Abstract
One of the largest contributions to biologically available nitrogen comes from the reduction of N2 to ammonia by rhizobia in symbiosis with legumes. Plants supply dicarboxylic acids as a carbon source to bacteroids, and in return they receive ammonia. However, metabolic exchange must be more complex, because effective N2 fixation by Rhizobium leguminosarum bv viciae bacteroids requires either one of two broad-specificity amino acid ABC transporters (Aap and Bra). It was proposed that amino acids cycle between plant and bacteroids, but the model was unconstrained because of the broad solute specificity of Aap and Bra. Here, we constrain the specificity of Bra and ectopically express heterologous transporters to demonstrate that branched-chain amino acid (LIV) transport is essential for effective N2 fixation. This dependence of bacteroids on the plant for LIV is not due to their known down-regulation of glutamate synthesis, because ectopic expression of glutamate dehydrogenase did not rescue effective N2 fixation. Instead, the effect is specific to LIV and is accompanied by a major reduction in transcription and activity of LIV biosynthetic enzymes. Bacteroids become symbiotic auxotrophs for LIV and depend on the plant for their supply. Bacteroids with aap bra null mutations are reduced in number, smaller, and have a lower DNA content than wild type. Plants control LIV supply to bacteroids, regulating their development and persistence. This makes it a critical control point for regulation of symbiosis.
Keywords: mutualism, nitrogen fixation, peas, symbiosis
The largest input of available nitrogen in the biosphere comes from biological reduction of atmospheric N2 to ammonium (1). Most of this comes from legume–Rhizobium symbioses, arising from infection of host plants and resulting in root structures called nodules (2). These symbioses are initiated by plant-released flavonoids and related compounds, which elicit synthesis of lipochitooligosaccharide Nod factors by rhizobia. Bacteria are trapped by curling root hairs that they enter via infection threads. These grow into the root cortex, into a zone of newly induced meristematic cells forming the origin of the nodule. Bacteria are released from infection threads by endocytosis and are surrounded by a plant-derived symbiosome membrane. In nodules of galegoid legumes (a clade in the subfamily Papilionoideae, such as Medicago, Pisum, or Vicia), bacteria undergo dramatic increases in size, shape, and DNA content (3) before they start to reduce N2. Plants provide differentiated bacteria (bacteroids) with dicarboxylic acids, which energize N2 reduction to ammonium for secretion back to the plant (4).
A simple exchange of dicarboxylates and ammonium is the classical model of nutrient exchange in nodules, but amino acid transport by bacteroids has also been shown to be essential (5). Rhizobium leguminosarum mutated in 2 broad-specificity amino acid ABC transporters (Aap and Bra) formed N2-fixing pea bacteroids that appeared morphologically normal in electron micrographs, but the plants were nitrogen-starved. In addition, rhizobial aspartate aminotransferase mutants did not fix N2 in alfalfa or pea nodules (5, 6). It was therefore proposed that glutamate, or a derivative of it, is donated by the plant and aspartate, or possibly alanine is secreted by the bacteroid (5). Although this is a radical departure from the classical model, it does not replace it, because dicarboxylic acids are the main carbon source for bacteroids, and ammonium is the main nitrogen secretion product (4, 5).
R. leguminosarum Aap (AapJQMP) and Bra (BraDEFGC) are ABC uptake systems containing periplasmic solute-binding proteins (AapJ and BraC), integral membrane proteins (AapQM and BraDE), and ATP-binding cassettes (AapP and BraFG) (7). Their broad specificity, for acidic, basic, and neutral amino acids obscures which amino acids move in bacteroids and leaves the amino acid cycling model unconstrained. Furthermore, a seemingly improbable alternative to this model was considered, where bacteroids become auxotrophic for an amino acid(s) in the nodule, even though aap bra null mutants are prototrophic in laboratory culture (5).
In this study, the solute specificities of Aap and Bra were constrained, and only branched-chain amino acids (LIV) must be transported into bacteroids to support development and, therefore, N2 fixation. Remarkably, bacteroids become symbiotic auxotrophs. This has major implications for our understanding of mutualism between partners in legume–Rhizobium symbioses.
Results
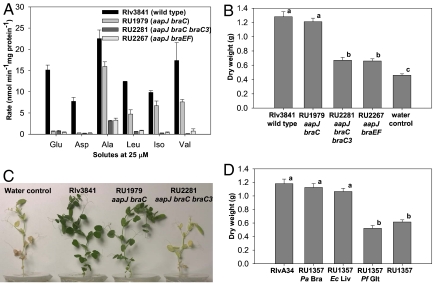
Analysis of uptake kinetics suggested that the BraDEFG membrane complex uses 2 solute-binding proteins (SBPs) with different specificities (7). Of these, BraC is well-characterized, with the gene in the braDEFGC operon, but the second SBP was unknown. By using the R. leguminosarum bv viciae strain 3841 (hereafter Rlv3841) genome sequence, 12 putative hydrophobic amino acid SBPs were identified. The protein with the highest identity (59%) to BraC is coded for by RL3540. This gene (braC3) was mutated by insertion of a kanamycin resistance cassette and recombined into an aapJ braC strain (RU1979) lacking both known SBPs of the Aap and Bra membrane complexes. With the 3 SBP genes mutated (RU2281; Fig. 1A), the uptake of all amino acids tested was reduced to similar background levels as in RU2267 (aapJ braEF), an aap bra null mutant which lacks detectable transport via Aap (SBP-mutated) or Bra (membrane complex-mutated). Thus, BraC and BraC3 are the 2 SBPs that interact with BraDEFG. In the double SBP mutant RU1979 (aapJ braC), transport occurred exclusively through BraDEFG via BraC3, and only LIV and alanine were transported (Fig. 1A).
Fig. 1.
The symbiotic phenotype of aap bra mutants is dependent on LIV transport. (A) Amino acid transport of Rlv3841 and aap bra mutants. (B) Shoot dry weights of pea plants inoculated with Rlv3841 and aap bra mutants (n ≥ 13). (C) Six-week-old pea plants inoculated with Rlv3841 and aap bra mutants. (D) Shoot dry weights of pea plants inoculated with RlvA34 and an aap bra null mutant (RU1357) complemented with heterologous narrow-solute specificity ABC transport systems from P. aeroginosa, E. coli, and P. fluorescens (n ≥ 14). Different letters above bars indicate significant differences (P < 0.01).
Plant Phenotype.
Peas inoculated with RU2267 (aapJ braEF) were yellow, had increased numbers of small, pale pink nodules, and had dry weights similar to uninoculated controls (Fig. 1B), and acetylene reduction was 30% of wild-type-inoculated plants. This is indistinguishable from the previously characterized aap bra null mutants (5). Plants inoculated with strain RU2281 lacking all 3 SBPs (aapJ braC braC3) were similarly defective in symbiosis (Fig. 1 B and C). However, peas inoculated with RU1979 (aapJ braC), which only transports LIV and alanine via BraC3, were indistinguishable from plants inoculated with wild-type Rlv3841 (Fig. 1 B and C). Thus LIV, and/or alanine transport alone is sufficient for effective nitrogen fixation.
Only LIV Transport Is Essential for Symbiosis.
Although LIV and alanine transport alone support effective N2 fixation, other amino acids may also do this. To resolve this, heterologous ABC transport systems of narrow solute specificity were expressed from a stable plasmid (pJP2) in an aapJQM braE null mutant (RU1357). This strain is derived from R. leguminosarum bv viciae A34 (RlvA34) and, unlike RU2267, allows complementation with the tetracycline-marked plasmid pJP2. The aap bra null mutants of RlvA34 and Rlv3841 behave identically on peas (5).
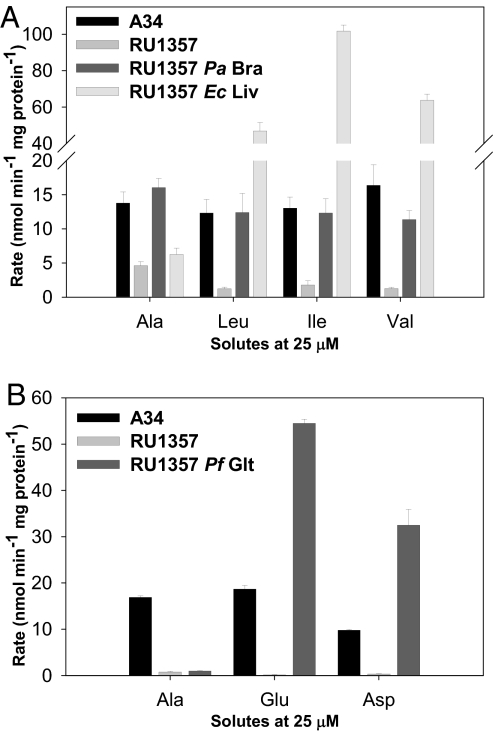
Pseudomonas aeruginosa strain PAO3012 braCDEFG (8) and Pseudomonas fluorescens SBW25 gltIJKL were fused to the Rlv3841 aapJ promoter and cloned in the plasmid pJP2. Escherichia coli K12 livKHMGF (9) was cloned into the same vector under control of the constitutive nptII promoter. P. aeruginosa BraCDEFG is specific for LIV and alanine transport (Fig. 2A), E. coli LivKHMGF is specific for LIV alone (Fig. 2A), and P. fluorescens GltIJKL is specific for aspartate and glutamate (Fig. 2B) when expressed in RU1357. The narrow specificity of the E. coli LivKHMGF system is consistent with a recent reinvestigation (9).
Fig. 2.
Amino acid transport of RlvA34 and an aap bra null mutant (RU1357) complemented with heterologous narrow-solute specificity ABC transport systems from P. aeroginosa and E. coli (A) and P. fluorescens (B).
Peas inoculated with RU1357 containing either P. aeruginosa BraCDEFG or E. coli LivKHMGF were healthy and green, with dry weights not significantly different from wild-type RlvA34 (Fig. 1D). However, plants inoculated with RU1357 P. fluorescens GltIJKL were yellow and stunted, and they had dry weights similar to RU1357 (Fig. 1D). Thus, the minimum transport requirement to complement the aap bra null mutant phenotype is LIV uptake alone. Mutualism in the legume–Rhizobium symbiosis requires exchange of a dicarboxylate, ammonia and an amino acid(s) (5), but the amino acid(s) moved is completely unexpected.
Bacteroids Specifically Require Branched-Chain Amino Acids.
Nitrogen assimilation via the glutamine synthase/glutamate synthetase system is largely inactive in bacteroids (10). The requirement for LIVs could be because they are the only amino acids that cross the plant-derived symbiosome membrane in significant amounts and are the nitrogen source for bacteroids. Consistent with this, amino acids such as glutamate do not cross the symbiosome membrane of soybean (11). It might therefore be possible to release bacteroids from their requirement for LIV transport by restoring glutamate synthesis. Expression of E. coli glutamate dehydrogenase (GdhA) from a nifH promoter resulted in significant glutamate synthesis in Rhizobium etli bacteroids (12). Therefore, gdhA was cloned in the plasmid pJP2 under the control of the Rlv3841 nifH promoter. Acetylene reduction by 3-week-old peas inoculated by RU1357 pJP2 nifHpGdhA (0.98 μmol h−1 plant−1 ± 0.07; n = 6) was not significantly different from RU1357 pJP2 (0.83 μmol h−1 plant−1 ± 0.12; n = 6) and was only 44% of RlvA34 pJP2 (2.22 μmol h−1 plant−1 ± 0.15; n = 6). This occurred despite high GdhA activity in RU1357 bacteroids containing nifHpGdhA (325 nmol min−1 mg protein−1; n = 2 plant harvests). Bacteroids require LIV transport, and this cannot be relieved by endogenous glutamate synthesis. Thus, although free-living cultures are prototrophic for LIV, bacteroids become auxotrophs. We propose to call this phenomenon “symbiotic auxotrophy.”
Symbiotic Auxotrophy Is Caused by Down-Regulation of LIV Biosynthesis.
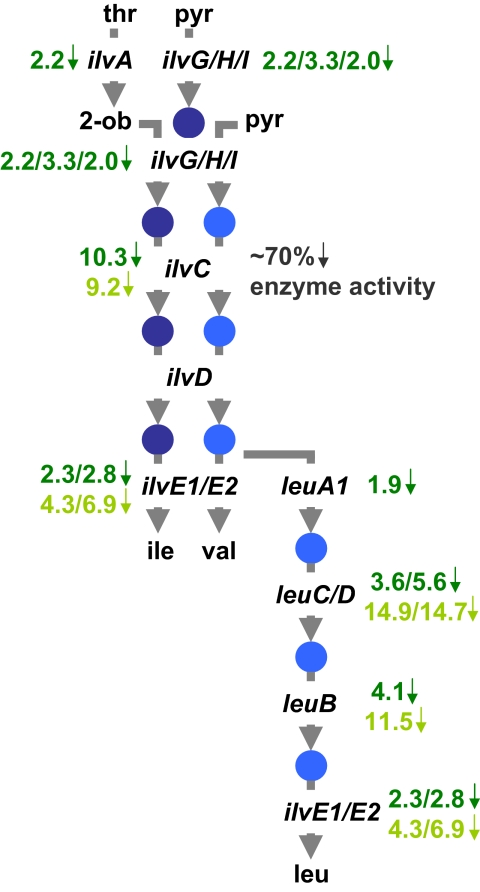
The carbon skeletons of leucine and valine are derived from pyruvate, whereas isoleucine is made from pyruvate and 2-oxobutanoate (Fig. 3). Both branches of LIV biosynthesis are coupled and dependent on a set of common genes, designated ilv and leu. In the final step of LIV synthesis, oxo acids are transaminated by using glutamate as a nitrogen donor. A microarray time course has been conducted on Rlv3841 bacteroids compared with cultured cells grown on minimal medium with 10 mM succinate and ammonium chloride (13), and we used this dataset to examine expression of ilv and leu genes. Although ilvD was constitutively expressed, ilvA, ilvE1, ilvGI, and leuA1 were moderately down-regulated (1.9- to 2.3-fold), and ilvC, ilvE2, ilvH, and leuBCD were strongly down-regulated (2.8- to 10.3-fold; Fig. 3 and Table S1). The down-regulation of ilvD, ilvE1/2, and leuBCD was confirmed by quantitative RT-PCR (4.3- to 14.9-fold; Fig. 3 and Table S1). Additionally, the enzyme activity of ketol-acid reductoisomerase (IlvC) decreased by ≈70% in bacteroids (2.53 nmol min−1 mg protein−1; n = 2 plant harvests) compared with cells grown in succinate ammonia minimal medium (7.82 ± 0.51 nmol min−1 mg protein−1; n = 3).
Fig. 3.
The biosynthetic pathway for branched-chain amino acids is down-regulated in bacteroids. The down-regulation of the Rlv3841 putative biosynthetic ilv and leu genes is shown in green (dark green, microarray; pale green, qRT-PCR; Table S1). The reduction in detectable ketol-acid reductoisomerase activity (IlvC) is given. Abbreviations are: threonine (thr), 2-oxobutanoate (2-ob), pyruvate (pyr), isoleucine (ile), valine (val), and leucine (leu).
The Symbiotic Performance of leuD Auxotrophs Requires LIV Transport by Aap or Bra.
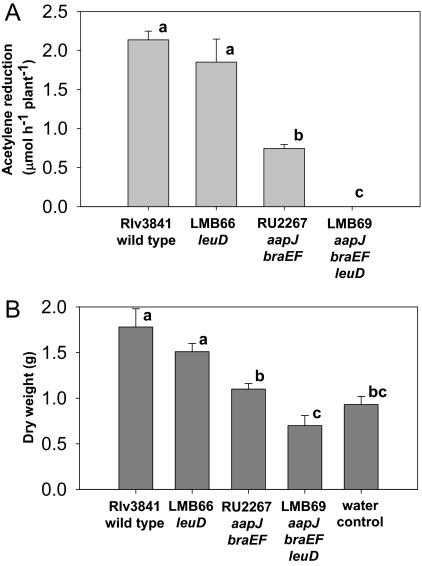
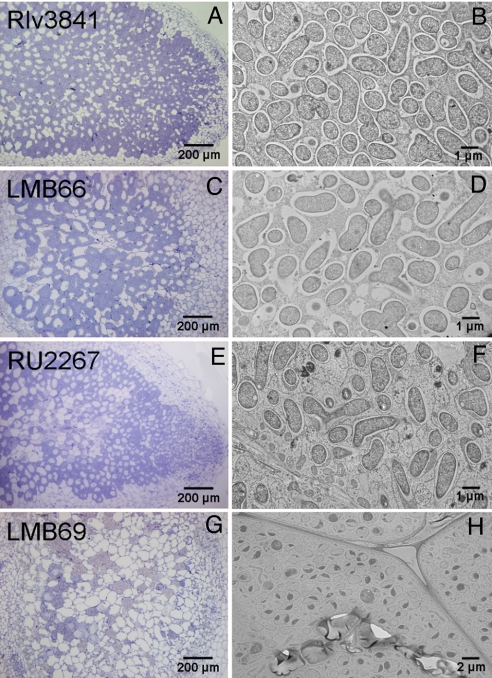
We wish to distinguish between symbiotic auxotrophy, where LIV biosynthesis is restricted specifically in planta, and auxotrophy, where a bacterial mutant cannot make LIV. In symbiotic auxotrophy, wild-type bacteroids reduce LIV synthesis and receive some or all of these amino acids from the plant. This implies that LIV auxotrophs might be unaltered in symbiotic performance, because the plant normally provides LIV. A caveat is that LIV mutants might be blocked in growth steps leading to bacteroid formation. To investigate this, both branches of the LIV biosynthetic pathway were mutated with interposons in ilvD (encodes dihydroxy-acid dehydratase) and leuD (encodes the small subunit of 3-isopropylmalate dehydratase). The Rlv3841 ilvD mutant (LMB65) did not nodulate peas, but the leuD mutant (LMB66) did so long as LIVs (all 1 mM) were added to plant growth medium. Leu (1 mM) alone also rescued nodulation but caused root deformation. Plants inoculated with LMB66 (leuD) formed fewer nodules (≈50–80%) relative to plants inoculated with Rlv3841, but both acetylene reduction and dry weights were indistinguishable between strains (Fig. 4). Although nodules infected by LMB66 were more spherical and contained more starch than wild type (Fig. 5C), electron micrographs showed that LMB66 bacteroids were visually normal (Fig. 5D). Although aap bra null mutants become starved for LIV, the leuD auxotroph was rescued by the plant as long as it was assisted in the early stage of interaction with roots and nodule initiation by addition of LIV.
Fig. 4.
The symbiotic phenotype of leuD mutants. (A) Acetylene reduction of pea plants inoculated with Rlv3841 and RU2267 and the respective leuD mutants LMB66 and LMB69 (n = 12); nodulation medium was supplemented with LIV at 1 mM. (B) Shoot dry weights of pea plants inoculated with the same strains (n = 8). Nodulation media were supplemented with LIV at 1 mM. Different letters above bars indicate significant differences (P < 0.01). Uninoculated control plants were the only ones for which the dry weight significantly increased on supplementing the growth medium with LIV.
Fig. 5.
Bacteroid development of aap bra and leuD mutants. (A) Light micrograph of a pea nodule formed by Rlv3841. (B) Transmission electron micrograph of Rlv3841 bacteroids. (C) Light micrograph of nodule formed by LMB66. (D) Transmission electron micrograph of LMB66 bacteroids. (E) Light micrograph of nodule formed by RU2267. (F) Transmission electron micrograph of RU2267 bacteroids. (G) Light micrograph of nodule formed by LMB69. (H) Transmission electron micrograph of LMB69-infected cortical nodule cells.
The leuD mutation was introduced into RU2267, forming LMB69 (aapJ braEF leuD). This strain grew, albeit poorly, in minimal medium, provided leucine was added at more than 0.5 mM. This suggests that Rlv3841 has an additional low-affinity leucine transport system sufficient for poor growth in culture. Peas inoculated with LMB69 plus 1 mM LIV had fewer (≈20–40% of wild type) white nodules and failed to reduce acetylene (Fig. 4A). At 6 weeks, dry weights of LMB69-inoculated and uninoculated plants were indistinguishable (Fig. 4B). Nodules from LMB69 were more spherical than wild type, containing mainly empty plant cells with elevated starch and no developed bacteroids (Fig. 5 G and H). Thus, a leuD auxotroph in planta is dependent on high-affinity LIV transport via Aap and Bra for bacteroid development.
The inability of the ilvD auxotroph to nodulate, despite addition of LIV to the medium, may mean peas never provide ile and val to rhizobia. However, it might result from a failure to provide ile and val at a particular step early in infection or development.
The aap bra Bacteroids Are Impaired in Development.
Electron micrographs show that aap bra null mutants form pleomorphic bacteroids; however, unlike wild type, they accumulate polyhydroxybutyrate (5). Furthermore, aap bra bacteroid protein per plant was only 38% of wild type (5). The severe limitation of aap bra null mutants for LIV suggests impaired development, leading to a drastic reduction in bacteroid number. Bacteroids from galegoid legumes increase their chromosome number via several cycles of endoreduplication (3). Therefore, information about size, quantity, and DNA content of RU2267 and Rlv3841 bacteroids was obtained by using flow cytometry. Bacteroids of RU2267 were smaller (50% reduced median forward scatter; Fig. S1A) than Rlv3841 and had a lower chromosome number of ≈5 compared with ≈8 (Fig. S1B). RU2267 had reduced bacteroid protein (42%) and cell number (48%) per plant compared with Rlv3841.
Discussion
The broad-solute range amino acid transporters Aap and Bra are essential for effective N2 fixation in pea nodules (5). Peas infected with Aap bra null mutants reduce N2 at ≈30–50% of wild-type rates, and plants become nitrogen-starved, with very low asparagine levels in the xylem sap. Because aspartate aminotransferase activity is also required for N2 fixation, an amino acid cycle was proposed (5, 6) to be essential for effective nitrogen assimilation by the plant. An alternative—that bacteroids are auxotrophic for amino acids—was considered but thought improbable because aap bra null mutants are prototrophic in laboratory culture. However, conclusive proof was lacking as a result of the broad specificity of Aap and Bra. The breakthrough in this work results from altering the specificity of Bra and showing that only LIV transport is essential for effective nitrogen fixation in peas. The seemingly improbable occurs, with bacteroids becoming symbiotic auxotrophs in planta. Symbiotic auxotrophy does not simply result from a lack of ammonium incorporation into amino acids, because overexpression of gdhA in bacteroids did not rescue aap bra null mutants. Instead, symbiotic auxotrophy is specific for branched-chain amino acids and is caused by significant down-regulation of LIV biosynthesis in bacteroids. This general transcriptional down-regulation of LIV biosynthesis also occurs in Sinorhizobium meliloti bacteroids in symbiosis with alfalfa (14, 15), and Bradyrhizobium japonicum with soybean (16). The dependence of the bacteroid on the plant for LIV suggests that a LIV transporter essential for symbiosis will be located on the peribacteroid membrane. In addition, LIV synthesis might be increased in the plant cytosol of nodule cells. However, this is not apparent in the Medicago truncatula gene expression atlas (http://bioinfo.noble.org/gene-atlas/).
The inability of aap bra null mutants to receive LIV from the plant results in fewer and less-developed bacteroids, as judged by overall bacteroid protein per plant, size, and DNA content. The presence of fewer bacteroids in plant cells infected by aap bra null mutants suggests a reduction in the total number of replication cycles before cell division ceases. However, it is not easy to distinguish between a decrease in replication cycles and a reduction in the number of bacteroids caused by earlier senescence.
Dependence of bacteroids on the plant for LIV has significant implications for our understanding of mutualism between symbionts. Bacteroids depend on their host for provision of all nutrients, but the reduction in bacteroid LIV biosynthesis to below the level required for effective nitrogen fixation is extraordinary. Why do bacteroids surrender control of LIV biosynthesis to the plant?
The first possibility is that plants induce symbiotic auxotrophy in bacteroids (i.e., the plant has control). Pseudoauxotrophy is known in E. coli, where provision of leu-containing peptides reduces expression of the first step in LIV biosynthesis (acetohydroxy acid synthase) (17), and bacteria require addition of ile and val. Legumes might impose a similar selection on bacteroids by provision of peptides and gain control over their development and persistence by regulating the supply of LIV.
If symbiosomes are considered as organelles, then a second possibility is that it is energetically favorable for the plant to provide LIV (i.e., mutualism dominates). This would generate a strong selective pressure if the overall result is more nitrogen-fixed for a given carbon/energy input. Put another way, an organelle that is efficient at N2 fixation but poor at biosynthesis relies on its host cell.
A third possibility is that metabolic precursors for LIV may become limited by bacteroid metabolism (i.e., the plant rescues). For example, the supply of pyruvate, which is the main oxo acid precursor for LIV biosynthesis, may be severely reduced by its use in metabolism to fuel N2 fixation. Another possibility includes the disruptive effect of peroxide on iron-sulfur cluster-containing enzymes, such as 3-isopropylmalate dehydratase (18). Peroxide is present in plant tissues during infection, bacteroid development, and senescence, and it requires detoxification by the bacterial symbiont for normal nodule function (19). However, a metabolic limitation would also need to explain the transcriptional down-regulation of the biosynthetic enzymes.
It is also relevant to ask why LIVs, rather than other amino acids, limit bacteroid development and persistence. Although there is no simple answer, LIVs are the most abundant amino acids in proteins (23% of Rlv3841 proteome) and are formed by a complex biosynthetic pathway subject to limitation. For example, the level of leu is a key indicator of amino acid availability in enteric bacteria sensed via the leucine-responsive protein (20).
Numerous rhizobial auxotrophs have been tested for N2 fixation (21–25). Although some are rescued in planta, suggesting that the host can provide the missing compound, it does not indicate that wild-type bacteroids require them. This contrasts with LIV, which plants must provide to wild-type bacteroids. The transport data do not permit distinction between leu, ile, and val with regard to which amino acid(s) need to be provided by the plant. However, although a leuD auxotroph formed effective nodules, an ilvD mutant did not, even though LIV was added to the external medium to rescue colonization. This suggests that although leu is provided in planta, ilv and val are not. Similarly, S. meliloti leu auxotrophs establish an effective symbiosis with alfalfa if rescued for nodulation by the addition of leu (21). However, nodules induced by ilv mutants do not contain bacteroids, again suggesting ile and val are not provided in planta (21). Overall, French bean, alfalfa, soybean, and pea are able to provide leu to invading rhizobia and developing bacteroids (refs. 21, 22, and 26, and the present study). These data are compatible with the provision of leu being indispensable to legume–Rhizobium symbioses.
It was originally proposed that amino acid cycling drives nitrogen fixation via a metabolic cycle, with glutamate or a derivative of it being imported into the bacteroid, and aspartate or alanine exported (5). However, we now demonstrate that aap bra null mutants lack LIV uptake rather than glutamate. Furthermore, amino acids do not need to cycle back to the plant, because LIVs are required for full development and persistence of bacteroids, rather than metabolic cycling. This raises the question of the role of aspartate aminotransferase, which is essential for N2 fixation and was proposed to be needed for return (cycling) of aspartate to the plant (5). Because LIV starvation occurs in mature bacteroids, aspartate aminotransferase may be needed early in bacteroid development to redeploy various amino acids. This fits with its role as the central transaminase in metabolism, and its mutation may cause widespread amino acid starvation in bacteroids. Consistent with this, although aatA bacteroids appear visually normal (5), flow cytometry showed their forward scatter is 50% of wild type, indicating they are smaller and not fully developed.
It has been shown that per bacteroid, aap bra null mutants fix nitrogen at the same rate as wild type (5). However, the failure of the mutants to fully develop and persist, with the consequent drop in total numbers of bacteroids per plant, explains the overall reduced N2 fixation rate. Even so, it is still necessary to explain the almost total collapse in amide export into the xylem. One possibility is that nodules retain a constant amount of nitrogen for their own needs before exporting amides. A plant fixing N2 at 30% of the wild-type rate would export a considerably lower percentage of amides. This may be particularly relevant to peas infected with aap bra null mutants, which induce an increased number of smaller nodules compared with the wild type.
Overall, this study shows that supply of LIV to bacteroids by the plant is essential for development, and ultimately for symbiotic N2 fixation by peas, and possibly legumes generally. Bacteroids are amino acid-starved and become symbiotic auxotrophs for LIV. This emphasizes that symbiosomes have complete metabolic dependence on their hosts and behave like organelles.
Materials and Methods
General Bacterial, Plant, and Molecular Analysis.
Peas (Pisum sativum cv Avola) were grown in 1 l pots in vermiculite for 3 weeks for acetylene reduction assays, or in 2 l pots in vermiculite for 6 weeks for dry weight experiments (27). Mutants and plasmids are described in SI Methods. RNA preparations, microarrays, and qRT-PCR were performed as described previously (28). A full description of the microarray results can be found elsewhere (13). R. leguminosarum uptake assays were performed with 25 μM (4.625 kBq of 14C) solute (7) and using cultures grown in acid minimal salts (AMS) with 10 mM glucose and 10 mM NH4Cl to an OD600 of ≈0.4.
Ketol-Acid Reductoisomerase Enzyme Assay.
Rlv3841 cultures were grown in AMS with 10 mM glucose and 10 mM NH4Cl to an OD600 of ≈0.4. Rlv3841 bacteroids were isolated from pea nodules and purified on Percoll gradients. Cell-free extracts obtained from bacterial culture pellets or bacteroids were disrupted in 40 mM Hepes, pH 7.2; 10 mM Mg2Cl; and 1 mM DTT (27). Ketol-acid reductoisomerase activity was assayed in 100 mM Tris·HCl, pH 7.5; 10 mM acetolactate; 2.5 mM NADPH; and 10 mM MgCl2 at 25 °C over 15 min with various amounts of crude extract. Acetolactate was freshly prepared from ethyl 2-acetoxy-2-metylacetoacetate following the protocol for Bacillus subtilis α-acetolactate decarboxylase (A7206; Sigma–Aldrich). Protein levels were determined by Bradford assay.
Flow Cytometry.
Bacteroids and free-living bacteria were stained with DAPI at 50 μg mL−1 and analyzed with an inFlux flow cytometer (Cytopeia). DNA content of stained bacteroids and bacteria was measured by quantifying DAPI fluorescence by using a photomultiplier tube with a 460/50-nm bandpass filter. Size information was obtained by measuring scatter characteristics on forward scatter. Data analysis was performed with FlowJo software (Tree Star).
Supplementary Material
Acknowledgments.
This work was supported by Biotechnology and Biological Sciences Research Council Grant BBS/B/02916.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903653106/DCSupplemental.
References
- 1.Newton WE. In: Nitrogen Fixation: From Molecules to Crop Productivity. Pedrosa FO, Hungria M, Yates MG, Newton WE, editors. Dordrecht, The Netherlands: Kluwer; 2000. pp. 3–8. [Google Scholar]
- 2.Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with Rhizobial infection in legumes. Ann Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 3.Mergaert P, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Lodwig EM, et al. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature. 2003;422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 6.Rastogi VK, Watson RJ. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in. Rhizobium meliloti. J Bacteriol. 1991;173:2879–2887. doi: 10.1128/jb.173.9.2879-2887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosie AHF, Allaway D, Dunsby HA, Galloway CS, Poole PS. Rhizobium leguminosarum has a second general amino acid permease with unusually broad substrate specificity and high similarity to branched-chain amino acid transporters (Bra/LIV) of the ABC family. J Bacteriol. 2002;184:4071–4080. doi: 10.1128/JB.184.15.4071-4080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoshino T, Koseterai K, Sato K. Solubilization and reconstitution of the Pseudomonas aeruginosa high-affinity branched chain amino-acid-transport system. J Biol Chem. 1992;267:21313–21318. [PubMed] [Google Scholar]
- 9.Koyanagi T, Katayama T, Suzuki H, Kumagai H. Identification of the LIV-I/LS system as the third phenylalanine transporter in Escherichia coli K-12. J Bacteriol. 2004;186:343–350. doi: 10.1128/JB.186.2.343-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patriarca EJ, Tate R, Iaccarino M. Key role of bacterial NH4+ metabolism in Rhizobium-plant symbiosis. Microbiol Mol Biol Rev. 2002;66:203–222. doi: 10.1128/MMBR.66.2.203-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udvardi MK, Salom CL, Day DA. Transport of L-glutamate across the bacteroid membrane but not the peribacteroid membrane from soybean root nodules. Mol Plant Microbe Interact. 1988;1:250–254. [Google Scholar]
- 12.Mendoza A, Valderrama B, Leija A, Mora J. NifA-dependent expression of glutamate dehydrogenase in Rhizobium etli modifies nitrogen partitioning during symbiosis. Mol Plant Microbe Interact. 1998;11:83–90. doi: 10.1094/MPMI.1998.11.2.83. [DOI] [PubMed] [Google Scholar]
- 13.Karunakaran R, et al. Transcriptomic analysis of Rhizobium leguminosarum b. v. viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J Bacteriol. 2009;191:4002–4014. doi: 10.1128/JB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capela D, Filipe C, Bobik C, Batut J, Bruand C. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: A transcriptomic dissection. Mol Plant Microbe Interact. 2006;19:363–372. doi: 10.1094/MPMI-19-0363. [DOI] [PubMed] [Google Scholar]
- 15.Barnett MJ, Tolman CJ, Fisher RF, Long SR. A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci USA. 2004;101:16636–16641. doi: 10.1073/pnas.0407269101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pessi G, et al. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact. 2007;20:1353–1363. doi: 10.1094/MPMI-20-11-1353. [DOI] [PubMed] [Google Scholar]
- 17.Lee CR, Cho SH, Yoon MJ, Peterkofsky A, Seok YJ. Escherichia coli enzyme IIANtr regulates the K+ transporter TrkA. Proc Natl Acad Sci USA. 2007;104:4124–4129. doi: 10.1073/pnas.0609897104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauly N, et al. Reactive oxygen and nitrogen species and glutathione: Key players in the legume-Rhizobium symbiosis. J Exp Bot. 2006;57:1769–1776. doi: 10.1093/jxb/erj184. [DOI] [PubMed] [Google Scholar]
- 20.Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Mol Biol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de las Nieves Peltzer M, et al. Auxotrophy accounts for nodulation defect of most Sinorhizobium meliloti mutants in the branched-chain amino acid biosynthesis pathway. Mol Plant Microbe Interact. 2008;21:1232–1241. doi: 10.1094/MPMI-21-9-1232. [DOI] [PubMed] [Google Scholar]
- 22.Ferraioli S, et al. Auxotrophic mutant strains of Rhizobium etli reveal new nodule development phenotypes. Mol Plant Microbe Interact. 2002;15:501–510. doi: 10.1094/MPMI.2002.15.5.501. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola TK, Kahn ML. Symbiotic phenotypes of auxotrophic mutants of Rhizobium meliloti 104A14. J Gen Microbiol. 1988;134:913–919. doi: 10.1099/00221287-134-4-913. [DOI] [PubMed] [Google Scholar]
- 24.Pain AN. Symbiotic properties of antibiotic-resistant and auxotrophic mutants of Rhizobium leguminosarum. J Appl Bacteriol. 1979;47:53–64. [Google Scholar]
- 25.Aguilar OM, Grasso DH. The product of the Rhizobium meliloti ilvC gene is required for isoleucine and valine synthesis and nodulation of alfalfa. J Bacteriol. 1991;173:7756–7764. doi: 10.1128/jb.173.24.7756-7764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kummer RM, Kuykendall LD. Symbiotic properties of amino acid auxotrophs of Bradyrhizobium japonicum. Soil Biol Biochem. 1989;21:779–782. [Google Scholar]
- 27.Allaway D, et al. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol Microbiol. 2000;36:508–515. doi: 10.1046/j.1365-2958.2000.01884.x. [DOI] [PubMed] [Google Scholar]
- 28.Fox AA, White JP, Hosie AHF, Lodwig EM, Poole PS. Osmotic upshift transiently inhibits uptake via ABC transporters in gram-negative bacteria. J Bacteriol. 2006;188:5304–5307. doi: 10.1128/JB.00262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.