Abstract
Nonhomologous end joining (NHEJ), a form of DNA double-strand break (DSB) repair, is conserved from bacteria to humans. One essential NHEJ factor is Ku, which consists of a heterodimer of Ku70 and Ku86. In a plethora of model systems, null mutations for Ku70 or Ku86 present with defects in DNA DSB repair, variable(diversity)joining [V(D)J] recombination, and/or telomere maintenance. The complete loss of Ku from bacteria to mice is, however, compatible with viability. In striking contrast, human patients with mutations of either Ku subunit have never been described. Here, we have used recombinant adeno-associated virus-mediated gene targeting to produce a human somatic cell line that expresses a conditionally null allele of Ku86. The induced loss of Ku86 results in cell death accompanied by massive telomere loss in the form of t-circles. Thus, Ku86 is an essential gene in human somatic cells because of its requirement, not in NHEJ or V(D)J recombination, but in telomere maintenance.
Keywords: NHEJ, rAAV, t-circles, DNA-PK, homologous recombination
The repair of DNA double-strand breaks (DSBs) is important for cellular survival, the maintenance of genomic integrity, and the prevention of tumorigenesis. DSBs can be caused by exposure to exogenous agents, including ionizing radiation and chemotherapeutic agents, and be generated by endogenous mechanisms, such as variable(diversity)joining [V(D)J] and class-switch recombination—processes required for the maturation of B and T lymphocytes. Moreover, the ends of linear chromosomes present cells with naturally occurring DSBs—i.e., telomeres—and these must be regulated to ensure the stable maintenance of the genome (1). Because of the importance of DNA repair, immune function, and genomic stability for organismal well-being, at least 2 mechanisms for the repair of DSBs have evolved: homologous recombination (HR) (2) and nonhomologous end-joining (NHEJ) (3).
In lower eukaryotes, HR, which requires regions of homology between the donor and the recipient DNAs, is the major pathway for general DNA DSB repair. In higher eukaryotes, HR is also important for meiosis, sister chromatid exchange, and the repair of stalled replication forks (2). Despite the importance of HR, the process of NHEJ, in which 2 DNA ends are joined together regardless of their DNA sequence homology, nonetheless predominates in mammals (3). The most upstream, and quite probably the most important NHEJ factor, is the Ku complex (4).
Ku is conserved from bacteria to humans. In bacteria, Ku exists as a homodimer, but in all other species it exists as 2 independent subunits, Ku70 and Ku86, that tightly heterodimerize (4). Importantly, in all organisms examined—with one glaring exception—mutations of either Ku subunit result in the expected deficits in DNA DSB repair and recombination. Intriguingly, humans are unique in that Ku has apparently evolved into an essential gene. This hypothesis is supported by the lack of documentation for even a single patient with a mutation in either Ku subunit. The absence of human Ku patients stands in contrast to some of the more downstream NHEJ factors, such as DNA-PKcs (5), Artemis (6), Cernunnos/XLF (7), and DNA LIGIV (8), for whom mutations in human patients have been described. Thus, it appears as if Ku—but not NHEJ—is essential. The bias that Ku may be essential in humans is also supported by the demonstration that the targeted disruption of both alleles of either Ku70 (9) or Ku86 (10) in human somatic cells was lethal. And although it was impossible to discern the mechanism of cell death in these studies, the fact that the inactivation of one allele of either Ku70 (11) or Ku86 by gene targeting (12) or the reduction of Ku86 levels by RNAi (13) resulted in telomere shortening suggested by extrapolation that the lethal event in a completely Ku-deficient human cell might be due to aberrant telomere maintenance.
Why human Ku-deficient cells should suffer from lethal telomere maintenance events when this is not observed in the cells/organisms of other Ku null species has been difficult to envision. What is clear, however, is that telomere maintenance is a species-idiosyncratic process. Ku and Ku mutants exemplify this. Thus, mutation of either Ku subunit in most species results in telomere defects. Despite this uniformity, however, there are some glaring incongruities with the phenotypes of the respective mutants. Thus, Saccharomyces cerevisiae Ku mutants show some telomere shortening (14) and a high temperature lethality associated with defective telomere maintenance (15). In contrast, Arabidopsis thaliana Ku mutants show massive telomeric expansions (16). Different yet from all these are chicken DT40 cells, where no telomere defects or only slight expansions have been reported (17). Finally, the mouse literature is conflicted, with slight telomeric expansions (18) or significant telomere shortening (19) being reported for similar mouse strains. Altogether, the diametrically opposed results generated from different model organisms combined with the apparent lethality of Ku-defective human cells has resulted in a conundrum concerning the impact of Ku mutations on human telomere maintenance. To scientifically address this issue, we constructed a human cell line that is conditionally null for Ku86. Upon expression of the Cre recombinase, the only functional allele of Ku86 is lost from the genome. The resulting cells are not viable, consistent with earlier observations (10). Importantly, it now can be demonstrated that cell death is associated with a telomere loss so rapid and extensive that it has no parallels in the mammalian literature. The telomere loss occurs nearly en masse in the form of extrachromosomal t-circles, and this is consistent with human Ku86 being an essential regulator for the suppression of rapid telomere loss.
Results
Construction of a Ku86 Conditionally Null Human Cell Line.
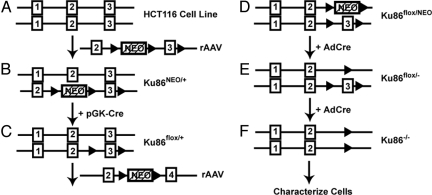
To generate a HCT116 cell line conditionally null for Ku86 expression, a gene-targeting scheme comparable to those used for making conditionally null mice was used except that in lieu of back-crossing, a second round of gene targeting was required (Fig. 1). For the first round of targeting, a recombinant adeno-associated virus (rAAV) vector (20) was constructed containing 3 LoxP recognition sites flanking the neomycin phosphotransferase (NEO) gene and exon 3 of human Ku86, respectively (Fig. 1A). Transient Cre expression was then used on one of the resulting clones (Ku86NEO/+; Fig. 1B and Fig. S1 A–C) to facilitate removal of the NEO selection cassette (Fig. 1C and Fig. S1 D–G). This cell line (Ku86flox/+) was then subjected to a second round of gene targeting using an rAAV knockout vector in which Ku86 exon 3 sequences had been replaced with the NEO gene (Ku86flox/NEO cells; Fig. 1D and Fig. S1 H–L). The Cre recombinase (AdCre) was once again used to remove the NEO gene, and the resulting cell line was designated as the conditionally null or Ku86flox/− cells (Fig. 1E and Fig. S1 M, N, and P). Ku86flox/− cells are viable, functional heterozygotes and could be expanded at will. When needed, these cells were infected yet again with AdCre to generate Ku86 null (Ku86−/−) cells, which could be biochemically characterized as they died (Fig. 1F and Fig. S1 N–P).
Fig. 1.
Scheme for functional inactivation of the human Ku86 locus. (A) A diagram of a partial Ku86 genomic locus. Exons are shown (not to scale) as numbered rectangles. In addition, a diagram of the rAAV targeting vector is shown. The filled triangles represent LoxP sites, and the hatched rectangle represents the NEO gene. (B) Diagram of the Ku86NEO/+ cell line generated by the first round of targeting. (C) Ku86NEO/+ cells were transiently transfected with a Cre expression plasmid (+pGK-Cre), and a clone of cells (Ku86flox/+) that had lost the NEO gene but retained the floxed exon 3 was identified. (D) Ku86flox/+ cells were then subjected to a second round of targeting using an rAAV exon 3 knockout vector. This generated the Ku86flox/NEO cell line. (E) The Ku86flox/NEO cell line was then infected with AdCre (+AdCre), and a clone of cells that were G418 sensitive (Ku86flox/−) was isolated. (F) These Ku86flox/− cells can be converted into Ku86 null (Ku86−/−) cells at the investigator's discretion by infecting them with AdCre.
Human Ku86 Null Human Cells Are Not Viable.
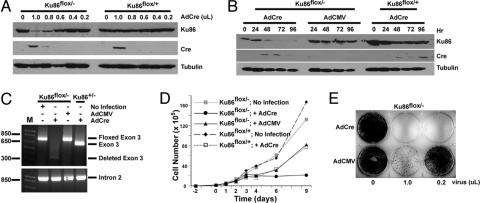
Ku86flox/− and Ku86flox/+ cells were infected with increasing concentrations of an AdCre virus stock, and 3 days later whole-cell extracts were prepared and analyzed by sequentially immunoblotting a single membrane for Ku86, Cre, and then tubulin (as a gel-loading control) expression. At high virus concentrations, Cre protein expression could be detected, and for Ku86flox/− cells this correlated with a reduction in Ku86 expression (Fig. 2A). The residual Ku86 expression (≈5% of wild type) is attributed to cells that were not infected by AdCre or that did not undergo a Cre-mediated LoxP deletion. To address the stability of Ku86 protein, Ku86flox/− and Ku86flox/+ cells were infected with 1 μL of either AdCre or AdCMV (an empty vector), and then whole-cell extracts were analyzed by immunoblotting. Ku86 expression was lost ≈72 h after infection only in those cells expressing Cre (Fig. 2B). To confirm these data, genomic DNA was isolated from Ku86flox/− cells 5 days after they had been infected with either AdCre or AdCMV or left untreated. This DNA was then subjected to PCR either using primers flanking Ku86 exon 3 or, as a loading control, primers residing within Ku86 intron 2 (Fig. 2C). As an additional control, genomic DNA isolated from a Ku86+/− cell line infected with AdCre was analyzed by using the same exon 3 primers. This latter sample showed the expected position for the wild-type exon 3 (Fig. 2C). In contrast, DNA isolated from Ku86flox/− cells left untreated or infected with AdCMV produced only a band that was 68 bp larger, an increase that corresponded to the size of the 2 LoxP sites flanking exon 3 (floxed exon 3; Fig. 2C). When DNA was isolated from Ku86flox/− cells that had been infected with AdCre, the floxed exon 3 band was no longer visible, and only a faint band corresponding to the size of the deleted exon 3 was observed (Fig. 2C). In toto, these experiments confirmed that the infection of Ku86flox/− cells with AdCre resulted in a nearly complete reduction of Ku86 protein expression due to the efficient excision of Ku86 exon 3. The effect of either AdCre or AdCMV infection on Ku86flox/− and Ku86flox/+ cell growth was next determined. Cells that were not infected grew exponentially (Fig. 2D). In contrast, all cell lines that had been infected with an adenoviral vector exhibited a brief cessation of growth for ≈3 days after infection. Eventually, by days 4 to 6, Ku86flox/− cells infected with AdCMV or Ku86flox/+ cells infected with AdCre resumed growth. In contrast, Ku86flox/− cells infected with AdCre never resumed growth, suggesting that Ku86 was required for proliferation (Fig. 2D). The essential nature of Ku86 was confirmed by infecting Ku86flox/− cells with AdCMV or AdCre and then fixing and staining the surviving cells 2 weeks after infection. The infection of Ku86flox/− cells with AdCre resulted in the death of nearly all of the cells (Fig. 2E). These experiments confirmed that Ku86 is an essential gene in the HCT116 human cell line (10).
Fig. 2.
Characterization of a human Ku86 conditionally null cell line. (A) Documentation of the loss of Ku86 protein. Cell lines were exposed to the indicated dose of AdCre, and 3 days later whole-cell extracts were prepared and analyzed by Western blot analyses using antibodies directed against Ku86, Cre, and tubulin. A single blot is shown that was sequentially probed, stripped, and reprobed. (B) Same as A except that a time course rather than a dose–response is shown. AdCMV is a control, empty vector. (C) Molecular evidence for the loss of the floxed exon 3. The indicated cell lines were exposed to no virus, AdCMV, or AdCre virus. Genomic DNA was isolated 5 days later and subjected to PCR using primers that flank exon 3. A control PCR was performed by using a primer set located within intron 2. Both sets of PCR reactions were electrophoresed on agarose gels and stained with ethidium bromide. (D) Ku86flox/− cells exposed to Cre stop growing. The indicated cell lines were plated out 2 days before infection (day 0). The growth of the cells was monitored by counting live (trypan blue-excluding) cells at subsequent days. (E) Ku86flox/− cells exposed to Cre recombinase die. Ku86flox/− cells were exposed to the indicated amounts of either AdCre or AdCMV virus and then fixed and stained with crystal violet ≈14 days later.
Human Ku86 Null Cells Accumulate DSBs.
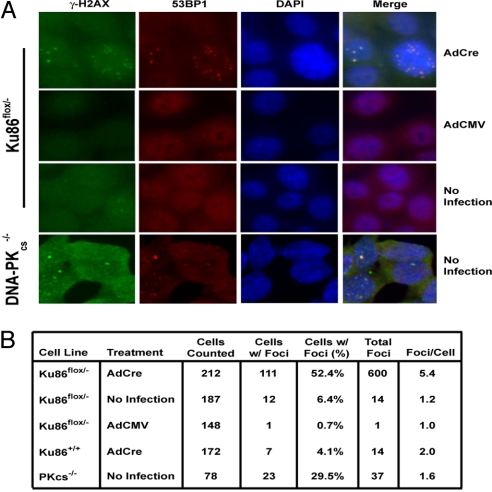
Ku86 plays an integral role in DNA DSB repair in every organism in which it has been examined (4). To determine whether human cells lacking Ku86 accumulated DNA DSBs spontaneously, an asynchronous population of Ku86flox/− cells was infected with AdCre, and 5 days later the cells were fixed and stained with fluorescently tagged secondary antibodies directed against primary antibodies for γ-H2AX, 53BP1 (both of which are markers for DNA DSBs), or Ku86. Only cells infected with AdCre produced significant γ-H2AX and 53BP1 foci (Fig. 3A), and this correlated precisely with the absence of Ku86 protein (Fig. S2). A total of 100 to 200 cells were scored for the presence and number of microscopically visible γ-H2AX foci. More than 50% of Ku86flox/− cells infected with AdCre contained at least one γ-H2AX focus, and the positive cells averaged 5.4 foci per cell (Fig. 3B). In contrast, noninfected Ku86flox/− cells, Ku86flox/− cells infected with AdCMV, or Ku86+/+ cells infected with AdCre had many fewer γ-H2AX-positive cells and fewer foci per positive cell (Fig. 3B). As an additional control, an isogenic DNA-PKcs−/− HCT116 cell line (21) that is known to be deficient in C-NHEJ and which accumulates spontaneous DNA damage was also scored. These cells showed detectable γ-H2AX and 53BP1 foci, but the frequency and especially the quantity were significantly less than the Ku86 null cell line (Fig. 3) Thus, human somatic cells accumulate DNA DSBs in the absence of Ku86, and this occurs at a frequency higher than is caused simply be a deficiency in C-NHEJ.
Fig. 3.
Human Ku86 null cells accumulate DNA DSBs. (A) Immunofluorescent detection of DNA DSBs. Ku86flox/− cells were either left untreated (No Infection) or were infected with AdCre or AdCMV. Cells were fixed 5 days later and stained with fluorescently tagged secondary antibodies specific for γ-H2AX (green) or 53BP1 (red) primary antibodies. DNA was identified by DAPI staining (blue), and all 3 panels were then overlaid (merge). (Magnification: 100×.) (B) Ku86flox/− cells have a higher incidence of γ-H2AX foci. A total of 100 to 200 cells from each experimental condition described in A along with Ku86+/+ cells infected with AdCre (as a Cre control) were scored for the percentage of cells containing at least one microscopically visible γ-H2AX focus and for the number of foci per positive cell.
Human Ku86 Null Cells Suffer Massive Telomere Loss.
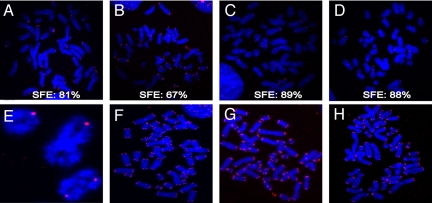
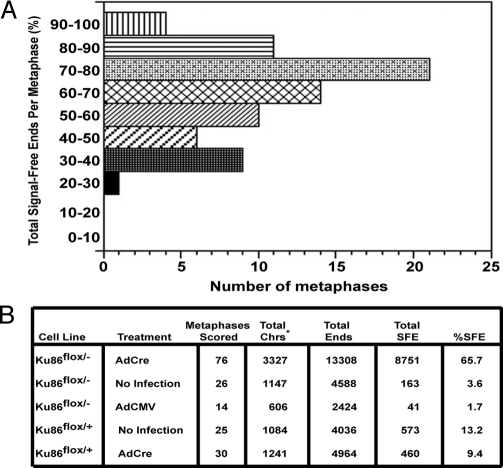
The telomere loss associated with the functional inactivation of a single allele of Ku86 in HCT116 cells suggested that Ku86 plays a role in human telomere length maintenance (12). A similar conclusion was arrived at by using RNAi to deplete Ku levels in HeLa, SAOS, and U2OS cells (13). To examine the effect of the complete loss of Ku86 expression on telomere length maintenance, Ku86flox/− cells were infected with AdCre, and 72 h after infection the cells were arrested in metaphase, hybridized with a telomere-specific probe, and analyzed by FISH. Strikingly, the majority of chromosomes in Ku86 null cells lacked a detectable telomeric signal (Fig. 4 A–D). On average, 65.7% of the chromosomes in Ku86 null cells were telomere-free (Fig. 5B), and no metaphase showed <20% of signal-free ends (SFEs; Fig. 5A). In contrast, any of the control cell lines examined, including Ku86flox/− cells infected with AdCMV (Fig. 4F), Ku86flox/+ cells infected with AdCMV (Fig. 4G), Ku86flox/+ cells infected with AdCre (Fig. 4H), and uninfected Ku86flox/− cells, showed a much lower incidence of SFEs (Fig. 5B). Because defects in telomere recognition factor 2 (TRF2) and DNA-PKcs lead specifically to dysregulation of leading-strand telomeres (22) and dysfunction of the Werner syndrome (WRN) protein causes defects only in lagging-strand telomeres (23), individual metaphases from Ku86 null cells were scored for the specific loss of leading or lagging-strand telomeres. The absence of Ku86, however, led to a random loss of telomeric ends, suggesting that it was affecting telomere maintenance in a manner discrete from TRF2, DNA-PKcs, and WRN (Fig. S3). Together, these data suggested that the high frequency of DNA DSBs and the lack of viability of Ku86 null cells are strongly correlated with—and likely due to—telomere loss.
Fig. 4.
Many of the chromosomes in human Ku86 null cells lack telomeres. FISH analyses of metaphase chromosomes with a telomere-specific Cy3-(C3TA2)3 protein–nucleic acid probe. Telomeres are seen as red dots, and metaphase chromosomes are stained blue. (A–D) Four independent metaphases observed in Ku86flox/− cells treated with AdCre. The percentage of SFEs is shown for each metaphase. (E) An enlargement of 3 chromosomes observed in a metaphase derived from Ku86flox/− cells treated with AdCre showing the apparent fusion of the sisters either with some residual telomere sequence (small red dots) or apparently lacking all telomeric sequence. Representative metaphases derived from: (F) Ku86flox/− cells treated with AdCMV, (G) uninfected Ku86flox/+ cells, or (H) Ku86flox/+ cells treated with AdCre are also shown. (Magnification: 120×.)
Fig. 5.
Significant telomere loss in Ku86 null cells. (A) A total of 76 metaphases were scored for the percentage of SFEs, and the number of metaphases containing 0–10%, 10–20%, etc. SFEs is shown. Every metaphase had at least 20–30% SFEs, and most metaphases exhibited 70–80% SFE. (B) Ku86 null cells undergo telomere loss. The indicated cell lines were subjected to the indicated treatments and scored for SFEs. Total Chrs*, total chromosomes. The number is smaller than the theoretical total because some metaphases contained <46 distinguishable chromosomes, and only chromosomes that could be unambiguously scored were included.
Interestingly, even though most Ku86 null chromosomes lacked telomeres, a higher frequency of chromosome–chromosome fusions was not observed in these cells versus any of the various heterozygous or wild-type cell lines. Instead, the vast majority of Ku86 null chromosomes seemed to have fused their sister chromatids with either the complete loss of telomeric sequence or with the retention of only small amounts of telomeric DNA (Fig. 4E). Despite this compelling visual evidence, repeated attempts to clone putative sister–sister fusion events have been unsuccessful, and thus these events may only be telomere associations.
Telomere Loss in Human Ku86 Null Cells Is Not Due to Defective Telomerase Activity.
The rapid and extensive telomere loss in Ku86 null human cells was unexpected, and we considered several possible mechanisms for this telomere attrition. Telomerase-negative, nonimmortalized diploid human cells undergo the attrition of ≈100 bp of telomeric DNA with each cell division because of the end replication problem. Although HCT116 is a telomerase-positive, immortalized cell line and presumably immune to this effect, human Ku interacts with telomerase, and it has been suggested that this interaction may be required for telomerase activity (24). Thus, a Ku86 null human cell could correspond to a telomerase activity null cell line and might passively lose its telomeres. Two facts argue strongly against this model. First, by using a standard telomere repeat amplification protocol, Ku86 null cells exhibited robust telomerase activity (Fig. S4). In addition, telomere loss in Ku86 null cells occurred over a period of 3 to 5 days, which for this cell line corresponds to only 3 to 5 (or fewer) cell divisions. Thus, passive telomere attrition due to the absence of telomerase activity is not a viable model.
Human Ku86 Null Cells Contain Elevated Levels of Extrachromosomal Telomeric FISH Signals.
Insight into the actual mechanism of telomere loss came from a more careful examination of the FISH experimental data (Fig. 4), where the elevated existence of extrachromosomal FISH signals (Fig. S5) struck our attention. In the Ku86 null metaphases, these extrachromosomal FISH signals costained with DAPI, indicating that they comprised telomeric fragments of DNA (Fig. S5 B–E). These data implied that at least some of the telomeric sequences that were lost from the ends of chromosomes in Ku86 null cells did not result from exonucleolytic degradation, but may have been generated by fragmentation of the telomere away from the body of the chromosome.
An indirect way that telomere fragmentation could occur would be as a consequence of apoptosis. However, Ku86 null cells showed no evidence of apoptotic induction, as indicated by PARP-1 cleavage (Fig. S6). Thus, we considered the possibility that the extrachromosomal FISH signals might correspond to t-circles and not to linear telomeric DNA fragments.
Human Ku86 Null Cells Contain Greatly Elevated Levels of t-Circles.
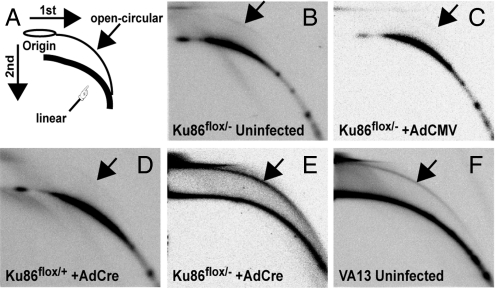
A handful of studies have demonstrated that telomeric DNA, when acted upon by HR, can be lost in the form of circles (“t-circles”). This phenomenon, termed “telomere rapid deletion” (TRD), was first observed in yeast (25). A similar, although perhaps not identical, process has subsequently been observed in a number of eukaryotic systems. Some of these systems include telomerase-negative human cancer cells that maintain their telomeres through an HR-mediated mechanism referred to as “alternative lengthening of telomeres” (ALT; reviewed in ref. 1), and cell lines and organisms in which telomere capping is disrupted such that HR is somehow up-regulated or derepressed (26, 27). To experimentally determine whether the extrachromosomal DNA observed in Ku86 null cells corresponded to t-circles, genomic DNA from these cells and control cell lines was subjected to neutral/neutral 2D gel electrophoresis. Before electrophoresis, the genomic DNA was digested to completion with the AluI restriction enzyme. Because t-circle DNA consists solely of repetitions of T2AG3, it should be refractory to AluI digestion. The restriction digests were then subjected to 2D gel electrophoresis. Open circular (t-circle) DNA should migrate slower in the second dimension and migrate as a retarded arc in the gel (Fig. 6A). In any of the negative control cell lines, only a faint t-circle arc was ever visible, and then usually only upon prolonged exposure of the autoradiograms (Fig. 6 B–D). In stark contrast, Ku86flox/− cells infected with AdCre exhibited a very prominent t-circle arc that comprised a significant fraction of the telomeric DNA (Fig. 6E). As a positive control, asynchronously growing WI-38 VA13 cells, which maintain their telomeres via ALT, were subjected to the same analysis. The expected t-circular arc was observed in this cell line (Fig. 6F). These experiments demonstrated that the loss of telomeric sequences in human Ku86 null somatic cells was mediated, at least in part, by the formation of t-circles.
Fig. 6.
Human Ku86 null cells contain elevated levels of extrachromosomal t-circles. Two-dimensional neutral/neutral gel electrophoresis analyses of AluI-restriction enzyme-digested genomic DNA from the indicated cell lines. After electrophoresis, each gel was transferred to nitrocellulose and hybridized with a 32P-(C3TA2)3 telomere-specific probe. (A) A diagram showing the expected arced migration of linear genomic DNA (finger) and the elevated arc expected for open-circular (“t-circle”) DNA (arrow). (B–F) Autoradiograms of the gels produced by using AluI-digested genomic DNA from the indicated cell lines: (B) uninfected Ku86flox/− cells; (C) Ku86flox/− cells treated with AdCMV; (D) Ku86flox/+ cells treated with AdCre; (E) Ku86flox/− cells treated with AdCre; and (F) uninfected ALT WI-38 VA13 cells. (Magnification: 1×.)
Discussion
Ku Is a Repressor of Telomere Loss in Human Somatic Cells.
We have constructed a unique and powerful reagent: a human cell line containing only 1 functional allele of Ku86, which has been engineered such that the allele becomes nonfunctional upon expression of the Cre recombinase. We used this cell line to confirm that Ku86 is an essential human gene, in stark contrast to every nonprimate system that has been investigated. Surprisingly, the absence of Ku86 precipitated a lethal telomere attrition accompanied by the extensive formation of t-circles.
Before the completion of these studies, our preferred model was that Ku was localized to the distal dsDNA end of telomeres and acted as a physical barrier to nucleases, which would otherwise exonucleolytically degrade the telomeres and make them palatable HR substrates. This possibility, for which there is ample support from a variety of model systems (28), was consistent with Ku86's deduced role during V(D)J recombination (4) as well our demonstration that Ku86+/− cells contained slightly elongated G overhangs (12). There was, however, no hyperelongation of G overhangs in Ku86 null cells (Fig. S7). This unexpected result, however, might still be consistent with Ku regulating end resection. Thus, if those telomeres that undergo extensive C strand resection are especially unstable, it might make them the most susceptible to HR-mediated recombination. This scenario might explain why the length of the G overhangs does not increase (because those that do are rapidly converted to HR substrates) while simultaneously providing an explanation for the existence of t-circles (which are the products of HR-mediated recombination; Fig. 6).
There are a variety of additional ways that Ku could regulate telomere loss. One possibility is that Ku directly competes with HR for repair/recombination events in human cells. Because Ku is an abundant protein, it generally outcompetes HR, whereas in a Ku-deficient cell, HR would presumably mediate most of the recombination events. This model is consistent with the demonstration that Ku-reduced human cell lines carry out gene targeting (an HR-mediated event) at increased frequencies (9), which implies that HR is up-regulated throughout Ku-reduced cells and not exclusively at telomeres. An alternative possibility is that Ku86 mediates its effects through the TRF2 or WRN proteins. TRF2 binds to the telomeric T2AG3 sequence and regulates t-loop formation. The expression of a mutant allele of TRF2, TRF2ΔB, promotes extensive telomeric t-circle deletion events (26). Thus, if a Ku deficiency phenocopied a TRF2ΔB mutation, it could explain the high frequency of TRD observed in Ku86 null cells. This hypothesis is attractive, given that TRF2 and Ku interact (29), although it is unknown whether that interaction is mediated through the TRF2B domain. It should be noted, however, that no alterations in TRF2 expression levels or cellular localization in Ku86 null cells has been detected (Fig. S5). Whether TRF2 activity is altered is an important future question to address. Alternatively, cells defective for WRN show an increased frequency of t-circles (30), a phenotype predicted if Ku positively regulates WRN. The fact that Ku and WRN proteins physically interact is supportive of this hypothesis. The fact that alterations in TRF2 (22) and WRN (23) result in leading-strand-specific and lagging-strand-specific defects, respectively, whereas the loss of Ku86 affected the telomere ends randomly (Fig. S3) is, however, difficult to reconcile with these models. In any case, there is precedent for Ku regulating telomere loss. In plants, deletion of Ku also results in the formation of t-circles (31). However, the parallel to plants is limited because Ku-deficient plants are viable and aphenotypic. Moreover, Ku deficiency in plants actually causes telomere hyperelongation, and not the shortening observed in humans and, in a somewhat evolutionarily perverse manner, the (re)expression of Ku70 in Ku70 null plants actually induced telomere shortening (32). Thus, the impact of Ku loss-of-function mutations seems to be species-specific. In humans, Ku86 is a potent negative regulator of telomere loss, and the absence of Ku86 function uniquely leads to lethal levels of telomere loss via t-circle formation. Whether this is an intrinsic function of Ku86 or whether it is mediated by TRF2, WRN, or another factor remains to be elucidated.
Where Are All of the Human Ku Mutants?
There are no known human Ku mutation syndromes. Our data predict that if a Ku patient is found, he or she will be carrying hypomorphic mutations and will have telomeric dysfunction, characteristics that describe individuals afflicted with dyskeratosis congenita (DC). DC patients have severely shortened telomeres and are predisposed to cancer. DC is a polygenic disorder, and although several complementation groups have been identified, the defective gene(s) for approximately half of DC patients remains unknown. Although DC patient cell lines are notoriously difficult to propagate, it would be interesting to determine whether they contain elevated levels of t-circles. Another likely candidate population for finding Ku mutations would be human ALT tumors. In plants, Ku suppresses telomere maintenance by ALT (31), and our data would correspondingly suggest that Ku mutations would promote ALT. The mechanism for the initiation of ALT is unknown, and an interesting possibility would be if reduced Ku activity (mutations?) were a prerequisite for this. Reduced Ku levels should facilitate the formation of t-circles, and these could then potentially facilitate an HR/rolling circle-based mechanism of telomere maintenance in the absence of telomerase as has been demonstrated for Kluyveromyces lactis (33).
Is Ku86-Regulated Telomere Loss in Human Somatic Cells Akin to Yeast TRD?
In yeast, TRD results in a sudden loss in telomeres that have become inappropriately long (25). TRD is inhibited by Ku (as is the telomere loss we have described here), and it requires HR activities. In this light, it is important to note that the formation of t-circles in human ALT cells requires the HR genes XRCC3 and NBS1 (34, 35) whereas the t-circles observed in WRN patient cells (30) and the telomere shortening events observed in plants (32) do not. This suggests that there are at least 2 discrete mechanisms for t-circle formation, only one of which parallels yeast TRD. Clearly, it will be important to determine whether Ku-mediated t-circle formation events require XRCC3 and NBS1 activities.
Methods
Construction of a Ku86 Conditionally Null Human Cell Line.
General rAAV vector construction and methodology were carried out as described (20, 21). The scheme and diagnostic intermediates for gene targeting are shown in detail in Fig. 1 and Fig. S1, respectively. The sequences of all PCR primers are available on request. To generate Ku86 null cells, 5 × 104 Ku86flox/− cells per well of a 6-well plate were routinely plated and allowed to attach for 18 h. Infection was carried out by adding 2 mL of fresh medium containing 5 × 108 adenoviral particles to each well. After 5 days of incubation, the cells were replated into 10-cm plates and allowed to incubate for another 48 h before the cells were harvested for experiments.
Immunoblot Analyses.
Whole-cell extracts were prepared, and total protein was quantitated by using a Bradford assay. For most experiments, 50 μg of total protein was applied to an SDS/PAGE and processed for immunoblot analysis (21). Anti-Ku86 (sc-5280) and anti-TRF2 (sc-9143) antibodies were purchased from Santa Cruz Biotechnology, and anti-PARP-1 (556362) antibody was purchased from BD PharMingen. Nuclear and cytoplasmic extracts were prepared by using the CelLytic NuCLEAR Extraction Kit (Sigma–Aldrich).
Immune Fluorescence Staining.
The indicated cell lines were grown on 4-well chamber slides at a density of 9 × 103 per well. Cre infections were carried out 18 h after plating. The cells were then fixed at 96 or 120 h after infection with 4% paraformaldehyde for 30 min at room temperature. Cells were subsequently permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. The indicated antibodies were incubated with the cells at 37 °C overnight. Cy3-labeled anti-mouse or Alexa 488-labeled anti-rabbit antibodies were used against the relevant primary antibodies. DAPI was used to stain the nucleus. Images were captured with a Zeiss Axiovert 2 Upright Microscope.
Neutral-Neutral 2D Gel Electrophoresis.
Neutral-neutral 2D gel electrophoresis was performed as described previously (26, 31) with the following modifications: precipitated DNA was digested with AluI in the presence of 30 μg/mL RNaseA. This AluI-digested DNA (8 μg) was separated in 0.4% agarose gels in 1× TBE buffer at 1 V/cm for 18 h at room temperature. The second dimension was performed in a 1.2% agarose gel in 1× TBE [90 mM Tris (pH 8.2), 90 mM boric acid, 2.5 mM EDTA] buffer containing 0.3 μg/mL ethidium bromide at 5 V/cm for 8 h at 4 °C. The DNA was then transferred onto a nitrocellulose membrane and hybridized with [32P] 5′ end radiolabeled (T2AG3)3 oligonucleotides. Autoradiographic images were captured with a Fujifilm FLA-5000 Phosphoimager after a 12-h exposure.
Telomere FISH and G Overhang Assays.
T-FISH and G overhang assays were performed exactly as described in ref. 12.
Supplementary Material
Acknowledgments.
We thank Drs. Anja-Katrin Bielinsky (University of Minnesota, Minneapolis, MN), Sang Eun Lee (University of Texas, San Antonio, TX), Kyungjae Myung (National Institutes of Health, Bethesda, MD), and Carolyn Price (University of Cincinnati, Cincinnati, OH), and members of our laboratory for their helpful comments on this manuscript. We are deeply indebted to Dr. Bert Vogelstein (John Hopkins University, Baltimore, MD) and members of his laboratory for all their advice and reagents concerning the rAAV gene targeting technology. This work has been supported in part by National Institutes of Health Grants HL079559 and GM069576 (to E.A.H.). Y.W. was supported in part by National Institutes of Aging training Grant T32-AG029796.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12217.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903362106/DCSupplemental.
References
- 1.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech Ageing Dev. 2008;129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: Relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson EA, Huffman JL, Tainer JA. Structural aspects of Ku and the DNA-dependent protein kinase complex. In: Seide W, Kow YW, Doetsch P, editors. DNA Damage Recognition. New York: Taylor and Francis; 2006. pp. 629–684. [Google Scholar]
- 5.van der Burg M, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moshous D, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 7.Buck D, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Riballo E, et al. Identification of a defect in DNA ligase IV in a radiosensitive leukemia patient. Curr Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 9.Fattah FJ, Lichter NF, Fattah KR, Oh S, Hendrickson EA. Ku70, an essential gene, modulates the frequency of rAAV-mediated gene targeting in human somatic cells. Proc Natl Acad Sci USA. 2008;105:8703–8708. doi: 10.1073/pnas.0712060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fattah KR, Ruis BL, Hendrickson EA. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair. 2008;7:762–774. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myung K, et al. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol Cell Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 14.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravel S, Wellinger RJ. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol Cell Biol. 2002;22:2182–2193. doi: 10.1128/MCB.22.7.2182-2193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riha K, Shippen DE. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:611–615. doi: 10.1073/pnas.0236128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei C, Skopp R, Takata M, Takeda S, Price CM. Effects of double-strand break repair proteins on vertebrate telomere structure. Nucleic Acids Res. 2002;30:2862–2870. doi: 10.1093/nar/gkf396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espejel S, et al. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.d'Adda di Fagagna F, et al. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 20.Rago C, Vogelstein B, Bunz F. Genetic knockouts and knockins in human somatic cells. Nat Protoc. 2007;2:2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 21.Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey SM, et al. Strand-specific postreplicative processing of mammalian telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- 23.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 24.Chai W, et al. Human Ku70/80 associates physically with telomerase through interaction with hTERT. J Biol Chem. 2002;277:47242–47247. doi: 10.1074/jbc.M208542200. [DOI] [PubMed] [Google Scholar]
- 25.Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 26.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 28.Fisher TS, Zakian VA. Ku: A multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol Cell Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Watson JM, Shippen DE. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol Cell Biol. 2007;27:1706–1715. doi: 10.1128/MCB.02059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 35.Zhong ZH, et al. Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J Biol Chem. 2007;282:29314–29322. doi: 10.1074/jbc.M701413200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.