Abstract
Little is known about how proteins begin to unfold. In particular, how and when water molecules penetrate into the protein interior during unfolding, thereby enabling the dissolution of specific structure, is poorly understood. The hypothesis that the native state expands initially into a dry molten globule, in which tight packing interactions are broken, but whose hydrophobic core has not expanded sufficiently to be able to absorb water molecules, has very little experimental support. Here, we report our analysis of the earliest observable events during the unfolding of single chain monellin (MNEI), a small plant protein. Far- and near-UV circular dichroism measurements of GdnHCl-induced unfolding indicate that a molten globule intermediate forms initially, before the major slow unfolding reaction commences. Steady-state fluorescence resonance energy transfer measurements show that the C-terminal end of the single helix of MNEI initially moves rapidly away from the single tryptophan residue that is close to the N-terminal end of the helix. The average end-to-end distance of the protein also expands during unfolding to the molten globule intermediate. At this time, water has yet to penetrate the protein core, according to the evidence from intrinsic tryptophan fluorescence and 8-anilino-1-naphthalenesulfonic acid fluorescence-monitored kinetic unfolding measurements. Our results therefore provide direct evidence for a dry molten globule intermediate at the initial stage of unfolding. Our results further suggest that the structural transition between the native and dry molten globule states could be an all-or-none transition, whereas further swelling of the globule appears to occur gradually.
Keywords: continuous transition, protein unfolding, single chain monellin, steady-state FRET
Two models are commonly invoked to describe the rate-limiting step during protein unfolding. The commonly accepted model is that the rate-limiting step is controlled by the extensive rearrangement of native structure upon the entry of water into the hydrophobic core (1–5). In the alternative model, based on the dry molten globule hypothesis (6), the rate-limiting step is an initial concerted rupture of the tight side-chain packing in the interior, without any entry of water. The free energy barrier arises because the loss of enthalpy in the dry globular transition state has not yet been compensated for by a gain in conformational entropy (7, 8). Experimental evidence in support of this hypothesis is, however, scarce and moreover, indirect (9–11). The dry molten globule model posits that the tight packing interactions are lost cooperatively when thermal fluctuations cause secondary structural elements to move marginally apart from each other (7). But small displacements of individual secondary structural units have not been detected as the initial steps of the unfolding of any protein. In particular, the detection of the rotation or translation of an α-helix or the fraying movement of a β-strand during the formation of a dry molten globule, which would constitute the most direct evidence in support of the dry molten globule hypothesis, has been difficult to capture in experiments.
Multisite FRET measurements allow determination of the displacements of specific segments of a protein structure, during folding or unfolding (12–19). In this study, 2-site FRET measurements of the unfolding of single-chain monellin (MNEI), in both equilibrium and kinetic unfolding experiments, have been carried out to determine how an intramolecular distance, spanning the N- and C-termini of the sole helix in the protein changes, compared with an intramolecular distance corresponding to the end-to-end distance of the protein. MNEI is an intensely sweet, small plant protein in which the sole α-helix is packed against a 5-stranded β-sheet in a β-grasp fold (Fig. 1A). In earlier studies, the folding and unfolding of MNEI have been characterized in detail (20–22). The folding and unfolding reactions of MNEI appear to be multistate, with multiple intermediates populating on parallel pathways (21). It is shown here that the unfolding of the protein begins with an initial expansion of the protein into a dry molten globular state, in which the sole helix has moved out of its place in the native structure. Furthermore, it is also shown that the initial structural transition between the native and dry molten globule states is likely to be an all-or-none transition, whereas further swelling of the globule appears to occur gradually.
Fig. 1.
FRET as a measure of intramolecular distances. (A) Structure of MNEI. The location of W4 and the residues (P97 and Q29) that were replaced by cysteine residues are shown. Both P97 and Q29 were mutated independently to C, to yield two different single cysteine-containing mutant proteins, Cys97 and Cys29, respectively. The structure was drawn from PDB file 1IV7 by using the program PyMOL (http://www.pymol.org). The sole thiol moiety in each protein was labeled with TNB (as described in SI Text) that quenches the fluorescence of W4 in a distance-dependent manner. The mutant proteins labeled with TNB are named as Cys97-TNB and Cys29-TNB. (B and C) Fluorescence emission spectra of unlabeled and TNB-labeled proteins. (B) Cys97 and Cys97-TNB; (C) Cys29 and Cys29-TNB. In both B and C, the solid blue line and the solid dark red line represent the fluorescence spectra of unlabeled and TNB-labeled native proteins, respectively. The dashed blue line and the dashed dark red line denote the fluorescence spectra of unlabeled and TNB-labeled unfolded protein, respectively. All of the fluorescence spectra in B and C were collected in an identical manner, with the excitation wavelength set to 295 nm.
Results and Discussion
The Quenching of Fluorescence of W4 by TNB Is Due to FRET.
Two different single cysteine-, single tryptophan-containing mutant forms of MNEI have been used (Fig. 1A), in each of which a single tryptophan residue, W4, serves as the donor fluorophore (D), and a thionitrobenzoate (TNB) adduct attached to a differently located thiol (of C29 or C97) serves as the FRET acceptor (A). W4 is located in the first β-strand at the N-terminus of the protein and is approximately at the N-terminal end of the helix (residues 9–28). C29 is located at the C-terminal end of this helix. C97 is the last residue in the protein sequence. For the TNB-labeled proteins, Cys97-TNB and Cys29-TNB, the FRET measurements probe how the end-to-end distance of the protein and the distance separating the N- and C-termini of the helix, respectively, change during unfolding.
Both the unlabeled mutant proteins, Cys97 and Cys29, exhibit identical fluorescence emission spectra of W4 in the N state and in the U state (Fig. 1 B and C). The fluorescence spectra are also similar to those of wild-type (WT) MNEI (21). The fluorescence of W4 is quenched dramatically in the N state of Cys97-TNB and Cys29-TNB, but to different extents (Fig. 1 B and C), and is also quenched to a lesser extent in the corresponding U states. The observation that the extent of quenching of W4 depends on the position of the C-TNB in the protein, and that the extent of quenching is more in the compact native state than in the expanded unfolded form, indicates that the quenching is distance-dependent and hence, due to FRET between the donor (W4) and the acceptor (TNB). The values of the Förster's distance (R0) and the D–A distances (R) for both the TNB-labeled proteins were determined (Figs. S1 and S2, and Table S1), as described in SI Text.
Unfolding of MNEI Begins by Formation of a Dry Molten Globule.
Before using site-specific FRET probes to study unfolding, it was important to use more traditional structural probes—far-UV circular dichroism (CD), near-UV CD, and intrinsic tryptophan fluorescence—to obtain a basic description of the unfolding of Cys97 and Cys29 (Fig. 2). For both proteins, the kinetic traces of far-UV CD monitored unfolding extrapolate at t = 0 to the signal of the native protein (Fig. 2 A and B). The kinetic amplitudes of unfolding match the equilibrium amplitudes (Fig. 2 A and B Insets), and the entire expected change in the far-UV CD signal (which represents the dissolution of hydrogen-bonded secondary structure) during unfolding, occurs in a single exponential phase.
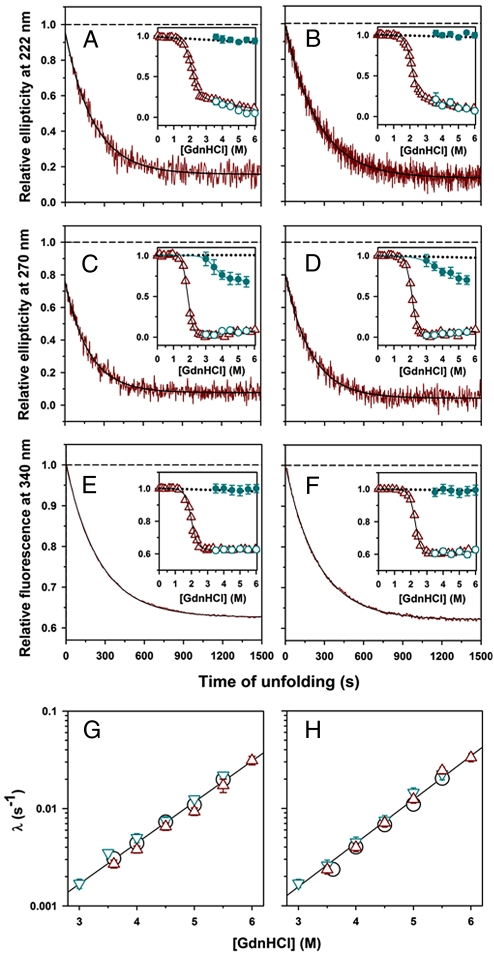
Fig. 2.
Unfolding kinetics of the unlabeled proteins. The unfolding of (A, C, E, and G) Cys97 and (B, D, F, and H) Cys29 at pH 8 and 25 °C was monitored by change in: (A and B) far-UV CD signal at 222 nm; (C and D) near-UV CD signal at 270 nm; and (E and F) fluorescence signal of W4 at 340 nm. In A–F, the solid dark red line shows the kinetic trace of unfolding in 4 M GdnHCl, and the solid black line through the data is a fit to a single exponential equation. The black dashed line represents the signal of native protein. Insets compare the kinetic versus equilibrium amplitudes of unfolding. The dark red triangles represent the equilibrium unfolding transition, and the continuous line through the data represents a fit to a two-state N⇌U model. The blue open circles represent the t = ∞ signal, and the blue filled circles represent the t = 0 signal, respectively, obtained from fitting the kinetic traces of unfolding to a single exponential equation. The black dotted line is a linear extrapolation of the native protein baseline. (C and D Insets) The blue line through the t = 0 values of the kinetic unfolding traces represents a fit to a two-state N⇌ I model. (G and H) Comparison of the observed rate constants of unfolding as monitored by change in far-UV CD signal at 222 nm (circles), near-UV CD signal at 270 nm (blue inverted triangles), and fluorescence of W4 at 340 nm (dark red triangles). In G and H, the black lines through the data are a straight line fit to the average of the rate constants from the 3 probes. The error bars, wherever shown, represent the standard deviations of measurements from at least 3 separate experiments.
However, the kinetic traces of near-UV CD-monitored unfolding do not extrapolate at t = 0 to the signal of the native protein at high denaturant concentrations (Fig. 2 C and D). The kinetic amplitudes are smaller than the equilibrium amplitudes (Fig. 2 C and D Insets), because part of the change in the near-UV CD signal (which represents the disruption of tertiary structure) occurs in the 10-s mixing dead time of the kinetic CD experiment, whereas the other part occurs in the observable single exponential phase. Hence, an unfolding intermediate, I, with native secondary structure but with partially disrupted tertiary structure, is populated within 10 s of unfolding, before the commencement of the major unfolding reaction. Because I is formed much faster than it subsequently unfolds, the denaturant dependence of its near-UV CD signal represents the denaturant dependence of the N⇌I equilibrium, if the N⇌I transition is an all-or-none “2-state” transition. I appears to be populated significantly only for unfolding in >3 M GdnHCl.
Fig. 2 E and F show that when the kinetics of unfolding of both the unlabeled mutant variants of MNEI was monitored by using intrinsic tryptophan fluorescence, a single exponential unfolding phase was observed that accounts for the entire expected change in fluorescence, and hence, the kinetic amplitudes match the equilibrium amplitudes (Fig. 2 E and F Insets). The unfolding kinetics of WT MNEI is similar. The rate constants of the observable change in signal as measured by tryptophan fluorescence and both the CD probes (described above) are similar at each denaturant concentration (Fig. 2 G and H). The quantum yield of tryptophan fluorescence decreases on hydration of W4, and the absence of any fast decrease in fluorescence accompanying the initial formation of I, indicates that even the exterior region of the protein, where W4 is located, remains as unhydrated in I as it is in N. Events that occur initially during unfolding can be expected to be similar to the final events during folding, and it is interesting that the final stage of folding too is silent to a change in intrinsic tryptophan fluorescence (21).
It should be mentioned here, that when the unfolding of MNEI was studied at pH 7, instead of at pH 8 as in this study, the unfolding traces extrapolated at t = 0 to values of fluorescence that exceeded that of N in the absence of denaturant (21). But unlike the native protein baseline of the equilibrium unfolding curve at pH 8, which has very little dependence on GdnHCl concentration, the native protein baseline at pH 7 has a steep positive slope. Thus, at pH 7, the extrapolated t = 0 fluorescence value of the kinetic trace of unfolding at any denaturant concentration was found to be less than that of the value expected for N in that denaturant concentration, on the basis of a linear extrapolation of the native protein baseline. It was therefore concluded that the fluorescence-monitored unfolding kinetics at pH 7, unlike those at pH 8 (Fig. 2 E and F), are biphasic (21).
An intact secondary structure and a disrupted tertiary structure are the defining characteristics of a molten globule (23, 24). Because the initial intermediate, I, possesses the secondary structure of N but a disrupted tertiary structure, and because it is as dry as N, it qualifies to be a dry molten globule. To confirm that I is not wet, its ability to bind the hydrophobic dye 8-anilino-1-naphthalenesulfonic acid (ANS) was studied. The fluorescence of ANS increases dramatically on binding to water-exposed hydrophobic patches in a wet molten globule, such as the one formed initially during the folding of MNEI (20, 21). It is expected that a hydrophobic probe like ANS would penetrate into the protein together with water, if there were any such penetration initially during unfolding. No transient increase in ANS fluorescence is, however, observed when the unfolding of Cys97 and Cys29 is carried out in presence of ANS, by using concentrations of protein and ANS similar to those with which an increase was observed in the folding studies (Fig. S3 A and B) (20, 21), or by using concentrations even 10-fold higher (Fig. S3 C and D). Hence, I is indeed a dry molten globule intermediate that is populated initially during the unfolding of MNEI.
It is commonly believed that only the hydrophobic interaction (exclusion of water) matters for the stability of folding (1–4). The significance of the observation of a dry molten globule is that it implies that only after close packing interactions are broken, can water enter and unfolding proceed. Thus, dispersion forces appear to play an important role in maintaining the integrity of structure of the native protein (25–27).
Expansion and Contraction of Intramolecular Distance During Unfolding.
For FRET measurements it was important to verify that the unlabeled and TNB-labeled proteins have similar biophysical properties. Fig. S4 shows that not only are the secondary structures and stabilities of Cys97, Cys29, Cys97-TNB, and Cys29-TNB similar, but so are the kinetics of unfolding (SI Text). In addition, like in the case of the unlabeled proteins, the kinetic traces of far-UV CD-monitored unfolding of Cys97-TNB and Cys29-TNB (Fig. S5 A and B) extrapolate at t = 0 to the signal of native protein: the kinetic amplitudes of unfolding match the equilibrium amplitudes (Fig. S5 A and B Insets). Moreover, for each TNB-labeled protein, the rate constant of the observable unfolding phase is similar, when measured by far-UV CD and FRET, at each concentration of GdnHCl (Figs. S5 C and D and S6). Hence, it was possible to use the fluorescence of the unlabeled protein (donor-only fluorescence) and the fluorescence of the corresponding TNB-labeled protein (donor fluorescence in the presence of acceptor), at different times of unfolding (Fig. S6) and in different concentrations of denaturant (Fig. S7), to determine how the FRET efficiency and the D–A distance change during unfolding (SI Text).
Fig. 3 A and B show that, during the unfolding of both TNB-labeled proteins, there is an initial decrease in FRET efficiency that occurs in an apparent burst phase (within the mixing dead-time of ≈6.2 ms), followed by a slower change. The slow change in FRET efficiency is a decrease for Cys97-TNB, but an increase for Cys29-TNB (SI Text). These results indicate that the average end-to-end distance of MNEI (the W4–C97TNB distance) expands during the unfolding reaction (Fig. 3C), as expected. However, the W4–C29TNB distance, separating the N- and C-termini of the α-helix, increases very rapidly and then decreases slowly, as the whole protein unfolds (Fig. 3D). Hence, the first response of MNEI, when it comes into contact with denaturant during unfolding, is to swell slightly in such a manner that the C-terminus of the helix and W4 move apart. Finally, as the whole structure dissolves slowly, the ends of the erstwhile helix come together, whereas the ends of the protein move apart.
Fig. 3.
Kinetics of unfolding of unlabeled and TNB-labeled proteins as monitored by FRET. (A) Cys97-TNB; (B) Cys29-TNB. In A and B, the changes in FRET efficiency during unfolding in 4 M GdnHCl (dark red continuous curve) and 6 M GdnHCl (dark green continuous curve) are shown, and the dashed black line shows the FRET efficiency estimated in the native protein. The data in A and B were converted into D–A distances by using Eq. S2, and the distances are shown for (C) Cys97-TNB and (D) Cys29-TNB at 4 M GdnHCl (dark red continuous curve) and 6 M GdnHCl (dark green continuous curve). The black dashed line shows the D–A distance estimated in the native protein. Identical kinetics of unfolding were seen at both 15 μM and 50 μM protein concentrations, indicating the absence of any transient aggregation during the unfolding reaction.
The slow observable phase of unfolding corresponds to the unfolding of the dry molten globule intermediate that is formed in the initial phase of unfolding. The dry molten globule intermediate possesses native-like secondary structure, but its buried side-chains have yet to become hydrated (Fig. 2; see above). The FRET studies show that a large expansion of the W4–C29TNB distance and a smaller expansion of the W4–C97TNB distance have occurred in the dry molten globule intermediate. One explanation for the initial expansion in distances is that the edge β-strand containing W4 has frayed and become detached from the rest of the β-sheet in the dry molten globule, leading to an increase in both measured distances. The other more likely (see below) explanation is that the α-helix has undergone a small movement away from the rest of the protein, thereby disrupting the internal hydrophobic core packing. It is possible that this movement of the helix may allow it to undergo partial unfolding at the ends (28, 29) or in the middle (30, 31), and thereby, to increase in length, although no partial unfolding is detected by far-UV CD (Fig. 2 A and B). It is likely that denaturant molecules enter into and bind to the dry molten globule, thereby facilitating subsequent unfolding (32). In a separate example of secondary structure motions versus overall unfolding, it was shown previously for phosphoglycerate kinase, again by using multisite FRET, that 2 regions of the protein move away from each other early during unfolding (12).
It is surprising that the W4–C29TNB distance is greater in the dry molten globule, than in the U state. The end-to-end distance of the 20-aa residue-long helix in MNEI is ≈30 Å. The distance measured by FRET in the N state is smaller (Table S1) because although the donor and acceptor are separated by 25 amino acid residues, the donor, W4, in the first β-strand, points toward the C-terminal of the helix (Fig. 1A). This distance is significantly smaller than the end-to-end distance of a 25-aa residue-long polypeptide chain in a random coil conformation, which is estimated to be ≈37 Å (33). Hence, a decrease in the D–A distance from the dry molten globule state to the U state appears possible only if some residual structure forms in the U state (34–36). Indeed, the observation that the W4–C97TNB distance, which separates 93 residues in the sequence, is only ≈ 26Å in the U state suggests that the U state is not a random coil but is compact.
Fig. 4 shows how the equilibrium and kinetic FRET efficiencies (Fig. 4 A and B), and the D–A distances derived from them (Fig. 4 C and D), change during the denaturation of Cys97-TNB and Cys29-TNB. For Cys97-TNB, the decrease in FRET efficiency, and the increase in the D–A distance, both have sigmoidal dependences on denaturant concentration (Fig. 4 A and C). For Cys29-TNB, however, the FRET efficiency shows a decrease-increase-decrease trend, and the D–A distance shows conversely an increase-decrease-increase trend, with an increase in denaturant concentration, but both still appear to change in a sigmoidal manner (Fig. 4 B and D). The observation of an initial expansion of the D–A distance confirms the results of the kinetic experiment, which suggest that unfolding starts at the helix ends, by the movement of the C-terminal of the helix away from W4. The observations that the helix collapses at ≈2 M GdnHCl (Fig. 4D), and that the collapse transition coincides with the overall unfolding transition of the protein (Fig. 2 A and B Insets), suggest that the 2 transitions are coupled. At 3 M GdnHCl, the whole protein begins to swell (Fig. 4C). Such a turnover in the end-to-end distance has been observed as a characteristic signature of a collapse transition in simulations of the formation or dissolution of helical structures in small peptides (37), and this observation therefore makes it unlikely that fraying of the edge β-strand instead of movement of the α-helix occurs during the formation of the dry molten globule (see above). The structural constraints imposed by stabilizing forces in a native protein, might be expected to significantly modulate the dynamics of a helix during unfolding (38). The observation of the turnover in the end-to-end distance of the helix (Fig. 4D) in the dry molten globule form of a protein, suggests that no such constraints are imposed by the structure of the dry molten globule.
Fig. 4.
Kinetic versus equilibrium amplitudes of the unfolding as monitored by FRET. (A) Cys97-TNB; (B) Cys29-TNB. In A and B, the changes in FRET efficiency during unfolding are shown. The data in A and B were converted into D–A distances by using Eq. S2, and the distances are shown in C and D for Cys97-TNB and Cys29-TNB, respectively. In A–D, the black circles represent the equilibrium unfolding data, and the black inverted triangles and the black triangles represent the t = 0 and the t = ∞ kinetic signals, respectively. The continuous black line represents a fit to a two-state N⇌U model, and the dotted black line is a fit of the native protein baseline and t = 0 kinetic data to a single exponential equation.
Interpreting the Native and Unfolded Protein Baselines of Equilibrium Unfolding Curves.
The native protein and unfolded baselines of the denaturant-induced equilibrium unfolding curve of a protein are typically taken to describe phenomenological dependences of the probe signal on the denaturant concentration, and are therefore ignored even when the dependences are pronounced (21). A previous time-resolved FRET study of the equilibrium unfolding of barstar (13) provided the first indication that in fact, native protein swells with increasing denaturant concentration in the native protein baseline regime, and that so does the unfolded protein with increasing denaturant concentration in the unfolded protein baseline regime. In this study too, both intramolecular distances in MNEI are seen to expand in the unfolded and native protein baselines regimes (Fig. 4 C and D). The observation that the native protein populates increasingly expanded forms present in the native state ensemble with increasing denaturant concentration is particularly important. It indicates that the native protein baselines of the D–A distance-monitored equilibrium unfolding transitions do not represent mere phenomenological dependences on denaturant concentration, but instead describe a true structural change: The swelling of the native state (13).
The denaturant dependence of each D–A distance in the dry molten globule intermediate appears continuous with that in the native protein, and the dependences are markedly different for the expansion of the 2 distances. The apparent continuity of the FRET-monitored transition from the N to I state is, however, not linear (Fig. 4), as is seen for the far-UV monitored transition (Fig. 2 A and B Insets), and the N to I reaction may appear to be continuous when studied by FRET, only because the swelling of N distorts the appearance of the transition. If the transition from the native to the dry molten globule state is indeed a continuous one, then the 2 states would be separated by many small distributed free-energy barriers (≤3 kBT), instead of 1 single dominant barrier (17, 39). Indeed, computer simulations have suggested that the initial fast transition during the unfolding of a protein may be a gradual structural transition (40). In fact, an early theoretical study had suggested that proteins undergo gradual transitions during denaturation (41). In this study, the sigmoidal nature of the near-UV CD-monitored N to I transition might suggest that it is an all-or-none transition (Fig. 2 C and D Insets) in which tight packing interactions are cooperatively disrupted. However, a sigmoidal transition does not necessarily indicate cooperative unfolding, but can instead arise from gradual unfolding (8, 13, 39). Hence, the possibility that the N to I transition is a continuous transition cannot be ruled out at the present time.
The Dry Molten Globule Hypothesis and Nature of the Initial Unfolding Transition.
According to a theoretically derived phase diagram of protein molecules in solution (6, 7), the native protein may denature into any 1 of 3 different globules: a dry molten globule into which water has not penetrated, a wet molten globule whose interior is hydrated, or a swollen globule that is also wet. In all cases, the denaturation reaction is all-or-none, and the transition state is dry, with the packing interactions not sufficiently perturbed for water to be able to enter. Transitions within and between the different denatured globules are continuous transitions, as is the swelling of the denatured globule to the final random coil state. In this study, MNEI is observed to unfold initially into a dry molten globule intermediate at all concentrations of denaturant used for unfolding (>3M GdnHCl). Because the initial unfolding intermediate is dry, the transition state of the all-or-none transition (see above) leading to it must necessarily also be dry. The results of this study provide direct experimental evidence for the existence of a dry molten globule state, and consequently of a dry globular transition state for a protein denaturation reaction.
Unfolding of the Dry Molten Globule Intermediate.
The major unfolding reaction of I to U (Figs. 2 and 3), in which large scale structural rearrangement occurs consequent to the entry of water into the interior of the protein, is much slower than the preceding transition leading to formation of I. According to the theoretically derived phase diagram (6), the large scale structural rearrangement of the initially formed denatured globule to the fully unfolded protein is a continuous swelling process that depends on denaturant concentration, and this prediction is supported by the observation that both D–A distances in I increase continuously with an increase in denaturant concentration (Fig. 4 C and D). The phase diagram (6) derives from a thermodynamic study of unfolding, which says nothing about the kinetics of the process. The kinetics of the unfolding of the dry molten globule is seen to be exponential (Figs. 2 and 3). Exponential kinetics are usually associated with an all-or-none process, but can also be produced by a gradual diffusive process occurring over many small distributed barriers (39). Indeed, a recent time-resolved FRET study of the unfolding of monellin strongly supports the latter possibility (42). The unfolding of the dry molten globule is very slow (Figs. 2 and 3); thus, it must be coupled to at least some activation barriers, such as those arising from the breaking of specific packing (van der Waal's) and other (H-bonding) intramolecular interactions still present in the dry molten globule (42). At least one of these activation barriers must necessarily be higher in free energy than that separating the native and dry molten globule states. It is possible that the transition from the dry to wet globule, in which some intramolecular interactions are broken on the entry of water, is an all-or-none transition under certain solvent and temperature conditions, as predicted by the theoretically derived phase diagram (6).
This study validates an important hypothesis of the dry molten globule model (6) by showing that the transition state of the initial transition from the native state to a denatured globule is dry (see above). But the initial denaturation step is also shown not to be the slowest step during the overall unfolding process. Unfortunately, this study provides no information about the barrier to the slowest step; hence, it neither validates nor invalidates another tenet of the dry molten globule model, namely that the transition state of the overall unfolding reaction is dry. Nevertheless, this study does validate another important prediction of the dry molten globule model (6, 7), which is that denaturation commences by the breakage of tight internal packing interactions, and that water plays no role in this initial transition. Moreover, because the initial events during unfolding are expected to be similar to the events that follow the rate-limiting step of refolding, which are very difficult to examine experimentally, this study also provides an insight into the nature of the final packing events that conclude the protein folding reaction.
Methods
Protein Expression, Purification, and TNB-Labeling.
The method for the purification of single chain monellin (WT MNEI) has been described in detail (21). WT MNEI contains a single tryptophan (W4) and a single cysteine (C42) residue. The mutant proteins, C42AP97C (Cys97) and C42AQ29C (Cys29), each with a single tryptophan residue (W4) and a single cysteine residue, were generated by site-directed mutagenesis, and the proteins purified as described earlier (21). The TNB-labeled proteins were obtained and quantified as described in SI Text.
Fluorescence and Far-UV CD Spectra.
Fluorescence spectra were collected on a Spex Fluoromax 3 spectrofluorimeter as described in ref. 21, with the excitation wavelength set to 295 nm. Far-UV CD spectra were collected on a Jasco J720 spectropolarimeter as described in ref. 21. The protein concentration used was 5–20 μM.
CD-Monitored Equilibrium and Kinetic Unfolding Experiments.
Equilibrium unfolding of all of the proteins using far- and near-UV CD was monitored as described in ref. 21. For kinetic experiments, the unfolding reaction was initiated by manually mixing the native protein with unfolding buffer, so that the final GdnHCl concentration at the time of unfolding was in the range of 3.0–5.5 M. The dead-time of the measurement was ≈8–10 s. The changes in the CD signals at 222 nm and at 270 nm were monitored by using the Jasco J720 spectropolarimeter.
Fluorescence-Monitored Equilibrium and Kinetic Unfolding Experiments.
The equilibrium unfolding reactions of the unlabeled and TNB-labeled proteins were carried out in an identical manner. The native protein was incubated in different concentrations of GdnHCl ranging from zero to 6 M for 6 h, and the equilibrium fluorescence signals were measured on the stopped-flow module (SFM-4) from Biologic. Sample excitation was carried out at 295 nm and emission was monitored at 360 nm, using a band-pass filter (Oriel).
The unfolding kinetics was also measured on the stopped-flow mixing module (SFM-4) from Biologic. The desired concentration of GdnHCl at the time of unfolding was obtained by appropriate dilution of the native protein, unfolding buffer, and refolding buffer inside the mixing module. The unfolding reactions of the unlabeled and TNB-labeled proteins were carried out in an identical manner. Sample excitation was carried out at 295 nm, and emission was monitored at 360 nm, by using a band-pass filter (Oriel). In all of the experiments, a mixing dead time of 6.2 ms was achieved by using an FC-15 cuvette with a path length of 1.5 mm and a total flow-rate of 5 mL/s. Unfolding reactions initiated by manual mixing gave identical kinetics.
The unfolding reactions of the unlabeled proteins were monitored also at 340 nm on excitation at 295 nm.
Supplementary Material
Acknowledgments.
We thank A.K. Patra for generating the mutant forms of MNEI used in this study, G. Krishnamoorthy for discussions, and R. L. Baldwin for comments on the manuscript. S.K.J. and J.B.U. are recipients of a SP Mukherjee Fellowship and a JC Bose National Fellowship, respectively, from the Government of India. This work was funded by the Tata Institute of Fundamental Research and by the Department of Science and Technology, Government of India.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905744106/DCSupplemental.
References
- 1.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:l–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–242. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 3.Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 4.Privalov PL. Stability of proteins: Small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 5.Kiefhaber T, Baldwin RL. Kinetics of hydrogen bond breakage in the process of unfolding of ribonuclease A measured by pulsed hydrogen exchange. Proc Natl Acad Sci USA. 1995;92:2657–2661. doi: 10.1073/pnas.92.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkelstein AV, Shakhnovich EI. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers. 1989;28:1681–1694. doi: 10.1002/bip.360281004. [DOI] [PubMed] [Google Scholar]
- 7.Shakhnovich EI, Finkelstein AV. Theory of cooperative transitions in protein molecules. I. Why denaturation of globular protein is a first-order phase transition. Biopolymers. 1989;28:1667–1680. doi: 10.1002/bip.360281003. [DOI] [PubMed] [Google Scholar]
- 8.Finkelstein AV, Ptitsyn OB. Protein Physics. London: Academic; 2002. Cooperative transitions in protein molecules; pp. 205–277. [Google Scholar]
- 9.Kiefhaber T, Labhardt AM, Baldwin RL. Direct NMR evidence for an intermediate preceding the rate-limiting step in the unfolding of ribonuclease A. Nature. 1995;375:513–515. doi: 10.1038/375513a0. [DOI] [PubMed] [Google Scholar]
- 10.Hoeltzli SD, Frieden C. Stopped-flow NMR spectroscopy: Real-time unfolding studies of 6-19F-tryptophan-labeled Escherichia coli dihydrofolate reductase. Proc Natl Acad Sci USA. 1995;92:9318–9322. doi: 10.1073/pnas.92.20.9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rami BR, Udgaonkar JB. Mechanism of formation of a productive molten globule form of barstar. Biochemistry. 2002;41:1710–1716. doi: 10.1021/bi0120300. [DOI] [PubMed] [Google Scholar]
- 12.Lillo MP, Szpikowska BK, Mas MT, Sutin JD, Beechem JM. Real-time measurement of multiple intra-molecular distances during protein folding reactions: A multisite stopped-flow fluorescence energy-transfer study of yeast phosphoglycerate kinase. Biochemistry. 1997;36:11273–11281. doi: 10.1021/bi970789z. [DOI] [PubMed] [Google Scholar]
- 13.Lakshmikanth GS, Sridevi K, Krishnamoorthy G, Udgaonkar JB. Structure is lost incrementally during the unfolding of barstar. Nat Struct Biol. 2001;8:799–804. doi: 10.1038/nsb0901-799. [DOI] [PubMed] [Google Scholar]
- 14.Lyubovitsky JG, Gray HB, Winkler JR. Mapping the cytochrome C folding landscape. J Am Chem Soc. 2002;124:5481–5485. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 15.Sridevi K, Udgaonkar JB. Surface expansion is independent of and occurs faster than core solvation during the unfolding of barstar. Biochemistry. 2003;42:1551–1563. doi: 10.1021/bi0268697. [DOI] [PubMed] [Google Scholar]
- 16.Sridevi K, Lakshmikanth GS, Krishnamoorthy G, Udgaonkar JB. Increasing stability reduces conformational heterogeneity in a protein folding intermediate ensemble. J Mol Biol. 2004;337:699–711. doi: 10.1016/j.jmb.2003.12.083. [DOI] [PubMed] [Google Scholar]
- 17.Sinha KK, Udgaonkar JB. Dependence of the size of the initially collapsed form during the refolding of barstar on denaturant concentration: Evidence for a continuous transition. J Mol Biol. 2005;353:704–718. doi: 10.1016/j.jmb.2005.08.056. [DOI] [PubMed] [Google Scholar]
- 18.Sinha KK, Udgaonkar JB. Dissecting the non-specific and specific components of the initial folding reaction of barstar by multi-site FRET measurements. J Mol Biol. 2007;370:385–405. doi: 10.1016/j.jmb.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 19.Huang F, Settanni G, Fersht AR. Fluorescence resonance energy transfer analysis of the folding pathway of Engrailed Homeodomain. Protein Eng Des Sel. 2008;21:131–146. doi: 10.1093/protein/gzm069. [DOI] [PubMed] [Google Scholar]
- 20.Kimura T, et al. Specific collapse followed by slow hydrogen-bond formation of β-sheet in the folding of single-chain monellin. Proc Natl Acad Sci USA. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patra AK, Udgaonkar JB. Characterization of the folding and unfolding reactions of single-chain monellin: Evidence for multiple intermediates and competing pathways. Biochemistry. 2007;46:11727–11743. doi: 10.1021/bi701142a. [DOI] [PubMed] [Google Scholar]
- 22.Kimura T, et al. Dehydration of main-chain amides in the final folding step of single-chain monellin revealed by time-resolved infrared spectroscopy. Proc Natl Acad Sci USA. 2008;105:13391–13396. doi: 10.1073/pnas.0801316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuwajima K, Nitta K, Yoneyama M, Sugai S. Three-state denaturation of α-lactalbumin by guanidine hydrochloride. J Mol Biol. 1976;106:359–373. doi: 10.1016/0022-2836(76)90091-7. [DOI] [PubMed] [Google Scholar]
- 24.Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 25.Privalov PL, Gill SJ. Stability of protein structure and hydrophobic interactions. Adv Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 26.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 27.Privalov PL, Makhatadze GI. Contribution of hydration to protein folding thermodynamics. II. The entropy and Gibbs energy of hydration. J Mol Biol. 1993;232:660–679. doi: 10.1006/jmbi.1993.1417. [DOI] [PubMed] [Google Scholar]
- 28.Schellman JA. The factors affecting the stability of hydrogen-bonded polypeptide structures in aqueous solution. J Phys Chem. 1958;62:1485–1494. [Google Scholar]
- 29.Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535. [Google Scholar]
- 30.Fierz B, Reiner A, Kiefhaber T. Local conformational dynamics in α-helices measured by fast triplet transfer. Proc Natl Acad Sci USA. 2009;106:1057–1062. doi: 10.1073/pnas.0808581106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz V, Ramanathan R. Waltzing α-helices. Proc Natl Acad Sci USA. 2009;106:1299–1300. doi: 10.1073/pnas.0812577106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua L, Zhou R, Thirumalai D, Berne BJ. Urea denaturation by stronger dispersion interactions with proteins than water implies a 2-stage unfolding. Proc Natl Acad Sci USA. 2008;105:16928–16933. doi: 10.1073/pnas.0808427105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldenberg DP. Computational simulation of the statistical properties of unfolded proteins. J Mol Biol. 2003;326:1615–1633. doi: 10.1016/s0022-2836(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Munoz V. Experimental identification of downhill protein folding. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 35.Pradeep L, Udgaonkar JB. Effect of salt on the urea-unfolded form of barstar probed by m value measurements. Biochemistry. 2004;43:11393–11402. doi: 10.1021/bi049320b. [DOI] [PubMed] [Google Scholar]
- 36.Saxena AM, Udgaonkar JB, Krishnamoorthy G. Characterization of intra-molecular distances and site-specific dynamics in chemically unfolded barstar: Evidence for denaturant-dependent non-random structure. J Mol Biol. 2006;359:174–189. doi: 10.1016/j.jmb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Kemp JP, Chen ZY. Formation of helical states in wormlike polymer chains. Phys Rev Lett. 1998;81:3880–3883. [Google Scholar]
- 38.Ihalainen JA, et al. α-helix folding in the presence of structural constraints. Proc Natl Acad Sci USA. 2008;105:9588–9593. doi: 10.1073/pnas.0712099105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha KK, Udgaonkar JB. Barrierless evolution of structure during the submillisecond refolding reaction of a small protein. Proc Natl Acad Sci USA. 2008;105:7998–8003. doi: 10.1073/pnas.0803193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung HS, Tokmakoff A. Temperature-dependent downhill unfolding of ubiquitin. II. Modeling the free energy surface. Proteins. 2008;72:488–497. doi: 10.1002/prot.22042. [DOI] [PubMed] [Google Scholar]
- 41.Poland DC, Scheraga HA. Statistical mechanics of non-covalent bonds in polyamino acids. IX. The two-state theory of protein denaturation. Biopolymers. 1965;3:401–419. [Google Scholar]
- 42.Jha SK, Dhar D, Krishnamoorthy G, Udgaonkar JB. Continuous dissolution of structure during the unfolding of a small protein. Proc Natl Acad Sci USA. 2009;106:11113–11118. doi: 10.1073/pnas.0812564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.