Abstract
Polyketides are among the major classes of bioactive natural products used to treat microbial infections, cancer, and other diseases. Here we describe a pathway to chloroethylmalonyl-CoA as a polyketide synthase building block in the biosynthesis of salinosporamide A, a marine microbial metabolite whose chlorine atom is crucial for potent proteasome inhibition and anticancer activity. S-adenosyl-l-methionine (SAM) is converted to 5′-chloro-5′-deoxyadenosine (5′-ClDA) in a reaction catalyzed by a SAM-dependent chlorinase as previously reported. By using a combination of gene deletions, biochemical analyses, and chemical complementation experiments with putative intermediates, we now provide evidence that 5′-ClDA is converted to chloroethylmalonyl-CoA in a 7-step route via the penultimate intermediate 4-chlorocrotonyl-CoA. Because halogenation often increases the bioactivity of drugs, the availability of a halogenated polyketide building block may be useful in molecular engineering approaches toward polyketide scaffolds.
Keywords: actinomycete, biological halogenation, marine natural product, proteasome inhibitor, Salinispora tropica
Polyketides are abundant microbial metabolites that possess a remarkable diversity in chemical structure and biological function. The enzymes that catalyze the assembly of these natural products, namely polyketide synthases (PKSs), belong to 3 protein families that similarly use small carboxylic acid building blocks as substrates. Polyketide biosynthetic pathways have evolved a myriad of ways to accommodate changes in the number and composition of their substrates, the manner in which they are assembled, and the further biochemical modification of the PKS product by tailoring enzymes to synthesize these often very complex organic molecules (1–4). The assembly line organization of modular type I PKSs in particular has facilitated their rational reengineering through combinatorial biosynthesis and mutasynthesis to yield new compound scaffolds that further extends their natural biosynthetic prowess (5).
Although PKSs exploit a wide assortment of priming carboxylic acid substrates to initiate the polyketide biosynthetic process (6), they are relegated to a small number of extending dicarboxylic acid units needed to elongate the growing polyketide chain via successive Claisen condensation reactions (7). The most common PKS extender units are malonyl-CoA, methylmalonyl-CoA, and, to a much lesser extent, ethylmalonyl-CoA that are selected by and attached to the PKS domain acyl carrier protein (ACP) by dedicated acyltransferases (ATs). These CoA-tethered PKS building blocks impart unreactive, aliphatic substituents (proton, methyl, and ethyl, respectively) to the polyketide backbone and contrast the second class of dedicated ACP-linked PKS extender units that instead harbor functionalized side chains. Methoxymalonyl-ACP, hydroxymalonyl-ACP, and aminomalonyl-ACP are relatively rare extender units that supply methoxy, hydroxyl, and amino groups, respectively, to the polyketide molecule (8). The programmed introduction of these ACP-bound extender units results in the strategic placement of functional groups that confers important structural and biological properties to the polyketide.
We recently proposed that the PKS extender unit chloroethylmalonyl-CoA was involved in the biosynthesis of the anticancer agent salinosporamide A in the marine bacterium Salinispora tropica (9). This CoA-linked halogenated metabolite provides the reactive chloroethyl side chain germane to salinosporamide A's irreversible binding mechanism against the 20S proteasome (10). The biosynthesis of salinosporamide's chlorinated building block is initiated by the S-adenosyl-l-methionine (SAM)-dependent chlorinase (11), which catalyzes the conversion of SAM to 5′-chloro-5′deoxyadenosine (5′-ClDA). Herein, we detail an 8-step biosynthetic pathway to chloroethylmalonyl-CoA and firmly establish this chlorinated metabolite as a PKS extender unit. Because halogen atoms not only favorably influence the bioactivity of drugs (12) but also offer chemically reactive handles for lead optimization by semisynthetic chemistry, this pathway to a halogenated PKS building block may facilitate the bioengineering of polyketide molecules for drug development.
Results
Analysis of the sal Gene Cluster.
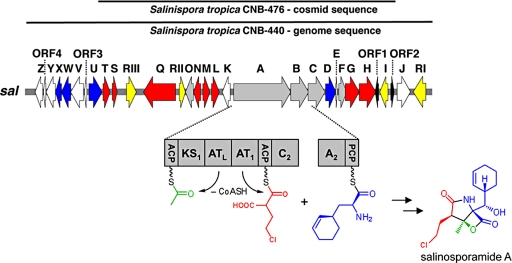
Complete genome sequence analysis of S. tropica CNB-440 revealed 19 secondary metabolic gene clusters (13), including a 41-kb hybrid PKS-nonribosomal peptide synthetase (NRPS) gene set consistent with salinosporamide A biosynthesis (Fig. 1 and SI Text, Table S1). Before genome sequencing of CNB-440 was completed, we attempted to clone and sequence the sal cluster from S. tropica strain CNB-476 by using PCR-amplified PKS, NRPS, and crotonyl-CoA carboxylase/reductase (CCR) gene fragments as probes. Library screening led to the identification of a pOJ446 cosmid clone containing a 33-kb genomic insert 99% identical in DNA sequence to strain CNB-440 (Fig. 1).
Fig. 1.
Organization of the sal biosynthetic gene cluster from Salinispora tropica. The sal DNA sequence in strains CNB-476 and CNB-440 is 99% identical. Genes putatively involved in the chloroethylmalonyl-CoA pathway (red), construction of the core γ-lactam-β-lactone ring system (gray), assembly of the nonproteinogenic amino acid l-3-cyclohex-2′-enylalanine (blue), regulation and resistance (yellow), unknown (white), and 2 partial transposases (black) are color-coded.
Central to the sal cluster is the salA gene that codes for a bimodular PKS of unusual domain organization harboring contiguous acyltransferase loading (ATL) and extender (AT1) (Fig. 1) domains rather than the standard ATL-ACPL-KS1-AT1-ACP1 assembly as observed in prototypical PKSs such as 6-deoxyerythronolide B synthase 1 (14). This noncanonical domain architecture is, however, found in several myxobacterial megasynthases such as those involved in stigmatellin, soraphen, and aurafuranone biosynthesis (15–17). Stable isotope feeding studies showed that salinosporamide A is biosynthesized from the building blocks acetate, the nonproteinogenic amino acid β-hydroxy-l-3-cyclohex-2′-enylalanine (CHA) and a sugar-derived chlorinated molecule that we hypothesized was a previously unknown PKS extender unit, namely chloroethylmalonyl-CoA (9).
We propose that the hexa-domained SalA is involved in the selection, attachment and condensation of acetyl-CoA and chloroethylmalonyl-CoA, to generate a β-ketothioester intermediate. The loading module is of the ACPL/ATL-type that recognizes monocarboxylic acid starter units such as acetyl-CoA rather than the KSQ-type that accepts dicarboxylic acids like malonyl-CoA (6). The possibility of chloroethylmalonyl-CoA as an extender unit prompted us to examine the phylogeny of AT1 in more detail (SI Text and Fig. S1), because AT domain divergence is a critical factor in the evolution of polyketide structural diversity (18). Malonyl-CoA- and methylmalonyl-CoA-specific AT domains share a common ancestor, diverging at some point of evolution to form 2 distinct groups, whereas the relatively rare ethylmalonyl-CoA and methoxymalonyl-ACP ATs appear to have evolved more than once because the known sequences reside in either clade (18). Our analysis with representative AT domains reconstructs that scenario and places SalA_AT1 in the methylmalonyl-CoA group, more closely related to some myxobacterial ATs although forming its own subclade (SI Text and Fig. S1). In addition, the distinct signature motifs (14) apparent from sequence alignments are also in agreement with AT1 accepting an unreported extender unit (SI Text and Fig. S1B).
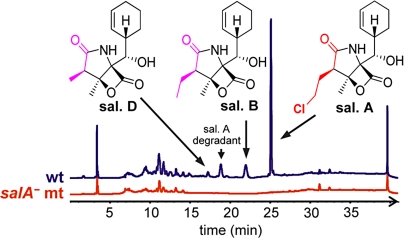
Moreover, the detection of salinosporamide analogs with different side chains at C2 (ethyl and methyl, Fig. 2), points to the promiscuity of AT1 in accepting not only chloroethylmalonyl-CoA (salinosporamide A), but also other substituted malonyl extender units such as ethylmalonyl-CoA (salinosporamide B) and methylmalonyl-CoA (salinosporamide D). To probe the central role of salA, we disrupted it via a single-cross-over homologous recombination event. Inactivation of salA abolished the biosynthesis of all salinosporamides, thereby confirming that this family of β-lactones is indeed derived from a PKS pathway (Fig. 2).
Fig. 2.
Inactivation of the PKS gene salA completely abolishes salinosporamide (sal.) production. HPLC chromatograms of culture extracts with detection at 210 nm. Mt, mutant; wt, wild-type.
The Chloroethylmalonyl-CoA Pathway.
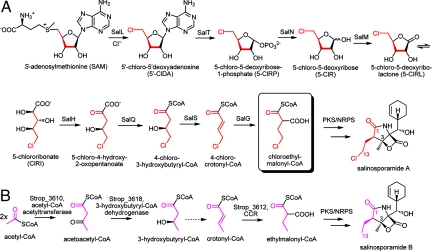
Based on the gene organization of the sal cluster, we propose a route to chloroethylmalonyl-CoA as illustrated in Fig. 3A. Biosynthesis of ethylmalonyl-CoA as a precursor of salinosporamide B is shown for comparison (Fig. 3B) and is not encoded in the sal locus but constitutes rather a primary metabolic pathway for acetate assimilation and a source of building blocks for secondary metabolite production (19, 20).
Fig. 3.
Comparison of chloroethylmalonyl-CoA and ethylmalonyl-CoA biosynthetic pathways. (A) Proposed pathway to chloroethylmalonyl-CoA as a PKS extender unit in salinosporamide A biosynthesis. (B) The corresponding ethylmalonyl-CoA moiety in salinosporamide B is derived from acetate (9). Crotonyl-CoA carboxylase/reductase (CCR) is a key enzyme in the ethylmalonyl-CoA pathway (19).
We recently reported that chlorine incorporation into salinosporamide A is catalyzed by the SAM-dependent chlorinase SalL in an orthogonal manner to other known enzymatic chlorination reactions, but analogous to fluorinase FlA of Streptomyces cattleya, a fluoroacetate and 4-fluorothreonine producer (11, 21). Earlier studies using 13C-labeled glucose showed an incorporation pattern in the chlorobutyrate moiety of salinosporamide A consistent with the ribose unit of SAM being the ultimate precursor (9). Interestingly, the sal cluster also harbors the FlB homolog SalT. FlB is the second enzyme in the pathway to fluorometabolites in S. cattleya, catalyzing the phosphorolytic cleavage of 5′-FDA to produce 5-fluoro-5-deoxy-d-ribose-1-phosphate (5-FRP) (22). Although SalT likely catalyzes the analogous conversion of 5′-ClDA to 5-ClRP (Fig. 3A), the 2 pathways appear to diverge at this point as no other structural homologous genes are shared between the sal and fl clusters (22). For fluorometabolites it has been speculated that an isomerase catalyzes the ring opening of 5-FRP to 5-fluoro-5-deoxy-d-ribulose-1-phosphate, in analogy to the known metabolism of 5′-methylthioadenosine (23). Indeed, the in vitro reconstitution of 4-fluorothreonine biosynthesis has been recently achieved by using besides FlA and FlB, 3 enzymes not coded in the fl cluster, that is, an isomerase, a surrogate aldolase and a pyridoxal (PLP)-dependent transaldolase (24).
For chloroethylmalonyl-CoA, we propose instead that the phosphatase homolog SalN and the dehydrogenase/reductase SalM catalyze the dephosphorylation of 5-ClRP to 5-chloro-5-deoxy-d-ribose (5-ClR) followed by oxidization to 5-chlororibonate (5-ClRI) possibly via the intermediate 5-chloro-5-deoxy-d-ribono-1,4-lactone (5-ClRL) (Fig. 3A). SalH, a dihydroxyacid dehydratase homolog, then putatively converts 5-ClRI to 5-chloro-4-hydroxy-2-oxopentanoate, which is then subjected to SalQ-catalyzed oxidative decarboxylation to 4-chloro-3-hydroxybutyryl-CoA. This product importantly results in a 4-carbon chlorometabolite consistent with the C1/C2/C12/C13 salinosporamide A fragment (9). SalQ shares >50% sequence identity with α-oxoacid ferredoxin oxidoreductases known to catalyze the oxidative decarboxylation of α-ketoacids with reduction of ferredoxin (Fd) to the corresponding CoA derivative, the prototype of which is pyruvate ferredoxin oxoreductase (25).
Domain analysis of the putative biosynthetic enzyme SalS suggests that it belongs to the hotdog fold superfamily that includes FabZ, a β-hydroxyacyl-ACP dehydratase involved in bacterial fatty acid biosynthesis (26). SalS's closest characterized homolog (47% amino acid identity) is Rv0130 from Mycobacterium tuberculosis, a 16-kDa protein shown to catalyze the reversible hydration of crotonyl-CoA to hydroxybutyryl-CoA in vitro (27). However, it has been shown that these enzymes function as dehydratases in vivo when coupled to a reductase (28). Hence, we propose that SalS catalyzes the reversible dehydration of 4-chloro-3-hydroxybutyryl-CoA to 4-chlorocrotonyl-CoA.
The final reaction toward chloroethylmalonyl-CoA is putatively catalyzed by SalG, which shows sequence identity (>60%) to crotonyl-CoA carboxylase/reductases (CCR). The biological function of CCR was recently revised to catalyze the last step of ethylmalonyl-CoA biosynthesis (19) (Fig. 3B), and similarly we hypothesize that SalG catalyzes the reductive carboxylation of 4-chlorocrotonyl-CoA to chloroethylmalonyl-CoA.
Gene Inactivation and Chemical Complementation.
To functionally identify the chloroethylmalonyl-CoA pathway enzymes, we inactivated each postulated gene by replacement with an apramycin resistance (aac (3)IV) marker using λ-Red recombination (11, 29). Inactivation led to the selective loss of salinosporamide A production in relation to salinosporamide B in case of the first and last committed pathway enzymes encoded by the chlorinase salL (11) and the 4-chlorocrotonyl-CoA carboxylase/reductase (Cl-CCR) salG, respectively. The remaining 6 mutants, however, all resulted in significant reductions in salinosporamide A yields, suggesting partial complementation by housekeeping enzymes (Table 1).
Table 1.
Salinosporamide production by S. tropica mutants compared to the wild-type
| Strain | Protein function | Sal. A, %† | Sal. B, %† | Strop homolog, sequence identity |
|---|---|---|---|---|
| wild-type | — | 100 ± 10 | 100 ± 13 | — |
| salA− | PKS | n.d. | n.d. | — |
| salL− | Chlorinase (11) | n.d. | 90 ± 20 | Strop_1405, 35% |
| salT− | Purine nucleoside phosphorylase | 50 ± 8 | 91 ± 10 | Strop_0986, 69% |
| salN− | Phosphatase | 16 ± 3 | 92 ± 9 | — |
| salM− | Dehydrogenase/reductase | 2.2 ± 0.2 | 120 ± 20 | Strop_2799, 35% |
| salH− | Dihydroxyacid dehydratase | 3.8 ± 0.7 | 70 ± 15 | Strop_1231, 36% |
| salQ− | α-ketoacid decarboxylase | 25 ± 6 | 98 ± 16 | Strop_1050‡ |
| salS− | Acyl dehydratase | 39 ± 14 | 95 ± 30 | — |
| salG− | Cl-CCR | n.d. | 94 ± 30 | Strop_3612, 53% |
| Strop_3612− | CCR | 112 ± 8 | 52 ± 6 | salG, 53% |
Sal., salinosporamide; n.d., not detected (<0.2%).
†Values are average of at least two independent experiments ± deviation.
‡Strop_1050 represents a partial duplication of salQ (894 of 3,450 nt).
salM deletion in particular led to a substantial loss in salinosporamide A production to approximately 2% wild-type levels. This low level of residual biosynthesis may result from biochemical cross-talk with homologous dehydrogenases such as Strop_2799 (35% sequence identity). We hypothesized that the absence of SalM might furthermore result in the accumulation of its putative substrate 5-ClR. To probe whether 5-ClR is indeed accumulated in the blocked mutant, we derivatized an organic extract of the supernatant with 2,4-dinitrophenylhydrazine and analyzed by LC/MS. The corresponding hydrazone of 5-ClR was detected in the salM but not in the salL mutant as expected (Fig. 4A). This result suggests that 5-ClR is indeed a key metabolite in the chloroethylmalonyl-CoA pathway and the substrate of SalM.
Fig. 4.
Chromatographic analysis of intermediates in the chloroethylmalonyl-CoA pathway. (A) 5-ClR as a key intermediate in the chloroethylmalonyl-CoA pathway. Scheme of the method used to derivatize 5-ClR for UV detection (352 nm) (Upper). LC/MS analysis of derivatized 5-ClR and water extracts of the salL (negative control) and salM mutants (Lower). (B) Feeding experiments with postulated intermediates (or their counterparts) in the chloroethylmalonyl-CoA pathway restore salinosporamide A production in the salL mutant. See Results for details.
Putative intermediates were next chemically synthesized and administered to the salL- and salG-defective mutants and salinosporamide A production analyzed. In accordance with the pathway proposed in Fig. 3A, 5′-ClDA (11), 5-ClR, 5-ClRL, ethyl 4-chloro-3-hydroxybutyrate (EtClHB) and 4-chlorocrotonic acid (ClCA) selectively restored salinosporamide A production in the salL mutant (Fig. 4B) but not in the salG mutant, providing further support that these compounds are intermediates either directly or indirectly in the biosynthesis of the presumed chloroethylmalonyl-CoA extender unit. The anticipated intermediate 5-ClRI, however, did not restore salinosporamide A biosynthesis, which may be a consequence of poor cellular uptake because of its polarity. Furthermore, saturated 4-chlorobutyric acid (ClBA) did not chemically complement either mutant and restore salinosporamide A production, which is consistent with the proposed in vivo role of SalG as a reductive carboxylase and not simply as a reductase.
Semiquantitative RT-PCR analysis was conducted to test the possibility of polar effects on downstream genes caused by each relevant gene replacement (SI Text and Fig. S2). Transcription of salS, salO, and salL in the salT, salN, and salM disruption mutants, respectively, was equivalent to the wild type. However, salG gene replacement by the apramycin resistance marker did have a negative effect on downstream salH transcript levels. The construction of a salG mutant in which salH was put directly under control of the putative salA promoter (see SI Text and Figs. S3 and S4) was able to revert the originally observed polar effect and restore salH transcription to wild-type levels. Nonetheless, salinosporamide A production was still not detected in this mutant, thereby confirming the dedicated role of SalG in 4-chloroethylmalonyl-CoA biosynthesis.
Characterization of SalG, a Chlorocrotonyl-CoA Carboxylase/Reductase.
To demonstrate the function of SalG as a 4-chlorocrotonyl-CoA carboxylase/reductase and establish the authenticity of chloroethylmalonyl-CoA as an intermediate, we biochemically characterized recombinant SalG, which was expressed as an octahistidyl-tagged protein in Escherichia coli. Chlorocrotonyl-CoA was chemically synthesized and converted by recombinant SalG to chloroethylmalonyl-CoA in a NADPH-dependent reductive carboxylation reaction as detected by LC/MS analysis. The apparent kinetic constants (Table 2) support a preference for the chlorinated substrate, whereas the enzyme also accepts crotonyl-CoA with 7-fold decrease in catalytic efficiency (kcat/KM). Moreover, the turnover (kcat) for the reductive carboxylation of chlorocrotonyl-CoA is ≈10-fold faster than for the reductive reaction in the absence of bicarbonate, which is consistent with the kinetic properties of the known CCR from Rhodobacter sphaeroides (19).
Table 2.
Apparent kinetic constants of SalG
| Substrate | Reductive carboxylation† |
Reduction only‡ |
||
|---|---|---|---|---|
| Crotonyl-CoA | Chlorocrotonyl-CoA | Crotonyl-CoA | Chlorocrotonyl-CoA | |
| kcat, min−1 | 15.4 ± 0.9 | 23.1 ± 2.6 | 4.7 ± 0.2 | 2.2 ± 0.2 |
| KM, μM | 20.7 ± 4.2 | 4.4 ± 1.8 | 9.8 ± 1.4 | 1.9 ± 0.7 |
| kcat/KM, min−1 μM−1 | 0.75 | 5.3 | 0.48 | 1.1 |
| Relative catalytic efficiency, % | 14 | 100 | 44 | 100 |
†(chloro)crotonyl-CoA + CO2 + NADPH → (chloro)ethylmalonyl-CoA + NADP+.
‡(chloro)crotonyl-CoA + NADPH → (chloro)butyryl-CoA + NADP+.
In addition to salG, the S. tropica genome harbors only one other CCR gene (Strop_3612), which likely encodes the housekeeping CCR responsible for ethylmalonyl-CoA biosynthesis. To test this hypothesis we carried out a separate deletion of Strop_3612. Although gene inactivation of Strop_3612 had no significant effect on salinosporamide A yields, salinosporamide B production fell 50% in this mutant (Table 1), which is consistent with its biosynthesis from a CCR-derived ethylmalonyl-CoA unit (Fig. 3B).
Discussion
We present in vivo and in vitro evidence for an exquisite pathway to a halogenated PKS building block composed of 8 enzymes in total, some of which are unique and others whose function can be replaced by primary metabolic counterparts. SalG, for instance, appears to have singularly evolved from a common CCR ancestor to accept a halogenated substrate. Its relaxed substrate discrimination, evident from the kinetic data presented from the fact that fluorinated (30) and brominated (31) salinosporamide analogs can be generated in vivo, is useful when attempting to engineer structural analogs. The residual biosynthesis of salinosporamide B from ethylmalonyl-CoA in the housekeeping CCR (Strop_3612) deletion mutant may derive from SalG-catalyzed reductive carboxylation of crotonyl-CoA. Alternatively, an orthogonal pathway for acetate assimilation may function in S. tropica (7, 20) as suspected in Streptomyces cinnamonensis in which genetic inactivation of its CCR likewise resulted in the reduced production of the ethylmalonyl-CoA-derived polyketide monensin A rather than abolishing it completely (32).
Inactivation of the chlorinase gene salL also abolished salinosporamide A biosynthesis (11) despite the presence of the salL homolog Strop_1405, which contains a domain of unknown function (DUF62, Table 1). We recently showed that a DUF62 ortholog from Salinispora arenicola (86% identity to Strop_1405) has SAM hydrolase but no halogenase activity in vitro (33), which was independently reported with an archaeal DUF62 (34). Together with in silico analysis, these data suggest divergent evolution of SAM-dependent halogenases from SAM hydrolases (33) and clarify the salL mutant phenotype.
Although the function of SalL and SalG appear unparalleled, the activities of the 6 other pathway-committed enzymes can be replaced to varying levels by primary metabolic counterparts (Table 1). For instance, another purine nucleoside phosphorylase involved in nucleotide metabolism (35), 5′-methylthioadenosine phosphorylase (Strop_0986, 69% identity to SalT), is encoded by the S. tropica genome and is likely the enzyme complementing the salT mutation. Similarly, S. tropica's chromosome codes for several short-chain dehydrogenases/reductases and dihydroxyacid dehydratases with sequence similarity to SalM and SalH, respectively (Table 1).
However, SalN and SalS have no clear homologs (<23% sequence identity to other Strop genes). In the case of SalS, primary metabolic enoyl thioester dehydratases such as the FabZ-like Strop_2797 and the unknown 3-hydroxybutyryl-CoA dehydratase (Fig. 3B) may include possible candidates to replace its function. It is noteworthy that FabZ enzymes, despite having a preference for ACP-bound substrates, also accept CoA analogs at lower rates, with crotonyl-CoA often used as a substrate to evaluate FabZ activity (28, 36, 37). Likewise, other phosphatases present in the S. tropica chromosome such as the putative alkaline phosphatase Strop_2394 may account for the salN mutant phenotype, because such enzymes have broad substrate specificity (38). Indeed a commercial alkaline phosphatase (EC 3.1.3.1; Sigma Chemical Co. A2356) catalyzes the phosphorylic cleavage of chemo-enzymatically prepared 5-ClRP to 5-ClR.
Although α-keto acid ferredoxin oxidoreductase SalQ is partially duplicated in the S. tropica genome as Strop_1050 (corresponding to 298 of 1,149 amino acids), its expression cannot explain the salinosporamide A levels observed in this mutant, because the C-terminal catalytic domain is not complete and the thiamine pyrophosphate binding site is missing in Stop_1050 (39). Alternatively, α-keto acid oxidative decarboxylation to form acyl-CoA can be mediated by 2-oxo acid dehydrogenases (OADH), multienzyme complexes consisting of 3 components, decarboxylase (E1), transacetylase (E2) and dehydrogenase (E3) (40). The S. tropica genome codes for a few OADHs, i.e., Strop_2097 (E1) to Strop_2099 (E3), Strop_0109 to Strop_0107 and Strop_3690 (E1-E3), putatively encoding pyruvate or branched-chain oxoacid and 2-oxoglutarate dehydrogenase complexes, respectively. The relaxed substrate specificity of OADHs (40) is a possible explanation for salQ's mutant phenotype.
To the best of our knowledge, the salinosporamide system is at present the only PKS machinery known to use chloroethylmalonyl-CoA as an extender unit. The discovery of chloroethylmalonyl-CoA expands our current knowledge of known PKS extender units and more significantly represents a functionalized CoA-tethered malonyl extender (7). In concert with the availability of an additional PKS building block, the divergence of AT domains is an important factor to the evolution of structural diversity in polyketides (18). Therefore, chloroethylmalonyl-CoA and SalA_AT1 extend the repertoire of tools to be used in polyketide assembly, significantly impacting our ability to generate polyketide scaffolds and possibly improve biological activity. Although the number of known extender units has expanded in recent years, a true increase in available building blocks may come from those biosynthesized by a CCR-like route (7), which could possibly lead to previously unknown extender units through conversion of a variety of α,β,-unsaturated acyl-CoA thioesters. The chloroethylmalonyl-CoA pathway presented here and SalG in particular is clearly representative of this scenario and points the way toward the genetic engineering of new polyketide scaffolds with strategically placed halogens that may serve key mechanistic roles or provide functional handles in the semisynthesis of derivatives.
Materials and Methods
Chemicals.
5′-ClDA, butyric acid and 4-chlorobutyric acid were obtained from Sigma. 5-ClR, 5-ClRL, 5-ClRI, ethyl 4-chloro-3-hydroxybutyrate and ClCA were synthesized according to modified literature preparations (see SI Text). Compounds were administered to the S. tropica mutant strains to a final concentration of 0.3 mM and products analyzed by HPLC-MS as described in ref. 11. The 2,4-DNP derivatization of synthetic 5-ClR and water extracts of 100-mL cultures of S. tropica salL and salM deficient mutants and monitoring by HPLC-MS followed literature precedence (41).
Inactivation of sal Genes.
salA was inactivated by homologous recombination via a single cross-over (42) after introducing pAEM3 into S. tropica CNB-440 by conjugation from E. coli ET12567/pUZ8002. Other genes were inactivated by using the PCR targeting system (29) with some modifications as described in ref. 11. See SI Text for details.
Characterization of SalG.
His8-tagged SalG protein was purified from E. coli BL21(DE3) by using Ni-affinity chromatography and gel-filtration (see SI Text for details) and stored in 50 mM phosphate buffer pH 7.2, 100 mM NaCl and 10% glycerol at −80 °C. Determination of apparent kcat and KM for each substrate and reaction was performed by continuous assay measuring NADPH consumption at 340 nm using a UV-1700 spectrophotometer (Shimadzu). Assay conditions involved SalG (47 nM) incubated with various concentrations of crotonyl-CoA and 4-chlorocrotonyl-CoA in reaction buffer (100 mM Tris-HCl, pH 7.9) under saturating levels of NADPH. Assays were carried out in the absence and presence of NaHCO3 (80 mM, final concentration) to determine the kinetics values for the reductive and the reductive-carboxylation reaction, respectively. Data were collected in triplicate and averaged. Confirmation of products was carried out by LC/(+)ESI-MS analysis using a water to methanol gradient containing 5 mM ammonium acetate. See SI Text for further methods.
For additional details see a list of gene sequences used in phylogenetic analysis (Table S2) and a summary of synthesized chlorinated substrates (Fig. S5).
Supplementary Material
Acknowledgments.
We kindly thank B. Gust (University of Tuebingen, Germany) and Plant Bioscience Limited (Norwich, UK) for providing the REDIRECT technology kit for PCR-targeting, A. Lapidus from the Joint Genome Institute (Walnut Creek, CA) for sal fosmids, and W. Fenical and P. R. Jensen (Scripps Institution of Oceanography, La Jolla, CA) for S. tropica strains. A.S.E. is a Tularik Postdoctoral Fellow of the Life Sciences Research Foundation, and L.L.B. was a National Sea Grant–Industry Graduate Fellow through the Washington Sea Grant program (R/B-47). This work was supported by National Oceanic and Atmospheric Administration (NA05NOS4781249 to B.S.M.) and the National Institutes of Health (CA127622 to B.S.M. and AI51629 to K.A.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.K. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. CP000667 (S. tropica CNB-440 complete genome sequence) and EF397502 (partial sal cluster from S. tropica CNB-476)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0901237106/DCSupplemental.
References
- 1.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: A tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 3.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat Prod Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 4.Shen B. Polyketide biosynthesis beyond the type I, II, and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–295. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 5.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 6.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 7.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan YA, et al. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc Natl Acad Sci USA. 2006;103:14349–14354. doi: 10.1073/pnas.0603748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beer LL, Moore BS. Biosynthetic convergence of salinosporamides A and B in the marine actinomycete Salinispora tropica. Org Lett. 2007;9:845–848. doi: 10.1021/ol063102o. [DOI] [PubMed] [Google Scholar]
- 10.Groll M, Huber R, Potts BC. Crystal structures of salinosporamide A (NPI-0052) and B (NPI-0047) in complex with the 20S proteasome reveal important consequences of beta-lactone ring opening and a mechanism for irreversible binding. J Am Chem Soc. 2006;128:5136–5141. doi: 10.1021/ja058320b. [DOI] [PubMed] [Google Scholar]
- 11.Eustáquio AS, Pojer F, Noel JP, Moore BS. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nat Chem Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural product biosynthesis. Chem Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Udwary DW, et al. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves CD, et al. Alteration of the substrate specificity of a modular polyketide synthase acyltransferase domain through site-specific mutations. Biochemistry. 2001;40:15464–15470. doi: 10.1021/bi015864r. [DOI] [PubMed] [Google Scholar]
- 15.Gaitatzis N, et al. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J Biol Chem. 2002;277:13082–13090. doi: 10.1074/jbc.M111738200. [DOI] [PubMed] [Google Scholar]
- 16.Frank B, et al. From genetic diversity to metabolic unity: Studies on the biosynthesis of aurafurones and aurafuron-like structures in myxobacteria and streptomycetes. J Mol Biol. 2007;374:24–38. doi: 10.1016/j.jmb.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Ligon J, et al. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene. 2002;285:257–267. doi: 10.1016/s0378-1119(02)00396-7. [DOI] [PubMed] [Google Scholar]
- 18.Ridley CP, Lee HY, Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erb TJ, et al. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: The ethylmalonyl-CoA pathway. Proc Natl Acad Sci USA. 2007;104:10631–10636. doi: 10.1073/pnas.0702791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akopiants K, Florova G, Li C, Reynolds KA. Multiple pathways for acetate assimilation in Streptomyces cinnamonensis. J Ind Microbiol Biotechnol. 2006;33:141–150. doi: 10.1007/s10295-005-0029-4. [DOI] [PubMed] [Google Scholar]
- 21.Dong C, et al. Crystal structure and mechanism of a bacterial fluorinating enzyme. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 22.Huang F, et al. The gene cluster for fluorometabolite biosynthesis in Streptomyces cattleya: A thioesterase confers resistance to fluoroacetyl-coenzyme A. Chem Biol. 2006;13:475–484. doi: 10.1016/j.chembiol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Deng H, O'Hagan D, Schaffrath C. Fluorometabolite biosynthesis and the fluorinase from Streptomyces cattleya. Nat Prod Rep. 2004;21:773–784. doi: 10.1039/b415087m. [DOI] [PubMed] [Google Scholar]
- 24.Deng H, Cross SM, McGlinchey RP, Hamilton JT, O'Hagan D. In vitro reconstituted biotransformation of 4-fluorothreonine from fluoride ion: Application of the fluorinase. Chem Biol. 2008;15:1268–1276. doi: 10.1016/j.chembiol.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 25.Ragsdale SW. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem Rev. 2003;103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 26.Kimber MS, et al. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 27.Johansson P, Castell A, Jones TA, Bäckbro K. Structure and function of Rv0130, a conserved hypothetical protein from Mycobacterium tuberculosis. Protein Sci. 2006;15:2300–2309. doi: 10.1110/ps.062309306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacco E, et al. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2007;104:14628–14633. doi: 10.1073/pnas.0704132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eustáquio AS, Moore BS. Mutasynthesis of fluorosalinosporamide, a potent and reversible inhibitor of the proteasome. Angew Chem Int Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- 31.Reed KA, et al. Salinosporamides D-J from the marine actinomycete Salinispora tropica, bromosalinosporamide, and thioester derivatives are potent inhibitors of the 20S proteasome. J Nat Prod. 2007;70:269–276. doi: 10.1021/np0603471. [DOI] [PubMed] [Google Scholar]
- 32.Liu H, Reynolds KA. Precursor supply for polyketide biosynthesis: The role of crotonyl-CoA reductase. Metab Eng. 2001;3:40–48. doi: 10.1006/mben.2000.0169. [DOI] [PubMed] [Google Scholar]
- 33.Eustáquio AS, Härle J, Noel JP, Moore BS. S-adenosyl-l-methionine hydrolase (adenosine-forming), a conserved bacterial and archeal protein related to SAM-dependent halogenases. Chembiochem. 2008;9:2215–2219. doi: 10.1002/cbic.200800341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng H, Botting CH, Hamilton JT, Russell RJ, O'Hagan D. S-adenosyl-l-methionine:hydroxide adenosyltransferase: A SAM enzyme. Angew Chem Int Ed. 2008;47:5357–5361. doi: 10.1002/anie.200800794. [DOI] [PubMed] [Google Scholar]
- 35.Pugmire MJ, Ealick SE. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem J. 2002;361:1–25. doi: 10.1042/0264-6021:3610001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, et al. Structural basis for catalytic and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) J Biol Chem. 2008;283:5370–5379. doi: 10.1074/jbc.M705566200. [DOI] [PubMed] [Google Scholar]
- 37.Sharma SK, et al. Identification, characterization, and inhibition of Plasmodium falciparum β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) J Biol Chem. 2003;278:45661–45671. doi: 10.1074/jbc.M304283200. [DOI] [PubMed] [Google Scholar]
- 38.Coleman JE. Structure and mechanism of alkaline phosphatase. Annu Rev Biophys Biomol Struct. 1992;21:441–483. doi: 10.1146/annurev.bb.21.060192.002301. [DOI] [PubMed] [Google Scholar]
- 39.Chabrière E, et al. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol. 1999;6:182–190. doi: 10.1038/5870. [DOI] [PubMed] [Google Scholar]
- 40.Bunik VI, Degtyarev D. Structure-function relationships in the 2-oxo acid dehydrogenase family: Substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins. 2008;71:874–890. doi: 10.1002/prot.21766. [DOI] [PubMed] [Google Scholar]
- 41.Karamanos NK, Tsegenidis T, Antonopoulos CA. Analysis of neutral sugars as dinitrophenyl-hydrazones by high-performance liquid-chromatography. J Chromatogr. 1987;405:221–228. [Google Scholar]
- 42.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: John Innes Foundation; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.