Telomeres, the specialized nucleoprotein structures present at the ends of linear chromosomes, function to prevent natural chromosomal termini from activating the DNA damage response and becoming substrates for inappropriate DNA repair. Telomeres are organized into lariat-like structures known as t-loops, which are formed by the invasion of the terminal G-rich 3′ telomeric overhang into the proximal duplex telomeric tract on the same chromosome (1) (Fig. 1). T-loops have been proposed to be a crucial means by which the telomeric end is hidden from DNA double strand break (DSB) repair pathways, with nonhomologous end joining (NHEJ) being the dominant mechanism of DSB repair in mammalian cells. Consequently, when telomeres become dysfunctional, and presumably no longer able to form t-loops, they can be engaged in NHEJ, giving rise to chromosome end-to-end fusions (2). Paradoxically, the Ku70/Ku86 heterodimer, a central component of NHEJ, is important at functional telomeres. There, instead of mediating NHEJ, Ku has been shown to contribute to various aspects of telomere structure and function. For example, Ku has been found to protect telomeres from inappropriate degradation and interchromosomal recombination and contribute to the tethering of telomeres to the nuclear periphery and the regulation of telomerase (3). Although Ku has a conserved role in NHEJ and appears to have at least one or more roles at the telomere across species, only in humans has it been shown to be an essential protein (4). The reason has been unknown, but it has been speculated that Ku's essential function lies in its telomeric, not NHEJ, role. In this issue of PNAS, Wang et al. (5) have not only confirmed this speculation but have revealed that Ku's essential role in human cells is to prevent dramatic telomere loss.
Fig. 1.
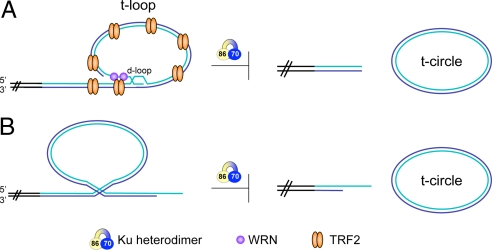
Possible scenarios by which Ku prevents telomere loss and t-circle formation. (A) Ku inhibits t-loop excision. This might occur (i) directly, independent of other proteins, via regulation of both TRF2 and WRN, or via recruitment of a protein/factor that represses recombination or (ii) indirectly, through free Ku sequestering away from telomeres a protein that promotes t-loop excision. (B) Ku prevents intramolecular recombination between telomeric repeats, independent of t-loops.
In their article, Wang et al. (5) demonstrated that the cell death that resulted from the conditional knockout of Ku86 in a human somatic cell line was associated with massive telomere loss. Telomere FISH showed that, on average, a striking 66% of chromosome ends were devoid of telomere signal after depletion of Ku. These so-called signal-free ends were associated with the appearance of extrachromosomal telomere circles (t-circles), which suggests that the telomere loss was the result of telomere rapid deletion, a previously described phenomenon that results in telomere shortening caused by intrachromosomal recombination (6). Additionally, ≈50% of Ku-deficient cells accumulated γ-H2AX foci, indicative of DNA ends being recognized as DSBs. This number is much greater than what was observed in a cell line that lacked DNA-PKcs, a protein that associates with DNA-bound Ku and is crucial for NHEJ-mediated repair. Moreover, the DNA-PKcs−/− cell line was viable and did not exhibit massive telomere loss (7). The results taken together, therefore, strongly suggest that the essential function of Ku86 in human cells is not to perform NHEJ, but rather to prevent the elimination of telomeres and the formation of t-circles.
A fair amount is known about t-circles already, because they have been observed in a number of settings. In many cases, t-circles are thought to arise from recombinational excision of t-loops (8). This mechanism makes sense when one considers that the base of the t-loop, known as a displacement loop (d-loop), resembles a Holliday junction (HJ) intermediate (Fig. 1). In contrast to what occurs in homologous recombination, branch migration and HJ resolution are suppressed at telomeric d-loops because these activities would result in the excision of a t-loop and a shortened telomere. The N-terminal basic GAR domain of TRF2, a component of the telomere maintenance complex shelterin, appears to play a major role in this suppression. This domain is required for TRF2 inhibition of resolvase cleavage of telomeric HJs in vitro and both mouse and human cells expressing high levels of TRF2ΔB, which lacks the domain, exhibit high levels of t-circles (9, 10). Additional factors implicated in t-circle formation in response to TRF2ΔB expression include XRCC3, which is associated with HJ resolvase activity in mammalian cells; Nbs1, a component of the Mre11 complex, which is involved in recombinational repair; and WRN, an helicase/exonuclease, which interacts directly with TRF2 (9, 11).
Of interest now is determining the mechanism by which Ku functions to prevent telomere loss and t-circle formation, and there are suggestions it may prove to be novel. Wang et al. (5) discuss that Ku has been shown to interact with both TRF2 (12) and WRN (13), supporting the notion that it might mediate its effects through one of these proteins. However, TRF2 deficiency or WRN dysfunction results in preferential loss of leading- or lagging-strand telomeres, respectively. In contrast, Wang et al. found that the telomere loss exhibited in Ku86-deficient cells was random. This result suggests that Ku prevents t-circle formation via either a combined disturbance in TRF2 and WRN function or a distinct mechanism like inhibiting directly or recruiting a repressor of recombination (Fig. 1A).
That Ku might function through a distinct pathway is also supported by findings in Arabidopsis, in which Ku was first implicated in the suppression of t-circle formation (14). T-circles arise in Ku-deficient plants; however, neither Mre11 nor any of the Rad51 paralogs, including XRCC3, are required for their formation. This finding raised the possibility that the factors that mediate t-circle formation in Ku-deficient plants may be distinct from those that arise as a result of TRF2ΔB expression. Whether this distinction will hold true for Ku-deficient human cells remains to be determined, but these findings highlight the notion that not all t-circles are created equally.
T-circles have also been observed in the setting of telomere hyperelongation, which has been shown to occur when telomerase activity is up-regulated in telomerase-positive human cell lines (15) or when telomeres are maintained by a telomerase-independent, recombination-based mechanism referred to as alternative lengthening of telomeres (ALT) (9, 16). An important difference between Ku in Arabidopsis and humans is that in plants Ku negatively regulates telomerase, such that telomeres are much longer in Ku-deficient cells compared with wild-type plant cells. Thus, telomere elongation may contribute to t-circle formation in Ku-deficient plants, but not Ku-deficient human cells where telomeres are significantly shorter (17, 18), again pointing to a potentially novel mechanism.
Although it is often envisioned that t-circles arise from excision of t-loops, it is possible that they arise from other forms of intrachromosomal recombination (Fig. 1B). In mouse cells, Ku itself has been shown to be important in the suppression of interchromosomal recombination between sister telomeres in a pathway that functions in parallel with TRF2 (19). Thus, in human cells, it is possible that its effects on suppressing intrachromosomal recombination are not localized to the base of the t-loop, but rather are distributed along the entire tract of telomeric chromatin.
A piece to the puzzle of how Ku suppresses telomere loss and promotes t-circle formation will likely come when it is determined exactly how Ku associates with the telomere. Ku has a high affinity for DNA ends independent of DNA sequence and could therefore load directly onto the telomeric end. This mode of association would require that the t-loop structure is disassembled at some point in the cell cycle, which is often suggested to occur during S phase, and that the telomere end is naked and available for Ku loading. Alternatively, Ku could associate with telomeres indirectly via protein–protein interactions, e.g., with TRF2, WRN, or other telomere-associated factors. Stepping back, it remains to be determined whether Ku's role in suppressing telomere loss in human cells is even direct as opposed to being mediated by the dysregulation of a Ku-interacting factor that is released by the absence of Ku.
Ku's essential role in human cells is to prevent dramatic telomere loss.
Regardless of the mechanism, what makes the work by Wang et al. (5) particularly important is that it will bring Ku more center stage in the study of human telomeres. For many years studies on mammalian telomere end protection have focused on the shelterin complex, which is a key player at mouse and human telomeres (8). Ku, however, has remained in the background, often left out of telomere models, most likely because a Ku deficiency alone in mouse cells does not lead to profound alterations in telomere length or structure or telomere dysfunction-induced foci (19). Instead, the field has focused on the potent role Ku has in mediating the fusion of dysfunctional telomeres. This work should intensify the interest in human Ku's contribution to normal telomere function.
Last, the finding of human Ku's essential telomeric function by Wang et al. (5) underscores the point that as much as there are common themes in how organisms maintain telomere structure and function there can also be significant differences even between two closely related organisms such as humans and mice. Prior analyses of Ku have indicated that its involvement at the telomere varies greatly among species (3). Why Ku is required for telomere maintenance in human but not mouse cells is unknown. It may relate to differences in their telomere lengths or how Ku interacts with the homologous recombination machinery or telomere-associated proteins. Determining the basis for human Ku's essential telomeric function will no doubt contribute to our general understanding of how telomeres differ between the two species.
Acknowledgments.
This work is supported by National Institutes of Health Research Grant 1R01GM077509-01A2.
Footnotes
The authors declare no conflict of interest.
See companion article on page 12430.
References
- 1.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 3.Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106:12430–12435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lustig AJ. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat Rev. 2003;4:916–923. doi: 10.1038/nrg1207. [DOI] [PubMed] [Google Scholar]
- 7.Ruis BL, Fattah KR, Hendrickson EA. The catalytic subunit of DNA-dependent protein kinase regulates proliferation, telomere length, and genomic stability in human somatic cells. Mol Cell Biol. 2008;28:6182–6195. doi: 10.1128/MCB.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 9.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Poulet A, et al. TRF2 promotes, remodels, and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2ΔB-mediated telomere shortening. Mol Cell Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 13.Li B, Comai L. Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J Biol Chem. 2000;275:28349–28352. doi: 10.1074/jbc.C000289200. [DOI] [PubMed] [Google Scholar]
- 14.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007;27:163–169. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 15.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009;28:799–809. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myung K, et al. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol Cell Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 19.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]