Abstract
Plant cells sense environmental nitrogen levels and alter their gene expression accordingly to survive; however, the underlying regulatory mechanisms still remains to be elucidated. Here, we identified and characterized a transcription factor that is responsible for expression of nitrogen assimilation genes in a unicellular red alga Cyanidioschyzon merolae. DNA microarray and Northern blot analyses revealed that transcript of the gene encoding CmMYB1, an R2R3-type MYB transcription factor, increased 1 h after nitrogen depletion. The CmMYB1 protein started to accumulate after 2 h and reached a peak after 4 h after nitrogen depletion, correlating with the expression of key nitrogen assimilation genes, such as CmNRT, CmNAR, CmNIR, CmAMT, and CmGS. Although the transcripts of these nitrogen assimilation genes were detected in nitrate-grown cells, they disappeared upon the addition of preferred nitrogen source such as ammonium or glutamine, suggesting the presence of a nitrogen catabolite repression (NCR) mechanism. The nitrogen depletion-induced gene expression disappeared in a CmMYB1-null mutant, and the mutant showed decreased cell viability after exposure to the nitrogen-depleted conditions compared with the parental strain. Chromatin immunoprecipitation analysis demonstrated that CmMYB1 specifically occupied these nitrogen-responsive promoter regions only under nitrogen-depleted conditions, and electrophoretic mobility shift assays using crude cell extract revealed specific binding of CmMYB1, or a complex containing CmMYB1, to these promoters. Thus, the presented results indicated that CmMYB1 is a central nitrogen regulator in C. merolae.
Keywords: nitrogen catabolite repression, red alga
Nitrogen is an essential and major component of every cell, and autotrophic organisms like plants predominantly use inorganic nitrogen sources such as ammonium or nitrate. Nitrate is reduced by nitrate reductase and nitrite reductase, and the resulting ammonium is usually assimilated by glutamine synthetase (GS) and glutamate synthase (GOGAT), which is known as the GS-GOGAT cycle (1). In land plants and green algal cells, the nitrate anion is imported into the cell by specific transporters (2), and reduced to nitrite by NAD(P)H-dependent nitrate reductase (NAR) in the cytosol. Nitrite is subsequently reduced to ammonium by ferredoxin-dependent nitrite reductase (NiR) in plastids. Plants have 2 types of GS isoenzymes that are localized in different compartments: 1 is located in the cytosol (GS1) and the other in the plastid/chloroplasts (GS2) (3, 4). As an intriguing exception, in Arabidopsis, a GS was recently found to be localized to the mitochondria (5).
Regulatory mechanisms for nitrogen-responsive gene expression have been most extensively studied among eukaryotes in the budding yeast Saccharomyces cerevisiae. Several studies have revealed that a GATA-type transcription factor (TF), Gln3, controls the gene expression in response to the nitrogen status (6–8). The intracellular localization of Gln3 is important for the gene expression and thought to be under control of the TOR (target of rapamycin) kinase signaling pathway (6–8). Under the nitrogen-replete conditions where the TOR pathway is active, phosphorylated Gln3 forms complex with Ure2, and is localized to the cytoplasm. However, under nitrogen-depleted conditions where the TOR pathway is inactive, Gln3 becomes dephosphorylated and is released from Ure2, which results in accumulation of Gln3 in the nucleus and activation of the nitrogen-responsive gene expression. The Gln3-controlled genes are regulated by nitrogen catabolite repression (NCR) mechanism (6, 7). During growth on a preferred nitrogen source, such as ammonium or glutamine, Gln3 is bound to Ure2, thus NCR is exerted on catabolic enzymes and transport systems devoted to nonpreferred nitrogen sources such as proline. It has also been reported that another GATA-type TF, Gat1, contributes to the nitrogen-responsive gene expression as Gln3 in S. cerevisiae (6–8). AREA and NIT2, also members of the GATA family of TFs, are required to stimulate transcription of genes controlled by NCR in Aspergillus nidulans and Neurospora crassa, respectively (9). Therefore, GATA-type TFs appear to play integral roles in nitrogen-related gene expression in fungi. In contrast, the regulatory mechanism of expression of the nitrogen-related genes in plants remains largely unknown.
Cyanidioschyzon merolae is a thermo-acidophilic unicellular red alga isolated from an Italian volcanic hot spring (10). C. merolae has an extremely simple cell structure and a minimally redundant small genome (11–14), therefore this alga is considered to be a suitable model to study the origin and evolution of photosynthetic eukaryotes. It is also useful for studying the fundamental transcriptional network because it possesses a small number of TFs: <100 for the 16.5 Mb of the nuclear genome (13, 14). Thus, identification of TFs and elucidation of the underlying mechanism for any given regulatory process should be far easier than in more complex higher plants.
In a series of studies dealing with the nitrogen assimilation process in C. merolae, we found a nuclear gene (CMI233C) that encodes GS and identified its localization in the cytosol (15). And recently, although no typical NiR was found in C. merolae, we revealed that 1 of 2 sulfite reductase-related proteins functioned as NiR (CmNIR) and was localized in the chloroplast. In this study, we attempted to clarify the underlying mechanism for nitrogen-responsive transcriptional regulation, and identified and characterized a relevant TF in C. merolae.
Results
Identification of Nitrogen-Responsive Transcription Factor.
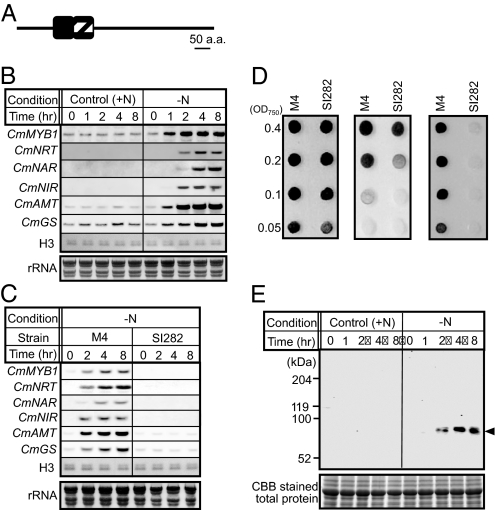
It has been reported that, in many cases, transcripts of a TF and its target genes are accumulated in a similar pattern (16). Therefore, we attempted to identify a nitrogen-responsive TF in C. merolae by transcriptome analysis after nitrogen depletion. Cells were grown to midlog phase and the cultivation medium was replaced with either nitrogen deplete (−N) or replete (+N) medium. Total RNAs from both cell cultures were isolated after 2 h from the medium exchange [T2; the subscript indicates the periods (hour(s) after the medium exchange)] and genome-wide DNA microarray analysis was performed to compare the transcriptomes of each condition. The result is shown in Table S1, where genes whose expression ratios (−N/+N) were >2.0 (mean folds) were defined as significantly increased under the nitrogen-depleted conditions (see SI Text). It was observed that transcripts of some key nitrogen assimilation genes [CmNRT (CMG018C, gene number in http://merolae.biol.s.u-tokyo.ac.jp/), CmNIR (CMG021C), CmAMT (CMT526C), and CmGS (CMI233C)] encoding nitrate/nitrite transporter, nitrite reductase (see Introduction), high affinity ammonium transporter, and glutamine synthetase, respectively, were increased in response to the −N condition as expected. With respect to expression of genes for TFs, CMJ282C (a MYB-related protein) and CMI163C (similar to TBP-associated factor TAF9) positively responded to the −N condition. We chose to further analyze the function of the MYB-related TF (hereafter referred to as CmMYB1) in relation to nitrogen regulation in C. merolae because of the following reasons: (i) the signal induction fold (−N/+N) of CmMYB1 transcripts was the highest among the TF genes (Table S1) and (ii) CMI163C was expected to be an RNA polymerase II general TF, and unlikely to function as a nitrogen specific TF. CmMYB1 was predicted to comprise 523 amino acids containing 2 MYB domains at the N-terminal region (positions are + 100 to + 149 and + 152 to + 200 where + 1 is the position of the first amino acid) (Fig. 1A). The 2 MYB domains are assigned as R2- and R3-type MYB domain, respectively, based on the result of phylogenetic analysis (Fig. S1), which shows that CmMYB1 is an R2R3-type MYB TF.
Fig. 1.
Domain structure of CmMYB1 and expression of CmMYB1 and nitrogen assimilation genes under nitrogen-depleted conditions. (A) Schematic representation of domain structure of CmMYB1. Black and hatched boxes indicate R2- and R3-type MYB domain, respectively. (B) Transcript levels of CmMYB1 and nitrogen assimilation genes under −N conditions. The C. merolae cells were harvested at the indicated time after nitrogen-depletion (−N) or control (+N) conditions and total RNAs were prepared from the cells. Total RNA (7 μg) was then subjected to Northern blot analysis with specific probes for the indicated genes. H3 (histone H3 gene, CMN176C) and rRNA stained with methylene blue are shown as the loading control. (C) Transcript levels of CmMYB1 and nitrogen assimilation genes under−N conditions in M4 and SI282 cells. Others are the same as in B. (D) Viability of M4 and SI282 cells. M4 and SI282 cells were exposed to −N (Middle) or control [+N (NH4+)] (Left) conditions for 8 h and diluted to an OD750 of 0.4, 0.2, 0.1, or 0.05 with medium containing NH4+, as indicated, and spotted onto a plate containing NH4+ as a sole nitrogen source. Logarithmic growth cells of M4 and SI282 cells were diluted with medium containing NO3− and spotted onto a plate containing nitrate as a sole nitrogen source the same as above (Right). All plates were incubated for 12 days. (E) Protein level of CmMYB1 under −N conditions. Aliquots containing 20 μg of total protein from the C. merolae cells harvested the same conditions as in B were separated by 7.0% SDS/PAGE and analyzed by immunoblot analysis with a CmMYB1 antibody. Arrowheads indicate the position of endogenous CmMYB1. The positions of molecular size markers are indicated in kilodaltons (kDa) at the Left. Total protein (≈55 kDa) stained with Coomassie Brilliant Blue (CBB) is shown as a loading control (Lower).
We then performed time course Northern blot analysis of the transcript levels of CmMYB1 after nitrogen depletion. The results indicated that CmMYB1 transcripts were significantly increased at T1, reached a peak at T2, and the level was maintained thereafter (Fig. 1B), which suggested that CmMYB1 is a nitrogen depletion-responsive TF in C. merolae. Under the same conditions, the transcripts of the key nitrogen assimilation genes, CmNRT, CmNAR (CMG019C for nitrate reductase), CmNIR, CmAMT, and CmGS, were also examined by Northern blot analysis (Fig. 1B). Transcripts of CmNRT, CmNAR, and CmNIR were detected at T2, whereas these transcripts were not detected at T0. In case of CmAMT and CmGS, although the transcripts were detectable even in nitrogen-replete conditions, the levels were increased at T1 after nitrogen depletion. All transcripts of the examined nitrogen assimilation genes were significantly increased at T4 compared with T2 after nitrogen depletion (Fig. 1B).
Next, to examine genetically whether CmMYB1 is involved in the expression of the key nitrogen assimilation genes, we constructed a null mutant strain lacking the CmMYB1 gene, named SI282, using a uracil auxotrophic mutant M4 (17) as the parental strain (see SI Text and Fig. S2). Results indicated that the nitrogen depletion-induced accumulation of the transcripts completely disappeared in SI282 (Fig. 1C). However, the basal levels of transcripts of CmAMT and CmGS were detected in SI282 irrespective of the nitrogen condition. We also transiently expressed CmMYB1 in SI282, and verified the complementation of the transcription of those nitrogen assimilation genes in response to the nitrogen status (Fig. S3). SI282 showed decreased cell viability after exposure to the nitrogen-depleted condition for 8 h compared with the parental strain (Fig. 1D, Left vs. Middle). Furthermore, as shown in Fig. 1D, SI282 could not grow under the nitrate growth conditions. These results clearly indicated that CmMYB1 plays a central role to directly or indirectly activate the nitrogen assimilation genes in response to the nitrogen status.
Protein Level of CmMYB1 Under Nitrogen-Depleted Conditions.
We then examined the CmMYB1 protein level in C. merolae cells under nitrogen-depleted conditions by immunoblot analysis. Antibodies raised against the recombinant CmMYB1 specifically recognized endogenous CmMYB1, apparent mass of which is 82 kDa in C. merolae cells (Fig. 1E, −N condition, see SI Text and Fig. S4). The CmMYB1 protein appeared at T2 after nitrogen depletion and reached a peak at T4, correlating with the expression of examined nitrogen assimilation genes (Fig. 1B). These results indicated that CmMYB1 is a nitrogen depletion-responsive TF. Although CmMYB1 transcripts were detected even under nitrogen-replete conditions (Fig. 1B), no CmMYB1 protein was detectable (Fig. 1E), implying a possible regulatory mechanism at the post-transcriptional level.
Intracellular Localization of CmMYB1.
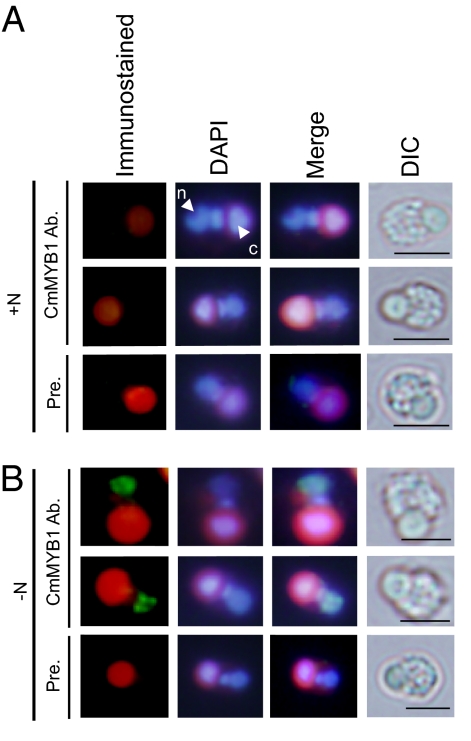
Intracellular localization of CmMYB1 in C. merolae cells was examined by indirect immuno-fluorescence microscopy analysis. At T4 after nitrogen depletion, the yellow-green fluorescence signal for CmMYB1 was observed in the nucleus (Fig. 2B). However, no signal was observed in the control experiment (+N condition) and also when the rabbit IgG purified from the preimmune serum was used (Fig. 2 A and B). These results are well consistent with the result obtained by immunoblot analysis shown in Fig. 1E, and indicated that the majority of the nitrogen depletion-induced CmMYB1 is localized in the nucleus.
Fig. 2.
Subcellular localization of CmMYB1. (A and B) Fixed cells under +N (A) or −N (B) conditions were reacted with rabbit anti-CmMYB1 antibody (CmMYB1 Ab.) or rabbit IgG purified from preimmune serum (Pre.), and the localization was detected with Alexa Fluor 488-conjugated goat anti-rabbit IgG antibodies (yellow-green). DAPI staining of cells (DAPI), merged image of immunostained and the nucleus DNA fluorescence (Merge), and differential interference contrast image (DIC) are shown. Positions of nucleus (n) and chloroplast (c) were indicated with arrowheads. (Scale bar, 2 μm.)
Specific Occupancies of CmMYB1 on the Promoter Regions of Nitrogen Assimilation Genes.
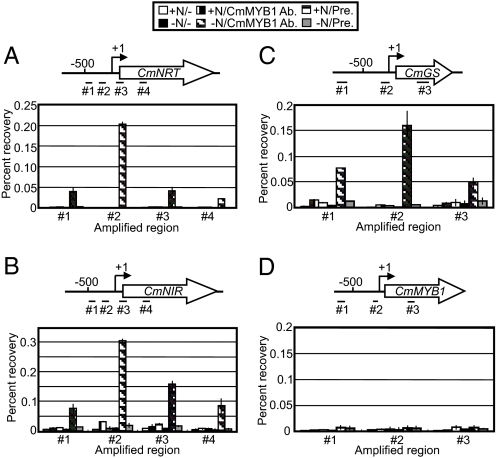
To examine a possibility that CmMYB1 is directly involved in transcriptional regulation of the nitrogen assimilation genes in the nucleus, we conducted chromatin immunoprecipitation (ChIP) analysis with the specific antibody against CmMYB1. As shown in Fig. 3 A–C, ChIP analyses indicated that DNA regions around the transcription start sites (TSSs) of the examined genes, which were determined by 5′-massively parallel signature sequence, primer extension, and the full-length EST analyses (see SI Text), were coimmunoprecipitated using the CmMYB1 antibody. In every case, the proximal upstream regions for each TSS showed the highest percentage recoveries: CmNRT, #2 (−254 to −112, +1 as the TSS); CmNIR, #2 (−219 to −89); and CmGS, #2 (−146 to + 20). These specific occupancies were not observed in the immunoprecipitations with preimmune serum or without serum under the nitrogen-depleted condition or by any combination of nitrogen status and antibodies under nitrogen-replete conditions. These results, taken together with the Northern blot analyses shown in Fig. 1, clearly indicated that CmMYB1 specifically occupies the promoter regions of the nitrogen assimilation genes, CmNRT, CmNIR, and CmGS, under nitrogen-depleted conditions as a positive regulator in vivo. We also examined the CmMYB1 occupancy on its own promoter region by ChIP analysis to determine the possibility of the autoregulation. The results indicated that no specific amplification of a promoter fragment of CmMYB1 was observed in any combination of nitrogen status and antibodies, implying that autoregulation is not involved in the CmMYB1 transcription (Fig. 3D). This result raises the possibility that other regulatory mechanism is responsible for the up-regulation of CmMYB1 expression in nitrogen-depleted conditions.
Fig. 3.
Specific occupancies of CmMYB1 on the promoter regions of nitrogen assimilation genes in vivo. (A–D) ChIP analysis. C merolae cells were exposed to −N or +N conditions for 4 h as in Fig. 1, and cells were subsequently fixed for ChIP analysis. Schematic diagrams above each image indicate the analyzed genes (A, CmNRT; B, CmNIR; C, CmGS, and D, CmMYB1) and positions that were amplified by quantitative real-time PCR following ChIP (+1 being the TSS). Minus (−), CmMYB1 Ab., and Pre. indicate the storage buffer for the antibodies used in this ChIP analysis, the affinity purified CmMYB1 antibody, and the rabbit IgG purified from the preimmune serum, respectively. Values are averages of at least 3 independent experiments and represent percentage recovery relative to the total input DNA.
Binding Region of CmMYB1 on the Promoters of the Nitrogen Assimilation Genes.
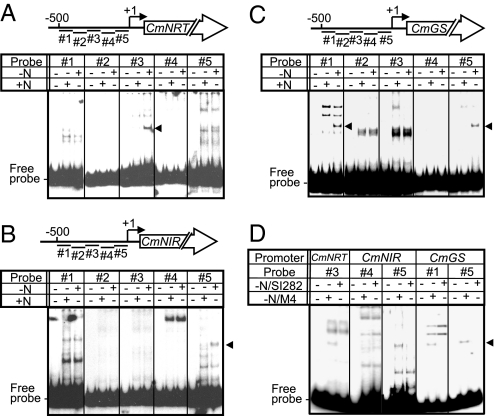
To verify whether CmMYB1 is able to bind directly to the promoter regions of nitrogen assimilation genes in vitro, electrophoretic mobility shift assay (EMSA) analysis was performed with purified recombinant CmMYB1 protein tagged with trigger factor (see SI Text). The results indicated that no specific band shift was observed for CmNRT, CmNIR, or CmGS promoter regions. These results may be because the trigger factor-tagged recombinant protein was not functional and/or requires post-translational modification or cofactor for DNA binding. Therefore, we next performed EMSA analysis using a C. merolae crude cell extract. The results shown in Fig. 4 A–C indicated that several specific shift bands were observed when CmNRT, CmNIR, or CmGS promoter regions were used as a probe DNA in the presence of high concentration of nonspecific competitor (see Materials and Methods). Some of them (indicated by arrowheads) were only detected with the crude cell extract prepared from nitrogen-depleted cells. To determine whether CmMYB1 is involved in the gel shift complex, we prepared a crude cell extract from SI282 cells and subjected to the EMSA analysis. The bands indicated by arrowheads in Fig. 4 A–C disappeared when a crude cell extract prepared from SI282 cells was used (Fig. 4D). However, the protein-NIR#4 complex, which was detected irrespective of the nitrogen status, was not changed with both crude cell extracts. We also confirmed that CmMYB1 was included in the gel shift complex by addition of CmMYB1 antibody to the reaction mixture (Fig. S5). These in vitro results are consistent with the data obtained by ChIP analyses and clearly indicated that CmMYB1, or a complex containing CmMYB1, specifically binds the promoter regions of nitrogen assimilation genes in a nitrogen status-dependent manner.
Fig. 4.
Electrophoresis mobility shift assay with C. merolae crude cell extract. (A–C) EMSA analyses with C. merolae crude cell extracts and the promoter regions of CmNRT (A), CmNIR (B), or CmGS (C). Schematic diagrams above each image indicate the analyzed genes and its probe positions (#1–#5) for EMSA. The TSSs (+1) were indicated with arrows. The presence or absence of the protein (12 μg) of crude cell extract that was prepared from cells harvested at 4 h after exposure to −N or +N conditions is indicated by + or −, respectively. Arrowheads indicate specific protein-DNA complexes that were specifically detected when proteins prepared from the −N condition were used. (D) EMSA analyses with crude cell extracts prepared from SI282 cells. EMSA analysis was performed under the indicated conditions as in A–C.
CmMYB1 Mediates NCR-Sensitive Transcription of the Nitrogen Assimilation Genes.
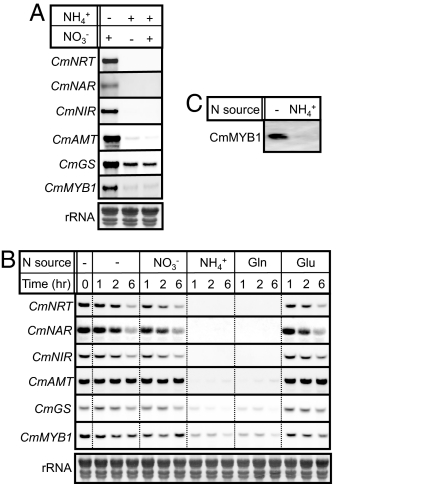
Our recent study indicated that the transcripts of CmNRT, CmNAR, and CmNIR were constantly expressed in the cells grown in culture containing nitrate as the sole nitrogen source, while hardly detected from cells grown with ammonium (Fig. 5A). In this study, we found that the levels of the transcripts of CmMYB1, CmGS, and CmAMT were also apparently increased under nitrate growth conditions compared with the ammonium growth condition (Fig. 5A). In A. nidulans and N. crassa, it was reported that NIRA and NIT4, GAL4-type TFs, were the specific activators for the nitrate assimilation pathway, respectively (9). Thus, to test the possibility of a nitrate specific regulatory pathway on the expression of the nitrogen assimilation genes in C. merolae, C. merolae cells were exposed to the −N condition for 4 h and supplied with nitrate, and total RNAs from the cells were sequentially isolated and analyzed. If a nitrate specific regulatory pathway is present, the −N-induced increases in those transcripts would show further enhancement. However, no transcriptional induction of those genes by addition of nitrate was observed [Fig. 5B, − (water) vs. NO3−]. However, the levels of those transcripts under cultivation with nitrate and ammonium were almost at the same levels as with ammonium (Fig. 5A, NO3− vs. NO3− + NH4+). Furthermore, these transcripts completely disappeared or decreased to the basal level after addition of ammonium to the −N treated C. merolae cells (Fig. 5B, − vs. NH4+). Reduction of the transcripts was similarly observed by addition of glutamine (− vs. Gln) but not by glutamate (− vs. Glu). These results suggested that C. merolae cells sense glutamine or ammonium as the nitrogen signal, but nitrate does not appear to be specifically sensed. With respect to glutamate, we do not have any evidence to discern if glutamate is not used as a nitrogen signal or not imported into C. merolae cells. In the case of the CmMYB1 transcript, the expression pattern was the same as those of the nitrogen assimilation genes, and the amount of −N induced CmMYB1 protein completely disappeared by addition of ammonium to the cells (Fig. 5 B and C). These observations indicated that CmMYB1 mediates the NCR-sensitive transcription of the nitrogen assimilation genes in response to nitrogen status.
Fig. 5.
Nitrogen catabolite repression-sensitive transcription of C. merolae nitrogen assimilation genes. (A) Transcript levels in the cells grown with nitrate and/or ammonium. Northern blot analyses were performed with total RNA (6 μg) isolated from cells grown with nitrate and/or ammonium as a nitrogen source using specific probes for the indicated genes. The presence (+) or absence (−) of ammonium (NH4+) or nitrate (NO3−) is indicated at the Top. The Lower shows rRNA stained with methylene blue as a loading control. (B) Transcript levels in the nitrogen starved cells after providing several nitrogen sources. C merolae cells were exposed to −N conditions for 4 h (set time 0), fed ammonium (NH4+), nitrate (NO3−), glutamine (Gln), glutamate (Glu), or water (−) at a final concentration of 5 mM, and sequentially harvested at the indicated time. After isolation of total RNAs from the cells, Northern blot analyses were conducted as in A. (C) CmMYB1 protein level. CmMYB1 protein level was investigated under the same conditions as in B, but the nitrogen source and time are NH4+ and 6 h, respectively.
Discussion
Transcriptional regulation of nitrogen assimilation genes is very important for plants, as assimilation of inorganic nitrogen is an essential biochemical process for growth. However, TF(s) responsible for such regulation have been elusive to date. One reason for the difficuty is the high proportion of gene redundancy in plant genomes. In the present study, we tried to identify a TF that contributes to the expression of nitrogen assimilation genes, using a primitive red alga C. merolae, making use of its minimally redundant small genome (see Introduction). Our in vivo and in vitro lines of evidence shown in this study clearly indicated that the R2R3-type MYB TF, CmMYB1, is a TF for the positive regulation of nitrogen assimilation genes in response to nitrogen status.
A possible regulatory mechanism for the nitrogen-depletion responsive gene expression by CmMYB1 is as follows: The CmMYB1 transcript increases first, and subsequently CmMYB1 protein accumulates in the nucleus and binds promoter regions of nitrogen assimilation genes. Consequently, expression of these nitrogen-responsive genes is derepressed to acclimate to nitrogen-limited environments. The accumulation of CmMYB1 in the nucleus seems to be sufficient for the derepression of those gene expression, since the level of CmMYB1 was proportional to that of those transcripts even in the nitrogen-replete conditions (Fig. S3). However, posttranscriptional modification or cofactor of CmMYB1 for the derepression could be required. CmMYB1 is likely to mediate NCR-sensitive transcription of the nitrogen assimilation genes as Gln3 and Gat1 in fungi (6, 7), because the expression of nitrogen assimilation genes was repressed when C. merolae cells were grown in the presence of glutamine or ammonium (Fig. 5). Under nitrate growth conditions, transcripts of CmNRT, CmNIR, and CmNAR also increased compared with cells grown with ammonium as in the case of nitrogen-depletion (Fig. 5A). The increased expression could be because of the decreased intracellular concentration of glutamine or ammonium, which may result in release from NCR, probably without sensing of nitrate levels.
As mentioned in the Introduction, GATA-type TFs function as nitrogen global regulators in fungi. Based on these cumulative studies, we first hypothesized the involvement of GATA-type TFs in the nitrogen regulation, and examined all 4 GATA-type TFs of C. merolae, CMB029C, CMD180C, CME102C, and CMR493C for this possibility. However, expression of those genes at transcription and protein levels were not significantly changed under nitrogen-depleted conditions. In addition, a specific gel-shift band was not observed in EMSA analysis with purified recombinant GATA-type proteins and nitrogen-responsive promoter regions. Recently, Bi et al. performed functional analysis of Arabidopsis GATA-type TFs by a reverse genetic approach (18). They obtained homozygous T-DNA insertion lines for 23 of the 30 Arabidopsis GATA-type TFs, and found 1 disruptant, which has an insertion in the GNC gene, revealed a reduction of the chlorophyll level. Although the expression of GNC was inducible by nitrate, transcript profiling experiments revealed that a considerable proportion of genes down-regulated in the loss-of-function mutants are involved in carbon metabolism but not nitrogen metabolism. Taking into account our results together with the previous report, it was suggested that GATA-type TFs are not involved in the expression of nitrogen assimilation genes in plant cells.
Given that an R2R3-type MYB TF is involved in nitrogen regulation in C. merolae, is regulation of nitrogen assimilation by an R2R3-type MYB TF conserved in higher plants as well? In Pinus sylvestris, PtMYB1 and PtMYB3, 2 R2R3-type MYB type TFs were shown to directly bind to the promoter region of the glutamine synthetase gene (19). In Lotus japonicus, 3 R2R3-type MYB TFs, LiMYB101, LiMYB102, and LiMYB103, were isolated, expressions of which were activated by nitrogen depletion in an organ specific manner (20). Lian et al. analyzed the expression profiles of rice seedlings under nitrogen-depletion stress using a microarray of 11,494 rice ESTs representing 10,422 unique genes. After 2 h from the shift to low nitrogen conditions, 5 TFs were activated, and 1 of them was an R2R3-type MYB TF (21). In Arabidopsis, a real-time reverse transcription-PCR platform for >1,400 TFs showed that transcripts of 14 TFs genes were increased in the nitrogen-starved seedlings compared with nitrogen-replete ones. Among them, PAP2 (At1g66390), which encodes an R2R3-type MYB TF, was drastically induced by 155.9-fold (other TF's rations were between 10.1 and 36.3) (22). PAP2 has known to be involved in the anthocyanin pigment metabolism (23), which is linked to nitrogen recycling pathway (24). These examples make it conceivable that for plant cells in general, regulation of nitrogen assimilation genes is mediated by R2R3-type MYB TFs in response to the nitrogen-depleted environments. MYB-type TFs comprise a large protein family in plant lineages compared with fungi and mammals (25, 26), suggesting the unique evolutional history of these TFs in plants.
Presently, the mechanism by which CmMYB1 activates the expression of nitrogen genes still includes much to be studied. In case of S. cerevisiae, the TOR pathway is involved in the intracellular localization of the GATA-type TFs, Gln3 and Gat1, in response to the nitrogen status as mentioned in Introduction (6–8). Recently, it was revealed that the TOR pathway also plays an important regulatory role in NCR sensitive transcription even after Gln3 is localized to the nucleus (27). Given the conserved structure and functions of TOR kinase among eukaryotes, it would be natural to postulate that the TOR pathway also plays a central role in the regulation of nitrogen assimilation genes through modification of the R2R3-type MYB TFs in plant cells. Further detailed study to CmMYB1 and the TOR pathway in C. merolae will give us important insights into the regulation and evolution of nitrogen assimilation systems in plant cells.
Materials and Methods
Strain and Growth Conditions.
Cyanidioschyzon merolae 10D was grown at 42 °C under continuous white light (100 μmol m−2 s−1) in liquid MA2 medium (28) at pH 2.5 bubbling with air supplemented with 2% CO2, supplemented with 0.5 mg/mL uracil if required. For nitrogen-depleted conditions, C. merolae cells were grown until 0.5–0.7 absorbance units at A750 and were collected by centrifugation (3,000 × g, room temperature, 5 min), and gently resuspended into the same volume of MA2 or nitrogen free medium, which has the same components as MA2 but 20 mM (NH4)2SO4, the solo nitrogen source, was substituted with 20 mM NaSO4. The cells were then continuously cultivated until the indicated times in the Fig. 1. In cases of nitrogen controlled media, 20 mM (NH4)2SO4 of MA2 was substituted with [5 mM NaNO3 and 20 mM Na2SO4], [2.5 mM (NH4)2SO4 and 17.5 mM Na2SO4], or [5 mM NaNO3, 2.5 mM (NH4)2SO4 and 17.5 mM Na2SO4] for NO3−, NH4+ or NO3−/NH4+ medium, respectively.
Microarray Analysis.
Oligonucleotide-based microarrays that cover 4,586 ORFs for >96% of all nuclear-encoded ORFs were designed and constructed (29). We used this array system with minor modification of the protocol. See SI Text for details.
Construction of CmMYB1-Null Mutant.
The transformation of C. merolae M4 strain (17) was performed as described in ref. 28. Details for the preparation of DNA for the transformation are provided in SI Text.
RNA Preparation and Northern Blot Analysis.
Total RNA and Northern blot analysis were performed as described in ref. 30. Gene-specific probes for Northern blot analyses were generated with specific primers (Table S2) and C. merolae genomic DNA as templates.
Preparation and Purification of Polyclonal Antibodies for CmMYB1.
Preparation and purification of polyclonal antibodies for CmMYB1 were carried out as described in ref. 30. See SI Text for details.
Immunoblot Analysis.
Immunoblot analysis was performed as described in ref. 30.
Indirect Immuno-Fluorescence Microscopy Analysis.
Immunostaining analysis was performed as described in ref. 30.
Chromatin Immunoprecipitation Analysis.
Chromatin immunoprecipitation analysis was performed as described in ref. 30. Primers used for real-time PCR are indicated in Table S3.
Preparation of C. merolae Crude Cell Extract and Electrophoretic Mobility Shift Assay (EMSA) Analysis.
C. merolae crude cell extracts were prepared in the same way as in the immunoblot analysis but the extraction buffer consisted of 12 mM Hepes-KOH, pH 7.9, 50 mM KCl, 3.5 mM MgCl2, 1 mM DTT, 15% glycerol, and Complete Mini, EDTA-free, protease inhibitor mixture tablet. Reactions (20 μL) were carried out in 10 mM Tris·HCl, pH 7.5, 50 mM KCl, 10% glycerol, 1 mM EDTA, 1 mM DTT, 1 μg poly(dI-dC)·poly(dI-dC) as a nonspecific competitor, 2.5 nM labeled probe, and/or 12 μg protein of C. merolae crude cell extract prepared from the +N or −N condition for 30 min at 30 °C. Detailed protocols for the preparation of DNA fragments used for EMSA analysis are provided in SI Text and Table S4.
Supplementary Material
Acknowledgments.
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) Grant-in-Aid for Creative Scientific Research 16GS0304 (to K.T.); Japan Society for the Promotion of Science (JSPS) Grant-in-aid for Scientific Research (B) 21370015 (to K.T.); and Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), Japan Society for the Promotion of Science (JSPS) Fellows Grant-in-Aid (S.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902790106/DCSupplemental.
References
- 1.Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galvan A, Fernández E. Eukaryotic nitrate and nitrite transporters. Cell Mol Life Sci. 2001;58:225–233. doi: 10.1007/PL00000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford NM. Nitrate: Nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- 5.Taira M, Valtersson U, Burkhardt B, Ludwig RA. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell. 2004;16:2048–2058. doi: 10.1105/tpc.104.022046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ter Schure EG, van Riel NA, Verrips CT. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- 7.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: Connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 9.Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroiwa T. The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays. 1998;20:344–354. [Google Scholar]
- 11.Ohta N, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10:67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- 12.Ohta N, Sato N, Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26:5190–5298. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 14.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terashita M, Maruyama S, Tanaka K. Cytoplasmic localization of the single glutamine synthetase in a unicellular red alga, Cyanidioschyzon merolae 10D. Biosci Biotechnol Biochem. 2006;70:2313–2315. doi: 10.1271/bbb.60174. [DOI] [PubMed] [Google Scholar]
- 16.Obayashi T, et al. ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45:667–671. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
- 18.Bi YM, et al. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44:680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-Maldonado J, et al. Functional interactions between a glutamine synthetase promoter and MYB proteins. Plant J. 2004;39:513–526. doi: 10.1111/j.1365-313X.2004.02153.x. [DOI] [PubMed] [Google Scholar]
- 20.Miyake K, et al. Isolation of a subfamily of genes for R2R3-MYB transcription factors showing up-regulated expression under nitrogen nutrient-limited conditions. Plant Mol Biol. 2003;53:237–245. doi: 10.1023/B:PLAN.0000009296.91149.34. [DOI] [PubMed] [Google Scholar]
- 21.Lian X, et al. Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol Biol. 2006;60:617–631. doi: 10.1007/s11103-005-5441-7. [DOI] [PubMed] [Google Scholar]
- 22.Scheible WR, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004;136:2483–2499. doi: 10.1104/pp.104.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allan AC, Hellens RP, Laing WA. MYB transcription factors that color our fruit. Trends Plants Sci. 2008;13:99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Razal RA, Ellis S, Singh S, Lewis NG, Towers GHN. Nitrogen recycling in phenylpropanoid metabolism. Phytochem. 1996;41:31–35. [Google Scholar]
- 25.Romero I, et al. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- 26.Riechmann JL, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 27.Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in. Saccharomyces cerevisiae. J Biol Chem. 2008;283:8919–8929. doi: 10.1074/jbc.M708811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnuma M, Yokoyama T, Inouye T, Sekine Y, Tanaka K. Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2008;49:117–120. doi: 10.1093/pcp/pcm157. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara T, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imamura S, Hanaoka M, Tanaka K. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 2008;27:2317–2327. doi: 10.1038/emboj.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.