Abstract
The Drosophila Toll receptor does not interact directly with microbial determinants, but is instead activated by a cleaved form of the cytokine-like molecule Spätzle. During the immune response, Spätzle is processed by complex cascades of serine proteases, which are activated by secreted pattern-recognition receptors. Here, we demonstrate the essential role of ModSP, a modular serine protease, in the activation of the Toll pathway by Gram-positive bacteria and fungi. Our analysis shows that ModSP integrates signals originating from the circulating recognition molecules GNBP3 and PGRP-SA and connects them to the Grass-SPE-Spätzle extracellular pathway upstream of the Toll receptor. It also reveals the conserved role of modular serine proteases in the activation of insect immune reactions.
Keywords: innate immunity, proteolytic cascade, insect immunity, antifungal, antimicrobial peptides
Sequential activation of extracellular serine protease (SP) cascades regulates important innate immune reactions, like blood clotting in arthropods and complement activation in vertebrates. In Drosophila, proteolytic cascades are also involved in the regulation of the Toll pathway, which mediates resistance to Gram-positive bacteria and fungi. Unlike mammalian Toll-like receptors, the Drosophila Toll receptor does not interact directly with microbial determinants and is instead activated by a cleaved form of the secreted cytokine-like molecule Spätzle (Spz) (1). The immune-induced cleavage of Spz is triggered by proteolytic cascades, conceptually similar to vertebrate blood coagulation or complement activation cascades. These proteolytic cascades have a functional core consisting of several SP that undergo zymogen activation, upon cleavage by an upstream protease (2, 3). Spätzle processing enzyme (SPE), an immune-regulated SP with a Clip-domain, has been identified as the terminal SP that maturates Spz (4, 5).
Genetic analysis supports the existence of several complex cascades of SP that link microbial recognition to activation of SPE. Pattern-recognition receptors (PRRs) are thought to be present in the hemolymph where they sense microbial-derived molecules (6). Detection of Gram-positive bacteria is mediated through the recognition of peptidoglycan by the peptidoglycan-recognition protein-SA (PGRP-SA) with the help of Gram-negative binding protein1 (GNBP1) (7–9). Activation of the Toll pathway by fungi is in part mediated by GNBP3 through the sensing of β-1,3-glucan (10). RNAi and loss of function analyses have shown that activation of SPE by either PGRP-SA/GNBP1 or GNBP3 requires Grass, a Clip-domain SP (4, 11).
Spores of entomopathogenic fungi such as Beauveria bassiana have the capacity to germinate on the fly cuticle and generate hyphae, which can penetrate the cuticle of insects and reach the hemolymph. It has been proposed that the presence of B. bassiana is detected, in a GNBP3-independent manner, through direct activation of the Toll pathway by a fungal protease PR1 (10). PR1 would directly cleave the host SP, Persephone (Psh), which triggers Toll pathway activation (10). Recently, this mode of activation was extended to the sensing of proteases produced by various bacteria (11). Surprisingly, tracheal melanization in mutant larvae lacking the serpin Spn77Ba also activates the Toll pathway in a Psh-dependent manner (12). This suggests that Psh-dependent Toll pathway activation is induced by a host factor derived from melanization. This also points to a possible cross-talk between the proteolytic cascades that regulate the Toll pathway and those regulating the melanization reaction.
Despite these recent studies, the extracellular events that lead to the activation of the Toll pathway remain poorly characterized. For instance, the apical protease linking recognition by PRRs to the cleavage of Spz is not known, leaving an important gap in our knowledge of Toll pathway activation. The in vitro reconstruction of a similar cascade with purified proteases in the beetle Tenebrio molitor, suggests an important role for a modular SP (Tm-MSP). In this insect, binding of peptidoglycan to the PGRP-SA/GNBP1 complex induces activation of the Tm-MSP zymogen that in turn activates another SP (Tm-SAE) to cleave Tm-SPE, the Tenebrio homolog of SPE, resulting in both activation of Spz and melanization (13, 14).
This result prompted us to investigate the role of the Drosophila homolog of Tm-MSP, which is encoded by the CG31217 gene. We generated a null mutation in CG31217 by homologous recombination and demonstrate its essential role in the activation of Toll by secreted PRRs.
Results
Generation of a modSP Mutant by Homologous Recombination.
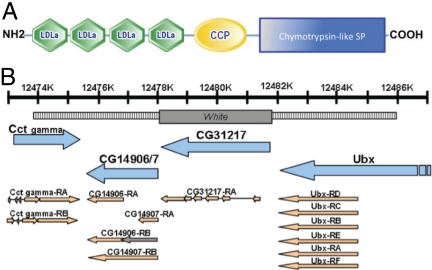
The CG31217 serine protease, named here ModSP (for modular serine protease), contains 4 low density lipoprotein-receptor class A (LDLa) domains and 1 complement control protein (CCP) module at its N terminus (Fig. 1A). ModSP shares 47% amino acid identity with the Tm-MSP (Fig. S1). Unlike several other Drosophila SPs involved in the immune response, modSP is not transcriptionally induced upon microbial infection and does not contain a Clip-domain (15, 16).
Fig. 1.
Structure of ModSP and generation of modSP mutants. (A) Domain organization of the ModSP protein. Symbols indicate the domains of LDLa, CCP, and SP respectively (adapted from ref. 14). The LDL receptor class A domain contains 6 disulphide-bound cysteines and a cluster of negatively charged amino acids that form the LDL receptor binding sites for LDL and calcium. The complement control protein (CCP) module, also known as Sushi domain, is found in a wide variety of complement like molecules and adhesion proteins. (B) Schematic representation of the modSP deletion. The gene map was adapted from FlyBase and includes modSP and the neighboring genes. The deleted segment replaced by the w gene (black box) and the flanking sequences used for recombination (dotted lines) are indicated.
To analyze the function of ModSP, homologous recombination was used to replace the entire modSP locus with a copy of the white gene (Fig. 1B). Two independent fly lines, modSP1 and modSP2, were obtained, and the absence of modSP transcript in homozygous mutants was confirmed by RT-qPCR (Fig. S2). We verified that the modSP deletion did not affect the expression of flanking genes. Both modSP1 and modSP2 mutants were perfectly viable and exhibited no visible developmental defects. Here, we only report data obtained with the modSP1 mutant, because both alleles produced identical results.
ModSP Mediates Toll Activation by Gram-Positive Bacteria.
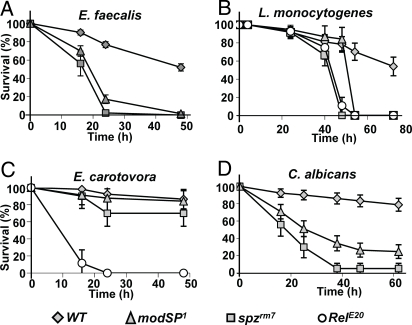
When challenged by a range of Gram-positive bacteria (Enterococcus faecalis, Listeria monocytogenes), modSP1-deficient flies rapidly succumbed to infection (Fig. 2A and B). The observed phenotypes were similar, albeit slightly weaker, to those generated by a null mutation in spz (spzrm7). The modSP1 mutation did not cause a general immune deficiency, because modSP1 flies exhibited a wild-type survival to a septic injury with the Gram-negative bacterium Erwinia carotovora, which is known to activate the Imd pathway (Fig. 2C).
Fig. 2.
ModSP is required for resistance to Gram-positive bacteria and fungal infections. The survival rates (%) of modSP1 flies infected with the Gram-positive bacteria E. faecalis (A), L. monocytogenes (B), the Gram-negative bacterium E. carotovora 15 (C), and the yeast C. albicans (D) were compared with wild-type flies and flies mutant in either the Toll pathway (spzrm7) or the Imd pathway (RelE20). Flies were infected by septic injury with a needle dipped in a concentrated pellet of each microbe (OD = 200, OD = 10 for E. faecalis). Bars represent confidence intervals at 5%.
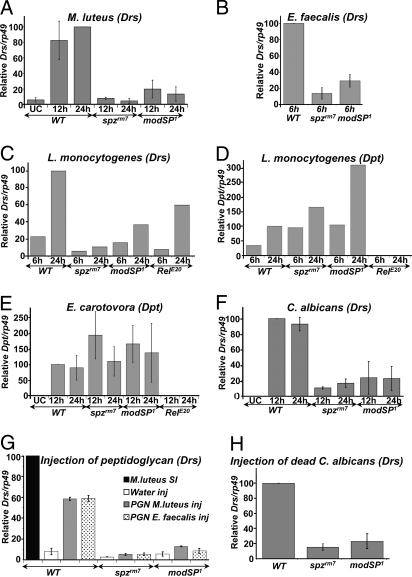
To test whether this immune susceptibility results from improper Toll activity, we monitored the expression of the antifungal peptide gene Drosomycin (Drs), a target of the Toll pathway. modSP1 mutants weakly induced Drs after infection with the Gram-positive bacteria M. luteus and E. faecalis (Fig. 3 A and B). Lysine-type peptidoglycan, a peptidoglycan form found in most Gram-positive bacteria including M. luteus and E. faecalis, is a potent inducer of the PGRP-SA-Toll pathway (17). The modSP1 mutation strongly reduced the induction of Drs in response to purified peptidoglycan extracted from M. luteus and E. faecalis as observed in spzrm7 mutants (Fig. 3G). This experiment demonstrates that ModSP is required for Toll pathway activity in a step downstream of the recognition of lysine-type peptidoglycan.
Fig. 3.
Analysis of antimicrobial peptide gene expression in modSP mutant. Drs (A–C and F–H) or Dpt (D and E) gene expression was monitored by RT-qPCR with total RNA extracted from wild-type, spzrm7, RelE20, and modSP1 adults collected at different time points after septic injury with M. luteus (A), E. faecalis (B), L. monocytogenes (C and D), E. carotovora (E), and C. albicans (F). (G and H) RT-qPCR analysis was performed with total RNA extracts from wild-type and mutant females collected 24 h after injection with 9 nL M. luteus peptidoglycan, E. faecalis PGN ([PGN] = 5 mg/mL) (G) or dead yeast (OD600 = 300) (H). The mean and standard deviation of 2 to 3 independent experiments is shown, except L. monocytogenes, for which 1 representative experiment among 2 is shown.
L. monocytogenes is a Gram-positive bacterium with diaminopimelic (DAP)-type peptidoglycan that can activate both Toll and Imd pathways (18). A strong reduction of Drs expression was observed in modSP1 flies upon infection with L. monocytogenes, whereas the level of Diptericin (Dpt, regulated by the Imd pathway) was similar or even higher to that observed in wild-type flies (Fig. 3 C and D). In addition, modSP1 did not affect the expression of the antibacterial peptide Dpt upon infection with the Gram-negative bacterium E. carotovora (Fig. 3E). The levels observed were similar or even higher than those observed in wild-type flies. In conclusion, our survival and antimicrobial peptide gene expression analyses strongly suggest a role of ModSP in the activation of the Toll pathway by Gram-positive bacteria.
ModSP Overexpression Activates the Toll Pathway.
We next performed a series of experiments to further assess that the modSP gene is responsible for the immune phenotype observed in modSP1 flies. First, similar phenotypes, both in terms of immune susceptibility and Drs expression, were observed in flies carrying the modSP1 mutation over deficiencies [Df(3R)P10, Df(3R)Spf] uncovering the modSP locus. This reduces the possibility that the phenotype is because of an associated mutation and indicates that modSP1 behaves as an amorphic mutation. We also generated fly lines overexpressing a full-length modSP cDNA under the control of an UAS element. Expression of modSP in whole organisms using the ubiquitous driver daughterless (da-Gal4) led to larval lethality. Importantly, a high level of Drs expression was observed in flies overexpressing modSP in fat body and hemocytes (genotype: c564-Gal4, UAS-modSP). This level reached 60% of that observed in wild-type flies collected 16 h after infection with M. luteus (Fig. S3A), indicating that overexpression of modSP is sufficient to activate the Toll pathway. Finally, 2 modSP RNAi fly lines from the Vienna Drosophila RNAi Center were used to specifically knockdown modSP expression in vivo. The levels of modSP transcripts were depleted to 30%–40% of the wild-type level in modSP-RNAi flies (genotype: da-Gal4, UAS-modSP-IR) (Fig. S2). Importantly, these flies were more susceptible upon septic injury with E. faecalis and had reduced Drs expression upon M. luteus infection (Fig. S3 B and C). Together, these experiments demonstrate that ModSP is essential for Toll activation by Gram-positive bacteria and that overexpression of full-length ModSP is sufficient to activate the Toll pathway.
ModSP Functions Between PGRP-SA and Grass in Activating the Toll Pathway.
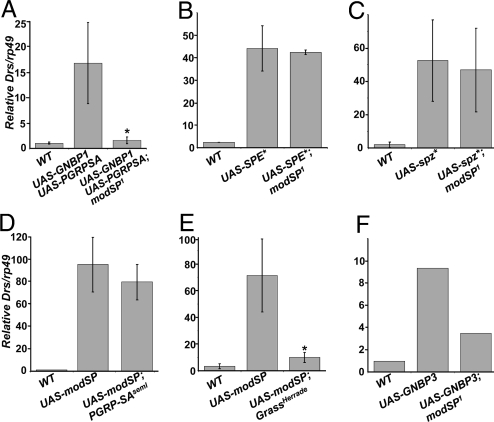
We next performed epistasis experiments to determine the position of ModSP in the cascade leading to Toll activation by Gram-positive bacteria. Detection of Gram-positive bacteria is mediated via the recognition of lysine-type peptidoglycan by PGRP-SA and GNBP1 (7–9). Simultaneous overexpression of GNBP1 and PGRP-SA triggers the Toll pathway, resulting in a constitutive expression of Drs in the absence of an immune challenge (8). The Drs expression induced by the overexpression of both GNBP1 and PGRP-SA was blocked by the modSP1 mutation (Fig. 4A). Conversely, the PGRP-SAseml mutation did not block the high expression of Drs induced by the overexpression of modSP (Fig. 4D). Together these experiments indicate that ModSP functions downstream of PGRP-SA. We further observed that modSP1 mutation did not block the induction of Drs expression upon overexpression of a matured and constitutively active form of Spz (UAS-spz*), or SPE (UAS-SPE*), which is in agreement with a function upstream of Spz and SPE (Fig. 4 B and C). Importantly, the Herrade mutation that affects the grass SP gene (11) fully blocked Drs expression induced by overexpression of modSP (Fig. 4E). Altogether, our epistatic analysis indicates that ModSP functions downstream of PGRP-SA and GNBP1 and upstream of Grass in the pathway that links Gram-positive bacterial recognition to Toll activation.
Fig. 4.
ModSP functions downstream of PRRs and upstream of Grass. (A) Drs expression induced by overexpressing GNBP1 and PGRP-SA was reduced by the modSP1 mutation. (B and C) Overexpression of UAS-SPE* (B), UAS-spz* (C), induced constitutive Drs expression that was independent of modSP. (D and E) Overexpression of UAS-modSP induced constitutive Drs expression that was independent of PGRP-SAseml (D) but dependent on Grass (E). (F) Drs expression induced by overexpressing GNBP3 was reduced by the modSP1 mutation. Drs was measured in the absence of immune challenge. To indicate the level of Drs obtained in the different conditions, the Drs/rp49 ratio was normalized to the levels observed in flies collected 16 h after septic injury with M. luteus (fixed to 100%). Results are representative of 3 experiments, except panel F that could be obtained only once. Values significantly different in a t test for P < 0.05 are denoted by *. The C564-Gal4 driver was used to drive UAS transgenes in these experiments.
ModSP Mediates Toll Activation by GNBP3 upon Fungal Infection.
Detection of yeast is mediated via the recognition of β-glucans by GNBP3. A loss-of-function mutation in GNBP3, GNBP3hades, induces a phenotype of reduced survival to the yeast Candida albicans and deficient Toll activation. Conversely, overexpression of GNBP3 triggers the Toll pathway, resulting in a constitutive expression of Drs in the absence of an immune challenge (10). We thus investigated whether ModSP could also function downstream of GNPB3 in the recognition of yeast. In support of this hypothesis, we observed that modSP1 deficient flies rapidly succumbed to C. albicans infection (Fig. 2D). In addition, modSP1 flies displayed reduced Drs induction upon septic injury with C. albicans (Fig. 3F). The modSP1 phenotype was similar, albeit weaker, to that generated by a null mutation in spz. The relevance of GNBP3 to Toll activation is particularly prominent when flies are injected with dead C. albicans, a condition limiting the activation of the Toll pathway through Psh (11). Drs expression was also strongly reduced by the modSP1 mutation upon injection of dead C. albicans (Fig. 3H).
We next investigated the interaction between GNBP3 and ModSP. Unfortunately, we were not able to reproducibly induce Drs by overexpression of GNBP3 as previously described (10). Nevertheless, we performed 1 successful experiment in which Drs was induced upon GNBP3 overexpression to 10% of the level reached in wild-type flies collected 16 h after infection with M. luteus. In this experiment, we observed a reduction of the GNBP3 mediated Drs induction by the modSP1 mutation (Fig. 4F).
Collectively, these experiments demonstrate a requirement of ModSP in the activation of the Toll pathway by both yeast and Gram-positive bacteria.
ModSP Is Not Involved in the Sensing of Protease Activity by the Psh Pathway.
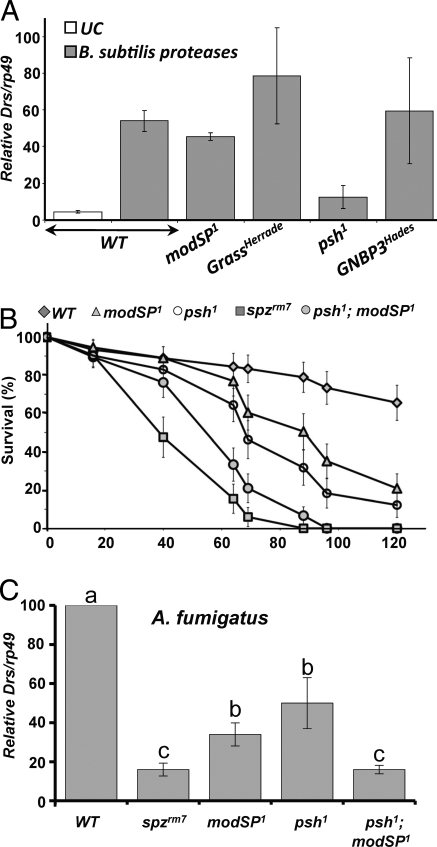
In Drosophila, injections of proteases of fungal or bacterial origin activate the Toll pathway in a Psh-dependent manner (10, 11). To investigate whether ModSP interferes with Psh-dependent Toll pathway activation, we first monitored the level of Drs expression upon injection of proteases derived from Bacillus subtilis or Aspergillus oryzae. Unlike the psh1 mutation, modSP1 did not impair Toll activation in these experimental conditions (Fig. 5A and Fig. S4).
Fig. 5.
ModSP and Psh define 2 distinct pathways that cooperatively activate Toll. (A) Drs gene expression was monitored by RT-qPCR with total RNA extracted from wild-type, GNBP3Hades, GrassHerrade, psh1, and modSP1 females collected 16 h after injection with B. subtilis derived proteases. Each graph represents the mean of 4 independent experiments with standard deviation. UC, unchallenged flies. (B and C) The survival rates (B) and the level of Drs expression (C) of modSP1 and modSP1; psh1 double mutants infected with A. fumigatus were compared with wild-type, psh1, and spzrm7 flies. Data were analyzed by ANOVA and means were separated for significance according to Fisher's protected LSD at P < 0.05; (F = 63.25, df = 4, P < 0.0001).
We next compared the response of modSP1 and psh1 flies to the injection of spores from A. fumigatus, a filamentous fungus known to produce proteases. Both modSP1 and psh1 flies exhibited a moderate increase of susceptibility to A. fumigatus infection compared with wild-type (Fig. 5B). In addition, the level of Toll activity in modSP1 flies and to a lesser extent in psh1 flies was reduced, although generally higher than levels observed in spzrm7 mutants (Fig. 5C). Importantly, in flies double mutant for psh1 and modSP1, the Toll pathway activation by A. fumigatus was reduced to a level comparable to the spzrm7 mutant. Correlatively, psh1; modSP1 double mutants exhibited an increased susceptibility to A. fumigatus compared with single mutant. These data indicate that the ModSP and the Psh branches of the Toll pathway contribute independently to the resistance to this fungus. We conclude that, like Grass (11), ModSP does not interfere with the Psh mediated activation of the Toll pathway.
ModSP Exhibits a High Level of Autoproteolysis and Does Not Cleave Grass.
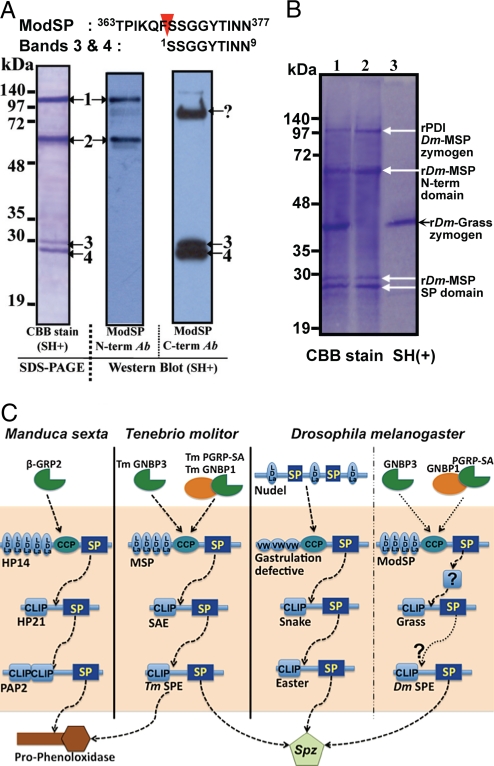
To gain insight into the activation mechanisms that regulate ModSP, we produced recombinant His-tagged ModSP and Grass proteins using the insect Sf9 cells. When we purified the recombinant ModSP-containing fractions and analyzed them by SDS-PAGE, we observed 4 major bands corresponding to ModSP. N-terminal sequencing showed that sequence of bands 1 and 2 matched the N terminus of ModSP, whereas the sequence of band 3 and 4 corresponded to the catalytic domain of ModSP (Fig. 6A). Western blot analysis using a polyclonal antibody against the N-terminal domain or the central domain of the protein confirmed that band 1 corresponded to full-length zymogen and that the other bands corresponded to either the N-terminal fragment (band 2) or the C-terminal part (band 3 and 4) resulting from the cleavage of ModSP. These results suggest that, as soon as it is expressed, ModSP is autoactivated. Of note, under the same conditions, T. molitor MSP did not show any autoactivation. Our results also reveal that Drosophila ModSP and Tm-MSP share the same cleavage site (Fig. 6A and Fig. S1). The presence of 2 processed C-terminal fragments of ModSP could either be because of the presence of a posttranslational modification or an additional cleavage at the C terminus of ModSP. We next investigated whether ModSP could cleave Grass in vitro. Fig. 6B shows that Grass was not cleaved when incubated with recombinant ModSP and CaCl2. In the same condition, Tm-SAE was cleaved by Tm-MSP. However, in absence of a known ModSP substrate, we cannot be fully sure that the 28 kDa form of ModSP we detected is active. Nevertheless, this result suggests that ModSP does not cleave the Grass zymogen. Additionally, the direct activation of Grass by ModSP is unlikely since the predicted cleavage of Grass requires a trypsin-like enzyme although ModSP exhibits a chymotrypsin-like specificity (Fig. S5).
Fig. 6.
ModSP exhibits a high level of autoproteolysis. (A) SDS-PAGE gel using purified recombinant rPDI-fusioned ModSP-containing fractions reveals 4 bands corresponding to ModSP as indicated by N-terminal sequencing (Top). Bands 1 and 2 matched the N terminus of ModSP, whereas the sequence of band 3 and 4 corresponded to the catalytic domain of ModSP. Western blot analysis using a polyclonal antibody against the N-terminal domain (Middle) or the central domain (Right) of the protein confirmed that band 1 corresponded to full-length zymogen and that the other bands corresponded to either the N-terminal fragment (band 2) or the C-terminal part (band 3 and 4) resulting from the cleavage of ModSP. Red arrow-head indicates the cleavage site of catalytic serine protease domain of ModSP (Top). The question mark (?) indicates a nonspecific band. (B) rModSP does not cleave rGrass in vitro. 1, rDm-Grass zymogen was incubated with rDm-MSP and CaCl2 at 30 °C for 1 h. 2, 3 indicate the purities of rPDI-fusioned ModSP, a mixture of active and zymogen forms, and rGrass zymogen under reducing conditions, respectively. The gel was stained by Coomassie brilliant blue R-250 (CBB). (C) Extracellular cascades of serine proteases in insects. Biochemical studies performed in T. molitor and M. sexta, and genetic analysis in D. melanogaster, reveal striking similarities in the mechanisms underlying serine protease activation by PRRs. In M. sexta, HP-14 is converted to a 2-chain active form in the presence of β-1,3-glucan and the β-1,3-glucan recognition protein 2 (βGRP2, a protein related to GNBP3) (23, 24). Binding of β-1,3-glucan and βGRP2 triggers the autoactivation of proHP14 into HP14 that then processes HP21. In turn, HP21 cleaves proPAP-2 into PAP2 that finally activates proPO. Hence, the mechanism of activation of HP14 in response to glucan is highly reminiscent of the mode of activation of Tm-MSP, with the exception that HP14 can be activated upon direct binding to peptidoglycan during bacterial infection (24). This suggests that the PGRP-SA/GNBP1 complex involved in sensing Gram-positive bacteria has been added on to a more ancient cascade core during evolution. Interestingly, a similar organization is also observed in the proteolytic cascade that regulates Toll during dorso-ventral patterning of the embryo.
Discussion
In this study, we have reported the generation of a null mutation of the gene encoding the SP ModSP and demonstrated its essential role in the activation of the Toll pathway by PRRs. This was illustrated by the observation that modSP deficient flies fail to activate the Toll pathway in response to either Gram-positive bacteria or yeast. Epistatic analyses demonstrated that ModSP acts downstream of PGRP-SA, but upstream of the SP Grass. Importantly, we showed that ModSP does not participate in the Psh-dependent branch of the Toll pathway as shown by the wild-type activation of this pathway in modSP1 flies upon injection of bacterial proteases. The modSP1 phenotype was very similar to that of a loss of function Grass mutant, suggesting that these SP act in a linear pathway connecting microbe recognition by PRR to the activation of Spz by SPE. The analysis of psh1; modSP1 double mutant flies indicates a synergistic action of the ModSP and Psh pathways in the response against filamentous fungi, which might be detected through both host PRRs and their virulence factors. Collectively, our results confirm the model of an activation of Toll by 2 extracellular pathways: A PRR-dependent pathway and a Psh-dependent pathway (11). We extend this model by showing that Grass and ModSP function in a common SP cascade. In addition, the apical position of ModSP suggests a direct branching of signals from secreted PRRs to the ModSP-Grass pathway. Biochemical analyses in T. molitor indicate that Tm-MSP interacts directly with the PRR complexes involved in the sensing of peptidoglycan (14) or glucan (30). Although it was not formally demonstrated in the present study, it seems likely that Drosophila ModSP acts as the most upstream SP directly activated by secreted PRRs.
ModSP has a number of structural features that make it unique among Drosophila SP and suggest its critical role in the initial events leading to the activation of the proteolytic cascade upstream of Toll. First, ModSP does not contain a Clip-domain in contrast to Grass and SPE. The function of the Clip-domain is still unknown but its presence in many SP that function in signaling cascades suggests a regulatory role (19). This indicates that ModSP activation differs from the chain reaction activating Grass or SPE. Second, ModSP contains additional domains such as the CCP and LDLa motifs in its N-terminal extremity. Consistent with the presence of the LDLa domain, a ModSP-GFP fusion protein was secreted and found bound to the surface of lipid vesicles that circulate in the hemolymph (Fig. S6). We speculate that the association of ModSP to vesicles is important to nucleate the activation of downstream SP.
In the beetle, T. molitor, Tm-MSP is activated upon formation of a tri-molecular complex composed of PGRP-SA, GNBP1, and lysine-type peptidoglycan (14). Although the possibility of a SP other than ModSP functioning as the initial protease in the Drosophila Toll pathway cannot be ruled out, our epistatic analysis suggests an apical role of ModSP. We show that overexpression of a full-length version of ModSP is sufficient to reach a high level of Toll activation, in contrast to other SP that generally require the overexpression of a preactivated form to fully induce the cascade (5, 20). These observations favor a model in which ModSP directly interacts with GNBP3 or GNBP1/PGRP-SA recognition complexes. Recruitment of ModSP by PRRs would increase its local concentration, a situation sufficient for its autoproteolysis. In agreement with this hypothesis, a recombinant form of ModSP produced in baculovirus appears to be unstable as a zymogen, presumably because of a high level of autoactivation (Fig. 6A). Unfortunately, this high level of autoproteolysis did not permit in vitro reconstitution experiments using ModSP, GNBP1, and PGRP-SA as performed with Tm-MSP (14). Nevertheless, we observed a higher level of ModSP activation when the protein was incubated with GNBP1 and PGRP-SA. The most parsimonious model is that ModSP would interact with GNBP1 or GNBP3, the common protein family members found in the 2 recognition complexes. The exact contribution of GNBP1 to sensing Gram-positive bacteria is currently debated. It was recently proposed that GNBP1 functions upstream of PGRP-SA by cleaving peptidoglycan, a step required for an optimal binding of PGRP-SA to peptidoglycan (21). In contrast, T. molitor GNBP1 is recruited subsequent to the binding of PGRP-SA to peptidoglycan and is required for the interaction with Tm-MSP (22). The implication of ModSP in sensing of fungi through GNBP3 and peptidoglycan through GNBP1/PGRP-SA suggests a similar mechanism of activation of this SP and Tm-MSP. This would favor a role of GNBP1 as a linker between PGRP-SA and ModSP. In accordance with this model, recombinant full-length Drosophila and Tenebrio GNBP1 did not exhibit any enzymatic activity toward peptidoglycan in vitro (Fig. S7).
The participation of ModSP and SPE in an extracellular pathway linking PRR recognition to Spz activation in both T. molitor (Coleoptera) and D. melanogaster (Diptera), which diverged ≈250 million years ago, demonstrates the conservation of this extracellular signaling module in insects. In the lepidopteran Manduca sexta, the hemolymph protein 14 (Ms-HP14) contains a domain arrangement very similar to that of ModSP (Fig. S1) and regulates the melanization cascade in response to microbial infection (23, 24). Collectively, biochemical studies performed in T. molitor and M. sexta, and genetic analysis in D. melanogaster, reveal striking similarities in the mechanisms underlying SP activation by PRRs (Fig. 6C). All involve the sequential activation of an apical modular SP (that displays a certain level of autoactivation) and clip-domain proteases. Interestingly, a similar organization is also observed in the proteolytic cascade that regulates Toll during dorso-ventral patterning of the embryo. Gastrulation Defective, the apical SP, contains von Willebrand domains and is also thought to be autoactivated, although the precise mechanism that triggers its activation has not been determined (25, 26). These features are also reminiscent of the complement activation by the lectin pathway in mammals in which the recognition of carbohydrate by the mannose binding lectin (MBL) leads to the autoactivation of MBL-associated serine proteases (MASPs). MASPs are also modular proteases with CUB, CCP, and EGF domains in their N terminus (27). Thus, genetic and biochemical analyses now reveal a similar level of organization for various proteolytic cascades in insects and increase our understanding as to how these cascades have evolved to fulfill diverse developmental or immune functions.
We are still far from a complete understanding of the precise sequence of events leading to Toll pathway activation, as the task is complicated by the high number of SP encoded in the Drosophila genome (16). Our analysis strongly suggests the existence of an intermediate SP acting between ModSP and Grass. Future works should identify this SP and determine the interaction between Grass and SPE. To date, no serpin regulating SP involved in the PRR-dependent pathways has been identified despite the critical role of this family in the negative control of proteolytic cascades. Further experiments combining genetics, biochemistry, and cell biology are required to identify additional components of this cascade and to clarify in vivo how and where proteolytic cascades downstream of PGRP-SA or GNBP3 are activated in the hemolymph compartment.
Materials and Methods
Fly Stocks.
A deletion of the modSP locus was obtained by homologous recombination (28). DNA sequences (of 4.2 kb and 4.8 kb) flanking the 5′ and 3′ parts, respectively, of the modSP locus (Fig. 1B) were cloned in the p(W25) vector (28). modSP sequence (3 kb; 3R: from nucleotide 12478339 to 12481351) was replaced by the white+ gene.
spzrm7, relE20, psh1, GNBP3hades, PGRP-SAseml, and GrassHerrade null alleles are described elsewhere (4, 7, 10, 11, 20). The c564 line specifically expresses GAL4 in adult fat body and hemocytes. OregonR flies were used as wild-type controls, if not indicated differently. Epistatic analyses were performed by standard genetic crosses. The UAS-GNBP3, UAS-PGRP-SA, UAS-GNBP1, UAS-SPE*, UAS-spz*, are described elsewhere (4, 5, 7, 8, 10, 11, 20); an asterisk (*) indicates that the constitutively activated form was used. A full-length cDNA of modSP (using the CG31217_cDNA gold LD43740 from DGRC) was placed downstream of UAS sequence using the pUASt vector. The UAS-ModSP-GFP transgene was obtained by a fusion of GFP to the C terminus of ModSP inserted in the pDONR221 Gateway entry clone (Invitrogen) and finally subcloned in the pTWG transgenesis vector. Drosophila stocks and crosses were maintained at 25 °C using standard fly medium. F1 progeny carrying both the UAS-construct and the GAL4 driver were transferred to 29 °C at late larval stage for optimal efficiency of the UAS/GAL4 system except for crosses involving UAS-SPE* or UAS-modSP that were performed at 20 °C.
Infection and Survival Experiments.
Bacterial and fungal infections were performed by pricking adults in the thorax with a thin needle previously dipped into a concentrated pellet of a microbial culture (OD = 200). Bacterial and fungal strains have been described in ref. 9. Flies were counted at different time points to monitor survival. Each experiment was performed with ≈80 flies for each genotype. Injection with M. luteus and E. faecalis peptidoglycan and its production were previously described (17). Injection of proteases from B. subtilis (>16 U/g, P5985; Sigma–Aldrich) or A. oryzea (>500 U/g, P6110; Sigma–Aldrich) was performed with a Nanoject apparatus using a concentration of 1/1500 as described in ref. 11.
RT-qPCR.
Drs, Dpt, ModSP, ModSP flanking genes and rp49 mRNA quantification by RT-qPCR was performed as described in ref. 9. Primers used to monitor mRNA expression can be obtained upon request.
Production of Recombinant ModSP Protein.
ModSP was expressed in Sf9 cells as a fusion partner with the Bombyx mori protein disulfide isomerase (bPDI) as previously described (29). Two rabbit polyclonal antibodies were raised using keyhole limpet hemocyanin (KLH)-conjugated synthetic peptide against the N-terminal and the C-terminal parts of ModSP respectively. Additional information is provided in the SI Text.
Supplementary Material
Acknowledgments.
We thank J.M. Reichhart, D. Ferrandon, the Bloomington Center, Ryu Ueda (National Institute of Genetic, Mishima), Vienna Drosophila Research Service for fly stocks, Pascale Cossart for the gift of the L. monocytogenes strain, J.P. Boquete for assistance, Carl Hashimoto, Nichole Broderick, and Marie Meister for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901924106/DCSupplemental.
References
- 1.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 2.LeMosy EK, Hong CC, Hashimoto C. Signal transduction by a protease cascade. Trends Cell Biol. 1999;9:102–107. doi: 10.1016/s0962-8924(98)01494-9. [DOI] [PubMed] [Google Scholar]
- 3.Jang IH, Nam HJ, Lee WJ. CLIP-domain serine proteases in Drosophila innate immunity. BMB Rep. 2008;41:102–107. doi: 10.5483/bmbrep.2008.41.2.102. [DOI] [PubMed] [Google Scholar]
- 4.Kambris Z, et al. Drosophila immunity: A large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–813. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Jang IW, et al. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Charroux B, Rival T, Narbonne-Reveau K, Royet J. Bacterial detection by Drosophila peptidoglycan recognition proteins. Microbes Infect. 2009 doi: 10.1016/j.micinf.2009.03.004. 2009 Apr 1. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 7.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 8.Gobert V, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–2130. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 9.Pili-Floury S, et al. In vivo RNAi analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- 10.Gottar M, et al. Dual detection of fungal infections in Drosophila by recognition of glucans and sensing of virulence factors. Cell. 2007;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang H, Kambris Z, Lemaitre B, Hashimoto C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev Cell. 2008;15:617–626. doi: 10.1016/j.devcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kan H, et al. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem. 2008;283:25316–25323. doi: 10.1074/jbc.M804364200. [DOI] [PubMed] [Google Scholar]
- 14.Kim CH, et al. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283:7599–7607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- 15.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross J, Jiang H, Kanost MR, Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: An initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- 17.Leulier F, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- 18.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 19.Piao S, et al. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J. 2005;24:4404–4414. doi: 10.1038/sj.emboj.7600891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–116. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, et al. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006;25:5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JW, et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104:6602–6607. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Jiang H. Interaction of beta-1,3-glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta. J Biol Chem. 2006;281:9271–9278. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Jiang H. Reconstitution of a branch of the Manduca sexta prophenoloxidase activation cascade in vitro: Snake-like hemolymph proteinase 21 (HP21) cleaved by HP14 activates prophenoloxidase-activating proteinase-2 precursor. Insect Biochem Mol Biol. 2007;37:1015–1025. doi: 10.1016/j.ibmb.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han JH, Lee SH, Tan YQ, LeMosy EK, Hashimoto C. Gastrulation defective is a serine protease involved in activating the receptor toll to polarize the Drosophila embryo. Proc Natl Acad Sci USA. 2000;97:9093–9097. doi: 10.1073/pnas.97.16.9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dissing M, Giordano H, DeLotto R. Autoproteolysis and feedback in a protease cascade directing Drosophila dorsal-ventral cell fate. EMBO J. 2001;20:2387–2393. doi: 10.1093/emboj/20.10.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan RC, Wijeyewickrema LC, Pike RN. The initiating proteases of the complement system: Controlling the cleavage. Biochimie. 2008;90:387–395. doi: 10.1016/j.biochi.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 28.Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goo TW, et al. Bombyx mori protein disulfide isomerase enhances the production of nuecin, an antibacterial protein. BMB Rep. 2008;41:400–403. doi: 10.5483/bmbrep.2008.41.5.400. [DOI] [PubMed] [Google Scholar]
- 30.Roh KB, et al. Proteolytic cascade for the activation of the insect Toll pathway induced by the fungal cell wall component. J Biol Chem. 2009 doi: 10.10741/jbc.M109.007419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.