNL1 and DCNL2, ectopic expression of DCNL3 and DCNL5 restored neddylation of Cdc53 in dcn1Δ cells, implying that these isoforms also possess Cul neddylation activity in this heterologous assay (Fig. S3D). Indeed, RNAi depletion of DCNL3 strongly affected the neddylation status of several Culs, including Cul3 (Fig. 1B and Fig. S3 B and C for statistics; and see Fig. S4 B and C for specific antibodies). Furthermore, similar to cells lacking Cul3 activity (19), DCNL3 depletion resulted in the accumulation of multinucleated cells and cells with butterfly nuclei (Fig. 1C) and increased levels of the Cul3-substrate Nrf2 (20) (Fig. 1D). Conversely, overexpression of HA-tagged DCNL3 but not a DAD-patch mutant (D241A-A265R-D271A; Fig. S3E) significantly increased Cul3 neddylation (Fig. 1E), whereas overexpression of Rbx1 had no effect (Fig. S5A). Together, these data suggest that DCNL3 influences Cul3 activity in human cells, implying that DCNLs may regulate Cul neddylation in a nonredundant manner.
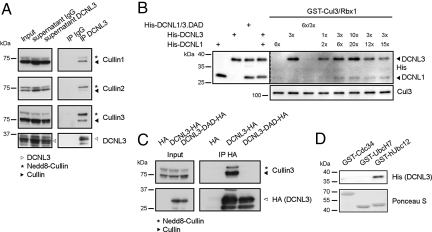
To corroborate these results, we immunoprecipitated endogenous DCNL3 and probed for specific association with endogenous Culs (Fig. 2A). Indeed, DCNL3 interacted with multiple Culs, including Cul3. Moreover, purified DCNL3 exhibited a higher affinity for GST-Cul3/His-Rbx1 complex purified from Sf9 cells compared with DCNL1 (Fig. 2B). As expected, in vitro and in vivo binding of DCNL3 and Cul3 depended on an intact DAD patch, confirming that the interaction is specific (Fig. 2 B and C). Finally, purified DCNL3 directly bound to GST-hUbc12 in vitro, but not to GST-UbcH7 or GST-Cdc34 controls (Fig. 2D), suggesting that DCNL3 specifically interacts with hUbc12 and Cul3 via a functional PONY domain.
Fig. 2.
DCNL3 directly binds Cul3 and hUbc12. (A) HeLa cell extracts were immunoprecipitated (IP) with specific DCNL3 antibodies or nonspecific control IgG and examined for associated Culs. An aliquot of total extract (input) and supernatants after immunoprecipitation were analyzed (Left). (B) The indicated ratio of purified His-DCNL3, His-DCNL1, and control His-DCNL-DAD mutants was incubated in vitro with Sf9 cell-expressed GST-Cul3/Rbx1 complexes. Bound proteins were visualized by immunoblotting with anti-His antibodies. Control input levels of DCNL1, DCNL3, and Cul3 are shown (Left and Lower). (C) HA-tagged wild-type and the DAD-patch mutant (D241A, A265R, D271A) of DCNL3 were immunoprecipitated (IP HA) from extracts prepared from stable 293T-Rex cell lines (input), and associated Cul3 was visualized with specific antibodies. (D) In vitro binding between recombinant 6×His-DCNL3 and GST-tagged hUbc12 or the ubiquitin-specific enzymes GST-Cdc34 and GST-UbcH7. The interaction was analyzed by immunoblotting with anti-His antibodies, while the input levels of the GST-tagged proteins were controlled by Ponceau S staining.
Localization of DCNL3 to the Plasma Membrane Is Mediated by a Conserved Amino-Terminal Motif and May Be Required for Cul3 Neddylation in Vivo.
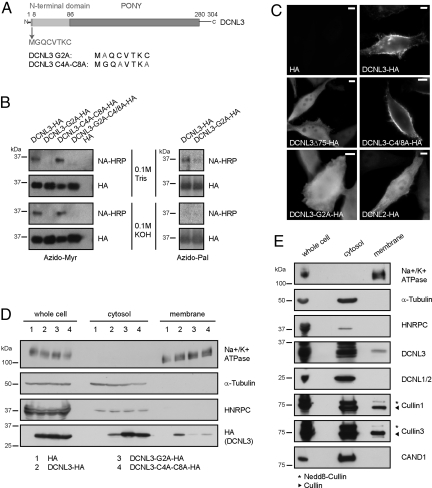
Bioinformatic analysis identified a conserved motif in the very N terminus of DCNL3-like proteins with the sequence M-G-(K/Q)-C-x-(S/T)-x-C (Fig. 3A and Fig. S5B), which strongly resembles the known acceptor sites for myristoylation (G2) and palmitoylation (C4 and C8) responsible for membrane targeting (SI Text and ref. 21). Indeed, HA-tagged DCNL3 expressed in COS-7 cells was specifically modified by the lipid analogues azido-myristate or azido-palmitate on the expected residues (Fig. 3B). Moreover, labeling of DCNL3-HA by acylbiotin exchange chemistry confirmed that the two cysteine residues are protected, indicating that they are palmitoylated in vivo (Fig. S5C).
Fig. 3.
Membrane localization of DCNL3 depends on lipid modifications of its N-terminal domain. (A) Schematic representation of DCNL3 highlighting the N-terminal domain of DCNL3 with the conserved membrane-targeting motif. The mutated amino acids in DCNL3-G2A and DCNL3-C4A-C8A used in the experiments are indicated. (B) COS-7 cells expressing wild-type or the indicated DCNL3-HA mutants were incubated with azido-myristate or azido-palmitate followed by immunoprecipitation with HA antibodies (HA). Conjugation of the lipid analogues to phosphine-biotin was detected by immunoblotting with neutravidin-HRP (NA-HRP). The membranes were treated for control with 0.1M Tris·HCl or 0.1 M KOH to remove thioester-linked palmitate but not N-linked myristate. Note that myristoylation at G2 is a prerequisite for palmitoylation of the Cys residues C4 and C8. (C) HeLa cells transfected with an HA-control plasmid (HA), HA-tagged DCNL2, wild-type, or the indicated DCNL3 mutants were analyzed by indirect immunofluorescence with HA-antibodies. (Bars: 5 μm.) (D) Whole-cell extracts were separated into membrane and cytosol fractions prepared from HeLa cells harboring an HA-control plasmid (lanes 1) or expressing DCNL3-HA (lanes 2), DCNL3-G2A-HA (lanes 3), or DCNL3-C4A-C8A-HA (lanes 4). The proteins were detected by immunoblotting with HA-antibodies. Antibodies specific for the following markers control the different fractions: Na+/K+ ATPase (plasma membrane), α-tubulin (cytosol), and HNRPC (heterogeneous nuclear ribonucleoprotein C, nucleus). One-tenth of the whole-cell extract was loaded compared with the cytosol and membrane fractions. (E) Whole cell, membrane, and cytosol fractions were prepared from HeLa cells, and the partitioning of endogenous DCNL3, DCNL1/2, Cul1, Cul3, and CAND1 was followed by immunoblotting with specific antibodies. The markers are described in D.
To examine the functional significance of these lipid modifications, we expressed HA-tagged DCNL3 and as a control DCNL2-HA in HeLa cells and analyzed the subcellular localization by immunofluorescence (Fig. 3C) and biochemical fractionation (Fig. 3D). Interestingly, DCNL3-HA was found at the plasma membrane, whereas DCNL2-HA was predominantly distributed throughout the cytoplasm and the nucleus. Moreover, cellular fractionation experiments revealed a significant portion of endogenous DCNL3 but not DCNL2 in the membrane fraction (Fig. 3E). In contrast to CAND1, a fraction of neddylated Cul1 and Cul3 was found at membranes (Fig. 3E), indicating that active Cul complexes may regulate membrane-associated processes. Interestingly, DCNL3 mutants deleted for its N-terminal domain (DCNL3-Δ75-HA) or harboring a specific point mutation that prevents myristoylation (DCNL3-G2A-HA) no longer accumulated at the plasma membrane, but localized throughout the cytoplasm (Fig. 3 C and D). The palmitoylation-defective DCNL3-C4A-C8A-HA mutant showed reduced membrane localization, suggesting that like in other cases palmitoylation may not be essential but contributes to stabilize DCNL3 at the plasma membrane (21).
DCNL3 Promotes Cul3 Neddylation at the Plasma Membrane.
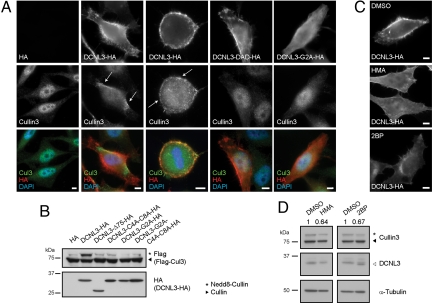
While endogenous Cul3 predominantly accumulated in the nucleus of untransfected cells or cells expressing an empty control vector (19), it was efficiently recruited to the plasma membrane in interphase and mitotic cells overexpressing DCNL3-HA (Fig. 4A). In contrast, no relocalization of Cul1 to the plasma membrane was observed (Fig. S5D), consistent with the preference of DCNL3 for Cul3 binding. Cul3 relocalization required an intact DAD patch in DCNL3-HA, implying that binding of Cul3 to DCNL3 triggered this process. Likewise, Cul3 only accumulated at the plasma membrane when DCNL3-HA contained a functional amino-terminal membrane-targeting motif (Fig. 4A and Fig. S5E), although a DCNL3 mutant protein lacking this domain (DCNL3-Δ75) efficiently interacted with Cul3 (Fig. S6A). Conversely, Cul3 fused to the first 11 aa of DCNL3 accumulated at the plasma membrane, indicating that the N-terminal motif of DCNL3 is not only necessary but also sufficient for membrane localization (Fig. S6B).
Fig. 4.
DCNL3 recruits endogenous Cul3 to the plasma membrane promoting Cul3 neddylation. (A) (Top) HeLa cells harboring an HA-control plasmid (HA) or expressing HA-tagged DCNL3, DCNL3-DAD, or DCNL3-G2A were analyzed by indirect immunofluorescence with HA-antibodies. (Middle) The localization of endogenous Cul3 was visualized with specific antibodies. (Bottom) The overlay pictures show DNA (DAPI, blue), DCNL (HA, red), and Cul (green) stainings. Arrows indicate the accumulation of Cul3 at plasma membranes. (Bars: 5 μm.) (B) HeLa cells were transfected with an HA-control plasmid (HA) or plasmids expressing HA-tagged wild type or the indicated DCNL3 mutants together with Flag-tagged Cul3. The neddylation state of Cul3 and DCNL3-HA expression was analyzed by immunoblotting. (C) HeLa cells were transfected with DCNL3-HA in the presence of different myristoylation and palmitoylation inhibitors (HMA and 2BP, respectively) or DMSO as a solvent control. Membrane localization of DCNL3 was detected by indirect immunofluorescence using HA antibodies. (Bars: 5 μm.) (D) The neddylation state of endogenous Cul3 in the presence of the myristoylation (HMA) and palmitoylation (2BP) inhibitors was quantified by LI-COR from immunoblots of 3 independent experiments. The ratio of neddylated over unnedddylated Cul3 in HeLa cells treated with the inhibitors was normalized to DMSO controls and the mean ratio is indicated (Fig. S6C for statistics). Immunoblotting for α-tubulin controls for equal loading.
To examine the functional importance of DCNL3 membrane localization for Cul3 neddylation, we compared the neddylation state of Cul3 in HeLa cells overexpressing wild-type or mutant forms of DCNL3-HA. Importantly, the potential to promote Cul3 neddylation in this assay correlated with the ability of DCNL3 to be targeted to membranes (Fig. 4B). To corroborate these findings, we quantified the ratio of neddylated and nonneddylated Cul3 in HeLa cells treated with different myristoylation [2-hydroxy-myristic acid (HMA)] and palmitoylation inhibitors [2-bromo-palmitate (2BP)], which as expected strongly reduced the accumulation of DCNL3-HA at the plasma membrane (Fig. 4C). Interestingly, the neddylated fraction of endogenous Cul3 significantly decreased after inhibitor treatment compared with solvent controls (Fig. 4D and Fig. S6C for statistics). Together, these results suggest that membrane localization of DCNL3 is essential for its function in Cul3 neddylation in vivo.
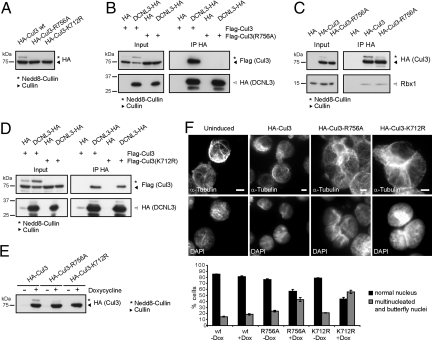
A Cul3 Mutant Unable to Interact with DCNL3 Is Not Neddylated and Inactive in Vivo.
To examine the functional relevance of the DCNL3–Cul3 interaction, we mutated the putative DCNL3 binding (R756) and the neddylation (K712) sites on Cul3 [Fig. S6D (14)]. Indeed, in contrast to wild-type HA-Cul3, no neddylated species of HA-tagged Cul3-K712R and Cul3-R756A were detected (Fig. 5A). As expected, the interaction of Cul3-R756A with DCNL3 was strongly reduced (Fig. 5B), whereas its interaction with Rbx1 was unaffected (Fig. 5C). Moreover, mutating the neddylation site on Cul3 did not affect binding to DCNL3 (Fig. 5D), indicating that DCNL3 binding to Cul3 may neither require Rbx1 nor Nedd8.
Fig. 5.
An intact DCNL-binding site is required for Cul3 neddylation and function in vivo. (A) The neddylation state of HA-tagged Cul3 and the Cul3-R756A and Cul3-K712R mutants expressed in HeLa cells was tested by immunoblotting of total cell extracts with HA antibodies. (B) Flag-Cul3 or Flag-Cul3-R756A were ectopically expressed in HeLa cells either together with a HA-control vector (HA) or a plasmid harboring DCNL3-HA. Cell extracts (input) were immunoprecipitated with HA-antibodies (IP HA) and associated Flag-Cul3 was detected with anti-Flag antibodies. (C) HA-Cul3, HA-Cul3-R756A, or an empty HA-vector control were expressed in HeLa cells and immunoprecipitated with HA-antibodies (IP HA; Upper). The association of endogenous Rbx1 was detected by immunoblotting (IP HA; Lower). Immunoblotting of an aliquot of the extract before immunoprecipitation controls for the expression of HA-Cul3 and HA-Cul3-R756A, respectively (Left). (D) Flag-Cul3 or Flag-Cul3-K712R were expressed in HeLa cells together with either an HA-control or DCNL3-HA, and coimmunoprecipitation was analyzed as described in B. (E) Extracts from 293T-Rex cells stably expressing HA-Cul3, HA-Cul3-R756A, or HA-Cul3-K712R were analyzed with HA-antibodies after induction with doxycycline (+) for 24 h. (F) 293T-Rex cells stably expressing HA-tagged Cul3, Cul3-R756A, or Cul3-K712R were grown for 10 days in the presence (+Dox) or absence (−Dox) of doxycycline. Cells were harvested onto coverslips by cytospin and analyzed for Cul3-like phenotypes by α-tubulin and DAPI staining. Multinucleated and butterfly nuclei were quantified from 3 independent experiments. (Bars: 5 μm.)
To test whether Cul3 neddylation is functionally important, we overexpressed the HA-tagged non-neddylatable Cul3 mutants Cul3-K712R and Cul3-R756A from the doxycycline-inducible promoter in 293T-Rex cells. As expected, HA-Cul3-K712R and HA-Cul3-R756A accumulated in their unmodified state when their expression was induced by the addition of doxycycline (Fig. 5E). Interestingly, a significant fraction of these cells accumulated with multiple nuclei or butterfly nuclei (Fig. 5F), indicative of reduced Cul3 function. These results suggest that non-neddylatable Cul3 is inactive and may function in a dominant-negative manner to trap endogenous Cul3 or Cul3-adaptor proteins.
Discussion
Our results demonstrate that DCNLs regulate CRL activation by promoting Cul neddylation in mammals. In particular, we uncovered a strong link between DCNL3 and Cul3 neddylation and found that DCNL3 localizes to the plasma membrane via myristoylation and palmitoylation of a conserved amino-terminal motif. Interestingly, membrane association of DCNL3 appears crucial for its neddylation function in vivo, revealing an uncharacterized function of Cul3-based ligases at membranes.
DCNL3 Functions in a Nonredundant Manner to Promote Cul3 Neddylation in Vivo.
Surprisingly, our results indicate that the human DCNLs function in a nonredundant manner. DCNL3 directly interacts with hUbc12 and human Culs, and RNAi depletion of DCNL3 reduced neddylation of several Culs including Cul3. Indeed, cells lacking DCNL3 exhibited defects characteristic for loss of Cul3 function, whereas overexpression of DCNL3 increases Cul3 neddylation in vivo. Moreover, ectopically expressed DCNL3 was able to recruit Cul3 but not Cul1 to the plasma membrane. These results reveal a general role of DCNL3 in Cul neddylation and suggest that DCNLs function as E3 ligases in an overlapping but nonredundant manner in vivo. Previous results indicate that the ring-finger protein Rbx1 directly catalyzes Cul neddylation. However, similar to yeast Cdc53, a Cul3 mutation that interferes with DCNL binding but retains the Rbx1 interaction abolished Cul3 neddylation in vivo, implying that Rbx1 may not be sufficient to promote Cul neddylation under physiological conditions. Interestingly, overexpression of such non-neddylatable mutant forms of Cul3 interfered with cytokinesis in a dominant-negative manner. Because Cul subunits and Cul-based complexes in general are known to form dimers (22–24), it is possible that the assembly of heterodimers between mutant and wild-type Cul3 abolishes its E3-ligase function (Fig. S6E). Consequently, these results imply that neddylation of Cul3 may be important for its E3-ligase function in vivo.
DCNL3 Functions at the Plasma Membrane Through a Distinct Amino-Terminal Domain.
Our results suggest that members of the DCNL3 family function at membranes. Indeed, the amino-terminal domain of DCNL3 contains a conserved motif, which is myristoylated and palmitoylated in vivo. Mutation of the potential myristoylation site (G2A) strongly reduced membrane association of DCNL3. However, biophysical studies revealed that binding of myristate is too weak to stably anchor a protein at the plasma membrane (21, 25), and thus a second fatty acid modification like palmitoylation (C16 S-acylation) is generally needed. Indeed, DCNL3 possesses 2 conserved cysteines (C4 and C8) close to the N-terminal myristoylation site, and mutation of these residues reduced membrane binding (Fig. 3D). Palmitoylation is known to be an important targeting signal for intracellular trafficking from the ER to the plasma membrane (e.g., N-Ras) but also for the localization of proteins to specific subdomains of the plasma membrane, mainly to cholesterol-rich microdomains such as lipid rafts (e.g., palmitoylation of Cys-184 of H-Ras) (26). Sequestration of DCNL3 to lipid rafts might therefore be required to activate DCNL3 and/or bring DCNL3 and membrane-bound Culs into close contact with important cofactors that facilitate Nedd8 modification. Indeed, we found a fraction of DCNL3 and Cul3, enriched for its neddylated form, in detergent-resistant membrane fractions (DRMs) (Fig. S7), and down-regulation of Cul3 function appears to impair endocytic trafficking.
In a more general context, our results support the notion that the different amino-terminal domains of DCNL family members may be important for their subcellular localization. Thus, the diversification of the DCNL proteins in mammalian cells may not only expand their functions toward additional substrates, but may also target their neddylation activity to specific subcellular compartments and tissues (Fig. S4A). Finally, our results imply that a subset of Cul3 and possibly also Cul1-type E3-ligases may function at membranes. We speculate that neddylation by membrane-associated DCNL3 locally activates these Cul-based E3-ligases, which in turn may ubiquitinate membrane-associated targets.
Experimental Procedures
Cell Culture, Immunofluorescence, and Transfection Experiments.
HeLa cells were grown as described (19) and transfected with Lipofectamine 2000 (Invitrogen). Small interfering RNA duplexes (Microsynth and Invitrogen; stealth siRNA duplexes; see SI Text) were applied for 72 h by using Oligofectamine (Invitrogen), and cells were lyzed for 30 min at 4 °C in Nonidet P-40 lysis buffer [20 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 0.2% Nonidet P-40, 10% glycerol, 1 mM NaF, 20 mM β-glycerophosphate, 1 mM DTT, complete protease inhibitor (Roche)], supplemented with 10 mM 1,10-Phenanthroline (Sigma). For quantitative analysis of Cul modification, fluorescent secondary antibodies were used (IRDye800CW goat anti-rabbit; LI-COR Biosciences), and the signal was quantified with the Odyssey Infrared Imaging system and ImageJ software from at least 4 independent experiments. Datasets were subjected to Student's t tests to assess their statistical significance. Cell lines stably expressing DCNL1-HA, DCNL1-DAD-HA, DCNL3-HA, DCNL3-DAD-HA, HA-Cul3, HA-Cul3-R756A, and HA-Cul3-K712R under the control of the inducible tetracycline promoter (CMV/Tet02) were generated by using Flip-In T-Rex 293 cells from Invitrogen. Protein expression was induced by the addition of 1 μg/mL of doxycycline (Sigma) for 24 h for coimmunoprecipitation experiments or 10 days for phenotypic analysis. For immunofluorescence, cells were directly fixed in 4% paraformaldehyde for 15 min and either permeabilized for 5 min with 0.5% Nonidet P-40 (HeLa cells) or attached to slides by cytospin and subsequently permeabilized (293T-Rex cells) and immunostained as described (19). At least 500 cells per experiment were counted for quantifications.
Detection of Azido Fatty Acylated Proteins and Lipid Inhibitors.
Labeling of starved COS-7 cells transfected with plasmids expressing HA-tagged wild-type or mutant DCNL3 with saponified azido-myristate (C12N3) and azido-palmitate (C14N3) was performed as described (27). After 4 h of labeling, cells were lyzed in RIPA buffer, and postnuclear supernatants were incubated with HA beads (Sigma) overnight at 4 °C. The beads were washed, resuspended in 100 μL of phosphine buffer [0.1 M Na-phosphate (pH 7.2), 1% SDS], and subjected to 80 °C heating for 20 min to release bound proteins. The supernatants were incubated for 2 h with 250 μM phosphine-biotin in the presence of 10 μM DTT at room temperature. The reaction was stopped by the addition of 5× sample loading buffer with 50 μM DTT. PVDF membranes were equilibrated in methanol and treated with either 0.1 M Tris pH 7.0 or 0.1 M KOH (both 1:9 in water/methanol) for 45 min. The membranes were washed extensively with PBS and 0.1% Tween20-PBS and blocked overnight in BSA blocking buffer (5% BSA, 1× Blotto, 0.1% Tween20). Biotinylated azido-fatty-acylated proteins were detected by using neutravidin-HRP (Pierce).
The inhibitors HMA (Sigma) or 2BP (Sigma) were saponified as described (27) and applied to starved cells for 24 h to a final concentration of 1 or 0.5 mM, respectively.
Supplementary Material
Acknowledgments.
HA-Cul3-pcDNA3 was provided by C. Chung (Seoul National University, Seoul, Korea), and the Cul1 and Rbx1 constructs were provided by W. Krek (Eidgenössiche Technische Hochschule). N.M.-S. is a member of the Molecular Life Science Zurich graduate program and was funded by the Boehringer Ingelheim Foundation. Y.-C.C. was supported by a Doctoral Research Scholarship from the Canadian Institutes of Health Research, and T.K. was supported by a Marie-Curie Intra-European Fellowship and the Roche Research Foundation. F.S. is a Research Scientist of the National Cancer Institute of Canada and was supported by Canadian Institutes of Health Research Grant MOP-57795 and the National Cancer Institute of Canada. D.D.O.M. was supported by graduate awards from the Alberta Heritage Foundation for Medical Research and the Canadian Institutes of Health Research. L.G.B. was supported by Canadian Institutes of Health Research Grant MOP-81248. The laboratory of M.P. is supported by grants from the Swiss National Science Foundation, Oncosuisse, and the Eidgenössiche Technische Hochschule.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812528106/DCSupplemental.
References
- 1.Hersko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Petroski MD, Deshaies RJ. Function and regulation of Cullin-Ring ubiquitin ligases. Nature. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 3.Rabut G, Peter M. Function and regulation of protein neddylation. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldenberg SJ, et al. Structure of the Cand1–Cul1–Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit Cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Kawakami T, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20:4003–4012. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duda DM, et al. Structural insights into NEDD8 activation of Cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamoah K, et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc Natl Acad Sci USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xirodimas DP, Saville MK, Bourdon J-C, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Gao F, Cheng J, Shi T, Yeh ETH. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFκB-dependent transcription. Nat Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 10.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8 modification of Cul1 is promoted by Roc1 as a Nedd8–E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 12.Megumi Y, et al. Multiple roles of Rbx1 in the VBC-Cul2 ubiquitin ligase complex. Genes Cells. 2005;10:679–691. doi: 10.1111/j.1365-2443.2005.00869.x. [DOI] [PubMed] [Google Scholar]
- 13.Kurz T, et al. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 14.Kurz T, et al. Dcn1 functions as a scaffold-type E3 ligase for Cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, et al. Structural basis for DCN-1's function in protein neddylation. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- 16.Sarkaria IS, et al. Squamous cell carcinoma-related oncogene is highly expressed in developing, normal, and adenomatous adrenal tissue but not in aggressive adrenocortical carcinomas. Surgery. 2004;136:1122–1128. doi: 10.1016/j.surg.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Ma T, et al. DCUN1D3, a novel UVC-responsive gene that is involved in cell cycle progression and cell growth. Cancer Sci. 2008;99:2128–2135. doi: 10.1111/j.1349-7006.2008.00929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim AY, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sumara I, et al. A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev Cell. 2007;12:887–900. doi: 10.1016/j.devcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resh MD. Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 22.Wimuttisuk W, Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chew E-H, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells: Evidence for cullin dimerization. Cell Signal. 2006;19:1071–1080. doi: 10.1016/j.cellsig.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Tang X, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 25.Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 26.Greaves J, Chamberlain LH. Palmitoylation-dependent protein sorting. J Cell Biol. 2007;176:249–254. doi: 10.1083/jcb.200610151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin DDO, et al. Rapid detection, discovery, and identification of posttranslationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.