Abstract
Prolyl cis/trans isomerizations determine the rates of protein folding reactions and can serve as molecular switches and timers. In the gene-3-protein of filamentous phage, Pro-213 trans → cis isomerization in a hinge region controls the assembly of the 2 domains N1 and N2 and, in reverse, the activation of the phage for infection. We elucidated the structural and energetic basis of this proline-limited domain assembly at the level of individual residues by real-time 2D NMR. A local cluster of inter-domain hydrogen bonds, remote from Pro-213, is stabilized up to 3,000-fold by trans → cis isomerization. This network of hydrogen bonds mediates domain assembly and is connected with Pro-213 by rigid backbone segments. Thus, proline cis/trans switching is propagated in a specific and directional fashion to change the protein structure and stability at a distant position.
Keywords: gene-3-protein, molecular timer, proline isomerization, protein folding, real-time NMR
The cis/trans isomerizations of peptidyl-prolyl bonds in proteins are intrinsically slow processes. They determine the rates of protein folding reactions and are used as slow switches to regulate biological processes (1–8). In unstructured protein chains, the trans form is favored over cis, and therefore proteins with cis prolyl bonds in the native state must undergo trans → cis isomerization during their folding (9, 10). In this case, folding typically starts with a particular proline still in the incorrect (trans) state, but when a certain extent of folding is reached, this trans proline acts as a barrier and blocks further folding (11–13).
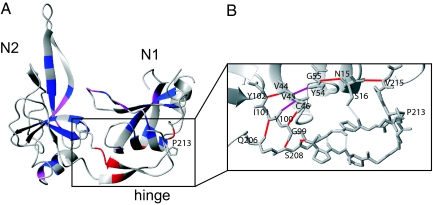
The coupling between conformational folding and prolyl isomerization has been studied for several small single-domain proteins (14–19). In the folding of the gene-3-protein (G3P) of phage fd, prolyl isomerization determines the rate of the final domain assembly step. G3P consists of 3 domains. The carboxy terminal (CT) domain anchors G3P in the phage coat (20), and the 2 amino terminal domains N1 and N2 form a functional entity that protrudes from the phage surface and mediates the infection of Escherichia coli cells (21). In the N1-N2 unit (Fig. 1A), N1 (residues 1–67) and N2 (residues 105–204) are linked by the hinge region, which forms numerous contacts with the N1 domain. The hinge region is formed by 2 noncontiguous chain regions (residues 89–104 between N1 and N2 and residues 205–220 after N2) and shows a well-ordered structure in the native protein.
Fig. 1.
Structure and folding mechanism of the gene-3-protein. (A) Crystal structure of N1-N2 G3P (21). The N1 domain is colored red, the N2 domain is blue, and the hinge region is green. The proline 213 side chain is indicated. (B) Refolding mechanism of G3P* (22). The domains are colored as in A. Time constants are given for refolding at 0.5 M GdmCl, 25 °C. The intermediate (trans P213) and the native (cis P213) state are labeled with I and N, respectively.
The time course of folding of G3P extends from milliseconds to hours (Fig. 1B). It starts with an extremely rapid folding reaction in the N1 domain with a time constant τ = 9.4 ms, followed by 2 folding reactions in N2, which show τ values of 7 and 42 s. In the final, very slow step (τ = 6,200 s), the 2 domains assemble in a reaction that involves further folding in the hinge region and is limited in rate by the trans → cis isomerization of Pro-213 (5, 22).
When G3P is fully folded, the phage is not infectious, because the binding site for its receptor is buried at the interface between the domains N1 and N2. The final, Pro-213-limited folding step must be reverted to unlock the domains and activate the phage for infection (5, 6, 23–25). This slow process is thus of dual importance. It represents the final rate-limiting step of folding of G3P as well as the transition between the partially folded, functionally active state and the fully folded but functionally inactive state. Therefore, G3P belongs to the increasing number of examples reported recently, where the isomeric state of a proline modulates activity, ligand binding, and physiological regulation (4, 8, 26–29).
Here we used real-time 2D NMR spectroscopy (30) and amide hydrogen exchange experiments (31, 32) to analyze the structure and the stability of the folding intermediate of G3P at the level of individual residues. In the final domain docking reaction a chain of interconnecting hydrogen bonds gets stabilized. This trace provides a molecular link between the isomeric state of remote Pro-213 and the stability of the inter-domain interface, which controls infectivity.
Results
We used a G3P* variant of the 2 N-terminal domains with 4 substitutions (T13I, T101I, Q129H, and D209Y), which is more stable than wild-type G3P and thus better suited for refolding studies (33). Phage carrying this stabilized version of G3P infect E. coli cells; the infectivity is, however, 10-fold reduced, because the locked, noninfectious form of G3P* is stabilized relative to the unlocked, infectious form (6). To study the folding mechanism of a protein by liquid state NMR spectroscopy at residue level, assignments of backbone amide resonance are required. G3P* consists of 226 residues and shows a well-dispersed 2D 15N-TROSY-HSQC spectrum. More than 90% of the amide cross peaks of G3P* could be assigned (see Fig. S1) by standard triple-resonance NMR experiments. These assignments together with the crystal structure of wild-type N1-N2 G3P (21) form the basis for the high resolution study of the structure and stability of the folding intermediate of G3P* with an incorrect trans Pro-213.
The Folding Intermediate of G3P* Shows Nonnative Interactions in the N1 Domain and in the Hinge Region.
The final folding reactions of the N1 and N2 domain (Fig. 1B) are complete within 5 min, but domain assembly is very slow (6, 34). The partially folded intermediate I, with an incorrect trans isomer at Pro-213 and incompletely assembled domains, therefore exists for an extended time (>8 h) during the refolding of G3P*. To identify those regions in the structure of I that are native-like and those that are not yet native, we used a kinetic 2D real-time NMR experiment (30, 31). In this experiment, refolding was started by a 4-fold dilution of the unfolded protein (in 4.0 M GdmCl) with 50 mM potassium phosphate buffer, pH 7.0, at 25 °C. After the completion of the folding reactions of the individual domains N1 and N2 (5 min), we measured a single 15N-TROSY-HSQC spectrum between 30 and 510 min (in 1.0 M GdmCl at 25 °C), a time window that covers the kinetics of the slow, proline-limited domain assembly reaction (I → N). This “kinetic spectrum” thus contains resonances from 2 populations of G3P*, the folding intermediate I, which decreases in concentration, and the folded protein N, which increases in concentration during the acquisition time of the kinetic spectrum (Fig. 2A). After the completion of refolding, a second NMR spectrum was recorded under identical conditions. It is referred to as the “reference spectrum” of the fully folded protein with a cis Pro-213 (Fig. 2B). Finally, the kinetic spectrum was subtracted from the reference spectrum to obtain the “difference spectrum” (Fig. 2C).
Fig. 2.
Structural properties of the folding intermediate with a trans Pro-213 of G3P* derived from real-time NMR spectroscopy. Sections of the kinetic (A), the reference (B), and the difference (C) 15N-TROSY-HSQC spectrum are plotted. Black contoured cross peaks present in C identify amide NH in a nonnative environment in I and are labeled red. Cross peaks that occur only in the reference, but not in the difference spectrum, identify amide NH in a native environment in I and are labeled blue in B. Red cross peaks in C result from the long-lived, but transiently accumulating, intermediate state I. (D) Distribution of amide NH with a native-like (blue) or a nonnative-like (red) chemical shift in the folding intermediate. The side chain of Pro-213 is indicated.
Two sets of signals (31, 35) were analyzed: The signals of set 1 originate from amide NH that are already in a native environment in the intermediate I. They show identical chemical shifts and identical intensities in I and N, and thus they vanish when the kinetic spectrum is subtracted from the reference spectrum. In the reference spectrum they show a positive peak. Gln-116 and Asn-182 provide examples for such native-like residues in I (Fig. 2).
The signals of set 2 originate from those amides that are in a non-native environment in the intermediate and become native during the rate-limiting folding reaction, which takes place during the acquisition of the kinetic spectrum. At native chemical shift positions, they show a reduced intensity in the kinetic spectrum, because only N, but not I, contributes to these signals. Therefore, a reduced, but positive signal is observed in the difference spectrum. Examples are provided by Thr-56 or Ser-208 (Fig. 2).
In Fig. 2D, the residues with native-like chemical shifts in the intermediate (set 1) are depicted in blue, those with a nonnative chemical shift (set 2) in red. A detailed listing of individual residues is given in Table S1. Almost all of the assigned residues of the hinge belong to set 2, evidently because the hinge is not native-like structured in the folding intermediate with the nonnative trans Pro-213. In the N1 domain, the residues that are close to the hinge (shown in red in Fig. 2D) also do not show native chemical shifts, whereas those distant from the hinge subdomain are in a native-like environment in the intermediate already (shown in blue in Fig. 2D). In the N2 domain, almost all residues are in a native conformation in the folding intermediate except Asn-138 and Asn-139. They form contacts with N1, which apparently cannot be fully established when Pro-213 in the hinge is still in the incorrect trans conformation. Together, the real-time NMR results suggest that the incorrect trans isomer of Pro-213 does not allow folding of the hinge to go to completion, and in turn, the hinge and the domains N1 and N2 cannot associate as in the fully folded state.
Folded G3P* Unfolds as a Cooperative Unit Under Native Conditions.
Chemical shift analyses such as those in Fig. 2 provide detailed, site-specific information about the structure but not the stability of the folding intermediate. Stability data can be derived from the protection of individual amide protons from exchange with the aqueous solvent (36). First, the fully folded form of G3P* was studied by amide hydrogen exchange (amide HX) measurements at pH 7.0, 25 °C. The protonated (NH) protein was transferred from 50 mM phosphate buffer in H2O to the same buffer in D2O, and the decrease in the intensities of the NH cross peaks were followed by a series of 15N-TROSY-HSQC spectra as a function of time for 7,000 min. We assumed that individual NH exchange by an EX2 mechanism (see Fig. S2) in a monoexponential reaction. Protection factors (P) were calculated by using the intrinsic exchange rates of corresponding amides in peptides as a reference (37). They are shown in Fig. 3A and in Table S1. Several NH of G3P* exchanged very slowly, and therefore, the NMR cross peak intensity decreased less than 5% during the HX experiment. For these NH, a lower limit for P of 2 × 106 is assumed, as indicated in Fig. 3A and 2 × 106 represents the highest protection factor of residues with well-defined exchange kinetics. It is equivalent to ΔGD ≥ 36 kJ/mol, which agrees with the ΔGD value of 37 kJ/mol, obtained from denaturant-induced unfolding transitions measured by fluorescence spectroscopy (33). No protection was observed for the glycine-rich unordered linker between N1 and the hinge, which extends from residue 68 to residue 88.
Fig. 3.
Amide NH protection factors of G3P* in various states. Protection factors are plotted as a function of the sequence position in the native state (A), in the folding intermediate (B), and in the isolated N1 domain (C). Domains are colored as in Fig. 1: N1 (red), N2 (blue), hinge region (green). Amide NH that showed no exchange in A were assigned to a value of 2 × 106, which is the highest protection factor that could be determined under the experimental conditions. They are marked by asterisks without error bars. All residues that could be analyzed are marked by a gray background. In A and C, these marked residues are all assigned nonprolyl residues. For the calculation of protection factors of the folding intermediate (B) only residues were analyzed that showed no exchange in the native state during the experiment. Black lines indicate error bars.
Protection factors of 106 or higher were observed for residues in all 3 structural units of G3P*: N1, N2, and the hinge region (Fig. 3A). This equally high protection suggests that unfolding of G3P* under native conditions is a cooperative reaction, in which domain disassembly and domain unfolding are coupled reactions.
For the isolated N1 domain (Fig. 3C), the most strongly protected amide NH show protection factors of about 2 × 105. This P is equivalent to an ΔGD value of 30 kJ/mol, in good agreement with the ΔGD value of 29.2 kJ/mol, as calculated from the GdmCl-induced unfolding transition of isolated N1 (33). The inter-domain interactions in G3P* mediated by the hinge thus increase the stability of the N1 domain at least 10-fold. A particularly strong stabilization is provided for amide NH around β strand 1 of N1 (residues 10–25), which contacts the hinge in folded G3P*.
HX experiments could not be performed with the isolated N2 domain, because it aggregates at high concentration under the exchange conditions (38). Urea-induced unfolding transitions measured at low protein concentrations gave an equilibrium constant of folding for isolated N2 of 180 at 15 °C. Therefore, isolated N2 is only marginally stable and at least 104-fold stabilized when it is assembled with the hinge and the N1 domain in fully folded G3P*.
A Competition Experiment Between Folding and Amide NH Exchange Reveals the Local Stability of the Folding Intermediate.
The conformational stabilities of the individual structural elements of G3P* in the folding intermediate were estimated from a competition experiment between refolding to the native state and amide HX (Scheme 1) (31, 32). In this experiment, unfolded G3P* (UH) was refolded for 5 min in H2O buffer to populate the protonated intermediate state (IH) with folded but unassembled domains and trans Pro-213 in the hinge. Next, the H2O-containing buffer was replaced by D2O buffer without GdmCl to initiate the competition between the folding of the intermediate to the native state and its amide HX (Scheme 1). After completion of the competition experiment (after 600 min), a 15N-TROSY-HSQC was recorded to determine the intensities of the remaining NH cross peaks (NH). In a reference experiment, native G3P* (NH) was equilibrated for the same time in the same buffer in D2O before measuring an identical 15N-TROSY-HSQC experiment (see Fig. S3). This reference spectrum provided the full intensity of the NH cross peaks in the native protein.
Scheme 1.
To facilitate the interpretation, the analysis was restricted to those 48 amide NH that are highly protected in the native protein and thus retained the full cross-peak intensity during the experiment (gray background in Fig. 3B). A decreased cross-peak intensity in the competition experiment relative to the reference experiment shows that partial amide HX must have occurred in the folding intermediate at a particular residue. Assuming that the amide HX from the folding intermediate follows an EX2 mechanism, the protection factor P of individual NH can be calculated with Eq. 1 (31). The rate of folding from the intermediate to the native state (kf = 1.6 × 10−4/s) is taken from our previous work (22), and the intrinsic exchange rates (kint) were taken from reference (37). S is the ratio of signal intensities as observed in the competition experiment (equivalent to NH in Scheme 1) and in the reference experiment (equivalent to NH + ND in Scheme 1).
The protection factors calculated for the folding intermediate by this procedure are shown in Fig. 3B and in Table S1. They are rather uniform for N1 and N2 and typically, in the range of ≥3 × 105, which is equivalent to a ΔGD value of ≈30 kJ/mol. In the C-terminal region of N1, the protection factors for the most highly protected NH (Fig. 3B) are similar in the folding intermediate and in the isolated N1 domain (Fig. 3C). In the N-terminal region, however, the protection factors are higher than for the isolated N1 domain, which indicates that this part is locally stabilized in the intermediate.
In the folding intermediate, the N2 domain is at least 103-fold stabilized compared with N2 in isolation (38) and only about 10-fold less stable than in the fully folded protein. This increased stability correlates well with the finding that N2 already shows native-like chemical shifts in the folding intermediate (Fig. 2). In the hinge subdomain, only 6 amide NH satisfied the criterion for our analysis, and they are not or only marginally protected in the folding intermediate (Fig. 3 A and B). For residues that cluster in the β-strands of N1 that point toward the hinge and in the β-strands of the hinge itself, up to 103-fold increases in the protection factors were observed (Fig. 4A).
Fig. 4.
Stability of hydrogen bonds in N and I. (A) Increase in amide protection during the domain docking reaction of G3P*. Blue residues have an equal protection in the native protein and in the folding intermediate or a <10-fold increase in protection. Magenta residues have a 10- to 100-fold increase in protection upon folding. Red residues have a >100-fold increase in protection against exchange during the domain docking reaction. (B) H-bonding network in the hinge and between the hinge and N1. All residues are shown in backbone representation; for Ser-16 and the Pro residues, the side chains are also shown. Residues that are involved in H-bonds and Pro-213 are labeled. H-bonds that gain protection in the I → N folding reaction are colored in red for >100-fold and in magenta for a 10- to 100-fold increase in protection. H-bonds that are not significantly protected in N are colored in gray.
Discussion
Trans Pro-213 Arrests N1 and the Hinge in Partially Folded Conformations and Maintains G3P in an Unlocked Form.
The N1 and N2 domains of G3P* start to fold individually, but their assembly is very slow, because it is limited in rate by the trans → cis isomerization of Pro-213 in the hinge between the domains (Fig. 1B). This isomerization retards folding more than 103-fold. When Pro-213 is cis, the 2 domains become tightly locked, which, in turn, retards unfolding, also ≈103-fold. Our real-time 2D NMR experiments in combination with amide HX revealed how Pro-213 trans → cis isomerization changes the structure of G3P* and the local stability during the transition from the unlocked folding intermediate, which is the infectious state, to the locked, fully folded state.
The trans isomer of Pro-213 in the unlocked folding intermediate affects the individual structural elements of G3P* differently. The N2 domain has virtually reached a conformation as in the fully folded protein already before the domain locking reaction (Fig. 2). However, in the hinge region and in parts of N1 facing the hinge, extended regions with non-native structure were detected. Folding of the hinge is thus coupled to Pro-213 trans → cis isomerization, and the N1 domain cannot associate with the hinge in a native-like fashion when Pro-213 is still trans. In folded G3P* (21), most inter-domain contacts are in fact localized between the hinge region and the N1 domain. Apparently, they are controlled by Pro-213 and cannot be established before Pro-213 trans → cis isomerization has occurred in the final step of folding. The loops Asp-24-Asp-28 in N1 and Phe-136-Asn-139 in N2 also change their structures during the slow domain locking reaction (Fig. 2D). These loops are remote from the hinge, but involved in the contacts between N1 and N2 in the folded protein. Evidently, the interdomain contacts in this remote area are also controlled by Pro-213 isomerization.
Pro-213 Isomerization Couples the N1 and N2 Domains to a Single Cooperative Unit.
The equilibrium unfolding of G3P* consists of 2 successive transitions (6, 22). In the first transition, the domains disassemble and N2 unfolds, but N1 remains native-like. It unfolds in the second transition, which occurs at higher temperature or denaturant concentration as confirmed by experiments with the isolated domains. N1 shows the same transition midpoints as part of G3P* and in isolation. N2, however, is much less stable in isolation (22, 33, 38).
The analysis of the amide HX experiments provided a residue-specific insight into the local stability of the fully folded protein with cis Pro-213. Highly protected amide NH with protection factors higher than 106 were found in all 3 parts of G3P* (Fig. 3A), suggesting that under native conditions N1, N2, and the hinge region form a cooperative unit. Apparently, domain assembly in the final folding reaction provides the major contribution to the stability of the entire protein and determines global unfolding under native conditions. This conclusion remains semiquantitative, because several NH in N1 and N2 were so strongly protected that they exchanged to a small extent only in our HX experiments. In wild-type G3P, the coupling between the domains in probably weaker, because two of the stabilizing substitutions in G3P* reside in the hinge that holds the domains together.
The N2 domain strongly benefits from the cooperative interactions in folded G3P*. Its equilibrium constant of folding is about 104-fold higher compared with the isolated N2 domain (38). In the folded protein, the chain ends of the N2 domain are kept together by the antiparallel β-sheet structure in the hinge, and the corresponding favorable change in entropy accounts most likely for a major part of the strong stabilization of N2. This entropic effect on the N2 domain is present to a large extent already when the hinge is only partially structured, as in the folding intermediate with a trans Pro-213.
Molecular Trace for the Pro-213-Controlled Domain Docking in the Final Folding Step.
Fig. 4A shows the locations of the 48 amide NH for which the protection factors could be determined in the intermediate state. Residues with similarly high protection factors in the intermediate and in the fully folded protein are colored blue. Here, the protection factor in I is already as high as in N or less than 10-fold smaller. These residues are mostly located in the N2 domain and in the part of N1 remote from the hinge. Their local stabilities are thus largely unaffected by the isomeric state of Pro-213, which agrees with the finding that most of them show native-like chemical shifts in the intermediate already (Fig. 2D)
Amide NH with a moderate (10- to 100-fold) increase in protection factor during the domain locking reaction are found in both N1 and N2 (magenta in Fig. 4A). Several of them are near the domain interface or in the core of N2, indicating that the already native-like folded structure of N2 is tightened during Pro-213-mediated domain assembly. Similar stabilizations are found in N1 near the tip and the base of the β hairpin that contacts the N2 domain in the folded protein. The amide NH that are more than 100-fold or even more than 103-fold stabilized in response to the Pro-213 trans → cis isomerization are all located in the hinge or in positions in the N1 domain that face the hinge.
Amide protons of a protein backbone are protected from exchange predominantly by H bonding with their acceptors, and increased protection factors indicate that the corresponding H bonds are strengthened. Fig. 4B provides an alternative representation of the NH protection results. It highlights directly those hydrogen bonds that become strongly stabilized when Pro-213 isomerizes from trans to cis. They form a chain of interactions that crosses the interface between N1 and the hinge twice. The chain initiates at Val-215 in the hinge and bridges over into the N1 domain at Ser-16. The protection of the backbone H bond between these 2 residues improves 650-fold upon Pro-213 trans → cis isomerization. H bonding then proceeds via Asn-15 to Gly-55 and on from Tyr-54 to Val-45. Its neighboring residues Cys-46 and Val-44 are engaged in backbone H bonds with Tyr-100 and Tyr-102, respectively, back in the hinge again. These 3 interdomain H bonds also become strongly stabilized in N. The chain of events is completed by the strong stabilization of 3 H bonds between the 2 β strands within the hinge. Remarkably, the Cys-46-Tyr-100 H bonds between N1 and the hinge, as well as the Gly-99-Ser-208 H bonds in the hinge, are stabilized more than 103-fold (Table S1). We propose that the strong stabilization of the cluster of hydrogen bonds between the residues 44–46 in N1 and the residues 99–102 and 206–208 of the hinge (Fig. 4B) provides the structural and energetic basis of the domain locking reaction that is triggered by the trans → cis switching at Pro-213.
The strongly stabilized backbone H bonds that are highlighted in Fig. 4B trace a molecular path that uses the spatial specificity of H bonds and differences in local stabilities to propagate the signal that is triggered by the Pro-213 switch. The cis/trans switching at Pro-213 is thus propagated over a Cα–Cα distance between 13.7 Å (to Gly-99) and 21.3 Å (to Gln-206). At present, we do not know how the Pro-213 isomerization signal itself is connected with the anchor points of the chain of H bonds at Val 215 and at Gly-99/Ser-208, because our technique probes the stabilization of backbone H bonds. It has, however, not escaped our attention that the chain regions between Pro-213 and the anchor points of the H bonded clamp are strongly enriched in prolines and in β-branched residues. These residues increase the chain rigidity and thus helps in propagating the Pro-213 isomerization signal to either side of the hinge.
During phage infection, the back reaction, Pro-213 cis → trans isomerization, is used to expose the binding site for its receptor TolA. This activation probably uses a similar mechanism. It reverses the last step of folding and thus weakens the aforementioned cluster of hydrogen bonds between N1 and the hinge region to unlock the domains of G3P. However, the present NMR folding studies had to be performed with a stabilized variant of G3P that contains 2 substitutions in the hinge, and therefore conclusions regarding the molecular mechanism of page infection remain tentative. Still, we now begin to understand how changes in the isomeric state at a proline residue can be propagated in a specific and directional fashion to other regions during folding or in a folded protein.
Materials and Methods
Expression and Purification.
G3P* was expressed and purified as described previously (22), using M9 minimal medium containing 15N-NH4Cl or 15N-NH4Cl, 13C-glucose, and D2O.
NMR Experiments.
All NMR spectra were recorded in 50 mM sodium phosphate pH 7.0 (or pD 6.6) at 298 K, containing 10% or 100% D2O if not stated differently. Spectra were processed using NMRpipe (39) and analyzed using NMRview (40). For the backbone resonance assignments, trHNCA, trHNCACB, and trHN(CO)CACB spectra of a 15N/13C/2H sample were acquired with a Bruker Avance 700 spectrometer equipped with a cryoprobe. The published backbone assignments of the N1-domain (25) were confirmed by 15N-NOESY-HSQC and 15N-TOCSY-HSQC spectra with a Bruker Avance 600 spectrometer.
Amide HX of native G3P* was followed by measuring 58 15N-TROSY-HSQC spectra over 5 days with a Bruker Avance 800 spectrometer equipped with a cryoprobe. Amide HX of native N1 was followed by measuring 100 15N-HSQC spectra within 3 days with a Bruker Avance 600 spectrometer. Both reactions were started by dissolving lyophilized protein in D2O buffer. Error bars in Fig. 3 A and C result from the fit of a single exponential function to the NMR intensities. The EX2 exchange regime was verified for G3P* by repeating the experiment at pH 8.0 (see Fig. S2).
For real-time NMR experiments, refolding of 15N G3P* was started by a 4-fold dilution of the unfolded protein (in 4.0 M GdmCl, 50 mM sodium phosphate, pH 7.0) to a final GdmCl concentration of 1.0 M in the same buffer. After a dead time of 30 min, 2 identical 15N-TROSY-HSQC experiments were acquired by a Bruker Avance 800 spectrometer equipped with a cryoprobe. Both spectra were measured for 8 h, the first (kinetic) spectrum during the folding reaction, and the second (reference) spectrum directly after the completion of refolding. Then the kinetic spectrum was subtracted from the reference spectrum to obtain the difference spectrum (31). The entire experiment was reproducibly repeated once.
For the competition experiment between refolding and amide HX, refolding of 15N G3P* was started by a 4-fold dilution of the unfolded protein (in 4.0 M GdmCl, 50 mM sodium phosphate, pH 7.0) to a final GdmCl concentration of 1.0 M in the same buffer. All buffers contained 100% H2O. After 5 min, when the refolding reactions of the individual domains were complete, the solvent was exchanged to a D2O-containing buffer without GdmCl using a NAP10 gel filtration column (GE Healthcare). Amide HX was allowed to proceed for 10 h at 298 K and then an 8-h 15N-TROSY-HSQC spectrum was measured with a Bruker Avance 900 spectrometer. For referencing, an identical amide HX experiment was performed starting with the same amount of folded G3P* in 1 M GdmCl. After the amide HX experiments, the protein concentrations in the NMR tubes were determined by absorption and used for the comparison of the NMR intensities observed in the 2 spectra after the exchange experiments. The calculation of the protection factors for the intermediate I according to Eq. 1 are based on end point analyses. With the current signal-to-noise ratio of the spectra, S, values between 0.99 and 0.03 can be used for a quantitative analysis. The lower limit is additionally determined by the presence of residual protons in the solvent (≈1%). Error bars for Fig. 3B were calculated for ln(P) by error propagation, because only differences in orders of magnitude are discussed.
Supplementary Material
Acknowledgments.
We thank Paul Rösch for NMR spectrometer time at 700 and 800 MHz, Hartmut Oschkinat and the FMP Berlin for NMR spectrometer time at 900 MHz, and Christian Löw for very helpful discussions. This research was supported by grants from the Deutsche Forschungsgemeinschaft (BA 1821/5-2 and SCHM 444/17-2) and the European Regional Development Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902102106/DCSupplemental.
References
- 1.Yaffe MB, et al. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 2.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat Struct Biol. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti AH. Native state proline isomerization: An intrinsic molecular switch. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 4.Fischer G, Aumüller T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev Physiol Biochem Pharmacol. 2003;148:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- 5.Eckert B, Martin A, Balbach J, Schmid FX. Prolyl isomerization as a molecular timer in phage infection. Nat Struct Mol Biol. 2005;12:619–623. doi: 10.1038/nsmb946. [DOI] [PubMed] [Google Scholar]
- 6.Eckert B, Schmid FX. A conformational unfolding reaction activates phage fd for the infection of Escherichia coli. J Mol Biol. 2007;373:452–461. doi: 10.1016/j.jmb.2007.07.060. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 9.Brandts JF, Halvorson HR, Brennan M. Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975;14:4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- 10.Schmid FX, Baldwin RL. Acid catalysis of the formation of the slow-folding species of RNase A: Evidence that the reaction is proline isomerization. Proc Natl Acad Sci USA. 1978;75:4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook KH, Schmid FX, Baldwin RL. Role of proline isomerization in folding of ribonuclease A at low temperatures. Proc Natl Acad Sci USA. 1979;76:6157–6161. doi: 10.1073/pnas.76.12.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid FX. Proline isomerization in unfolded ribonuclease A. The equilibrium between fast-folding and slow-folding species is independent of temperature. Eur J Biochem. 1982;128:77–80. [PubMed] [Google Scholar]
- 13.Balbach J, Schmid FX. In: Mechanisms of Protein Folding. Pain RH, editor. Oxford: University Press; 2000. pp. 212–237. [Google Scholar]
- 14.Goto Y, Hamaguchi K. Unfolding and refolding of the constant fragment of the immunoglobulin light chain. J Mol Biol. 1982;156:891–910. doi: 10.1016/0022-2836(82)90146-2. [DOI] [PubMed] [Google Scholar]
- 15.Nall BT. Proline isomerization and protein folding. Comments. Mol Cell Biophys. 1985;3:123–143. [Google Scholar]
- 16.Schreiber G, Fersht AR. The refolding of cis- and trans-peptidylprolyl isomers of Barstar. Biochemistry. 1993;32:11195–11203. doi: 10.1021/bi00092a032. [DOI] [PubMed] [Google Scholar]
- 17.Lilie H, Rudolph R, Buchner J. Association of antibody chains at different stages of folding: Prolyl isomerization occurs after formation of quaternary structure. J Mol Biol. 1995;248:190–201. doi: 10.1006/jmbi.1995.0211. [DOI] [PubMed] [Google Scholar]
- 18.Pappenberger G, et al. Kinetic mechanism and catalysis of a native-state prolyl isomerization reaction. J Mol Biol. 2003;326:235–246. doi: 10.1016/s0022-2836(02)01373-6. [DOI] [PubMed] [Google Scholar]
- 19.Schmid FX. In: Handbook of Protein Folding. Kiefhaber T, Buchner J, editors. Weinheim: Wiley-VCH; 2005. pp. 916–945. [Google Scholar]
- 20.Boeke JD, Model P. A prokaryotic membrane anchor sequence: Carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc Natl Acad Sci USA. 1982;79:5200–5204. doi: 10.1073/pnas.79.17.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holliger P, Riechmann L, Williams RL. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 A: Evidence for conformational lability. J Mol Biol. 1999;288:649–657. doi: 10.1006/jmbi.1999.2720. [DOI] [PubMed] [Google Scholar]
- 22.Martin A, Schmid FX. The folding mechanism of a two-domain protein: Folding kinetics and domain docking of the gene-3 protein of phage fd. J Mol Biol. 2003;329:599–610. doi: 10.1016/s0022-2836(03)00433-9. [DOI] [PubMed] [Google Scholar]
- 23.Levengood SK, Beyer WF, Jr, Webster RE. TolA: A membrane protein involved in colicin uptake contains an extended helical region. Proc Natl Acad Sci USA. 1991;88:5939–5943. doi: 10.1073/pnas.88.14.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Click EM, Webster RE. Filamentous phage infection: Required interactions with the TolA protein. J Bacteriol. 1997;179:6464–6471. doi: 10.1128/jb.179.20.6464-6471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holliger P, Riechmann L. A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure. 1997;5:265–275. doi: 10.1016/s0969-2126(97)00184-6. [DOI] [PubMed] [Google Scholar]
- 26.Pletneva EV, Sundd M, Fulton DB, Andreotti AH. Molecular details of Itk activation by prolyl isomerization and phospholigand binding: The NMR structure of the Itk SH2 domain bound to a phosphopeptide. J Mol Biol. 2006;357:550–561. doi: 10.1016/j.jmb.2005.12.073. [DOI] [PubMed] [Google Scholar]
- 27.OuYang B, Pochapsky SS, Dang M, Pochapsky TC. A functional proline switch in cytochrome P450cam. Structure. 2008;16:916–923. doi: 10.1016/j.str.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min L, Fulton DB, Andreotti AH. A case study of proline isomerization in cell signaling. Front Biosci. 2005;10:385–397. doi: 10.2741/1536. [DOI] [PubMed] [Google Scholar]
- 29.Aumüller T, Fischer G. Bioactivity of folding intermediates studied by the recovery of enzymatic activity during refolding. J Mol Biol. 2008;376:1478–1492. doi: 10.1016/j.jmb.2007.12.057. [DOI] [PubMed] [Google Scholar]
- 30.Balbach J, et al. Protein folding monitored at individual residues during a two-dimensional NMR experiment. Science. 1996;274:1161–1163. doi: 10.1126/science.274.5290.1161. [DOI] [PubMed] [Google Scholar]
- 31.Steegborn C, Schneider-Hassloff H, Zeeb M, Balbach J. Cooperativity of a protein folding reaction probed at multiple chain positions by real-time 2D NMR spectroscopy. Biochemistry. 2000;39:7910–7919. doi: 10.1021/bi000270u. [DOI] [PubMed] [Google Scholar]
- 32.Koide S, Dyson HJ, Wright PE. Characterization of a folding intermediate of apoplastocyanin trapped by proline isomerization. Biochemistry. 1993;32:12299–12310. doi: 10.1021/bi00097a005. [DOI] [PubMed] [Google Scholar]
- 33.Martin A, Schmid FX. Evolutionary stabilization of the gene-3-protein of phage fd reveals the principles that govern the thermodynamic stability of two-domain proteins. J Mol Biol. 2003;328:863–875. doi: 10.1016/s0022-2836(03)00359-0. [DOI] [PubMed] [Google Scholar]
- 34.Martin A, Schmid FX. A proline switch controls folding and domain interactions in the gene-3-protein of the filamentous phage fd. J Mol Biol. 2003;331:1131–1140. doi: 10.1016/s0022-2836(03)00864-7. [DOI] [PubMed] [Google Scholar]
- 35.Balbach J, Steegborn C, Schindler T, Schmid FX. A protein folding intermediate of ribonuclease T1 characterized at high resolution by 1D and 2D real-time NMR spectroscopy. J Mol Biol. 1999;285:829–842. doi: 10.1006/jmbi.1998.2364. [DOI] [PubMed] [Google Scholar]
- 36.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakob RP, Schmid FX. Energetic coupling between native-state prolyl isomerization and conformational protein folding. J Mol Biol. 2008;377:1560–1575. doi: 10.1016/j.jmb.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.