Abstract
Rationale: Phenotypic and genotypic heterogeneity of lung cancer likely precludes the identification of a single predictive marker and suggests the importance of identifying and measuring multiple markers.
Objectives: We describe the use of a fluorescent protein microarray to identify and measure multiple non–small cell lung cancer–associated antibodies and show how simultaneous measurements can be combined into a single diagnostic assay.
Methods: T7 phage cDNA libraries of non–small cell lung cancer were first biopanned with plasma samples from normal subjects and patients with non–small cell lung cancer to enrich the component of tumor-associated proteins, and then applied to microarray slides. Two hundred twelve immunogenic phage-expressed proteins were identified from roughly 4,000 clones, using high-throughput screening with patient plasmas and assayed with 40 cancer and 41 normal plasma samples. Twenty patient and 21 normal plasma samples were randomly chosen and used for statistical determination of the predictive value of each putative marker. Statistical analysis identified antibody reactivity to seven unique phage-expressed proteins that were significantly different (p < 0.01) between patient and normal groups. The remaining 20 patient and 20 normal plasma samples were used as an independent test of the predictive ability of the selected markers.
Main Results: Measurements of the 5 most predictive phage proteins were combined in a logistic regression model that achieved 90% sensitivity and 95% specificity in prediction of patient samples, whereas leave-one-out statistical analysis achieved 88.9% diagnostic accuracy among all 81 samples.
Conclusion: Our data indicate that antibody profiling is a promising approach that could achieve high diagnostic accuracy for non–small cell lung cancer.
Keywords: biomarkers, fluorescent array, phage display, tumor- associated autoantibodies
Circulating tumor markers could impact non–small cell lung cancer (NSCLC) patient outcomes through improved screening, diagnosis, staging, and management (1–3). The search for biomarkers for the early detection and diagnosis of NSCLC has met with little success. Much emphasis has been placed on the discovery and characterization of unique tumor markers (4–11). Yet, no marker has been identified that has adequate sensitivity or specificity to be clinically useful, although a combination of multiple markers has been shown to increase diagnostic accuracy. Tumor-associated antibodies may expand the number of available markers (10–16). Our previous work clearly shows that some proteins from T7 phage display NSCLC libraries are recognized by antibodies in cancer patient plasma but not in normal plasma (17). Although additional investigation may identify a single highly predictive marker of NSCLC, heterogeneity of this disease will likely require a panel of markers to achieve the sensitivity and specificity required for clinical application.
In the context of developing a diagnostic assay for NSCLC, we have adapted fluorescent microarray technology to the task of identifying immunogenic phage-expressed proteins and assessing the presence of their corresponding antibodies. Robotic microarray spotters that allow grouping of thousands of proteins, in replicate, onto a single glass slide make this feasible, efficient, and reproducible. Automated spotting of the array also allows production of hundreds of identical chips, thus making this technology a logical tool for this application. To develop an assay for detecting NSCLC, we employed a high-throughput method of isolating immunogenic phage-expressed proteins from T7 phage NSCLC tumor libraries, using antibodies in NSCLC patient plasmas. We then combined multiple phage-expressed proteins onto a single protein “diagnostic chip” and evaluated our ability to predict disease.
METHODS
Human Subjects
After informed consent was obtained, 50 plasma samples (10 used for selection and 40 for analysis) were obtained from individuals with histologically confirmed NSCLC (Table 1). Normal control subjects included 41 individuals with a minimum smoking history of 20 pack-years, ages 50–75 years (considered “high-risk”), and 5 nonsmoking control subjects used for selection and analysis.
TABLE 1.
CHARACTERISTICS OF PATIENT SAMPLES*
| Sample | Age (yr) | Sex | Smoking Status | Tumor Type | Tumor Stage |
|---|---|---|---|---|---|
| Selection (n = 10) | 55–77 (average, 63) | Male (n = 7); female (n = 3) | Active (n = 6) | Adeno (n = 4) | I/II (n = 2) |
| Former (n = 4) | SCC (n = 3) | III (n = 4) | |||
| Never (n = 0) | NSCLC† (n = 3) | IV (n = 4) | |||
| Training (n = 20) | 50–79 (average, 64) | Male (n = 13); female (n = 7) | Active (n = 12) | Adeno (n = 7) | I/II (n = 6) |
| Former (n = 8) | SCC (n = 7) | III (n = 9) | |||
| Never (n = 0) | NSCLC† (n = 6) | IV (n = 5) | |||
| Testing (n = 20) | 50–88 (average, 71) | Male (n = 16); female (n = 4) | Active (n = 10) | Adeno (n = 8) | I/II (n = 5) |
| Former (n = 10) | SCC (n = 7) | III (n = 10) | |||
| Never (n = 0) | NSCLC† (n = 5) | IV (n = 5) |
Definition of abbreviations: Adeno = adenocarcinoma; NSCLC = non–small cell lung cancer; SCC = squamous cell carcinoma.
Samples in analysis were divided randomly into training and testing groups. Case subjects were not stringently matched with control subjects for age (average age for patients with cancer, 68 years; average age of control subjects, 55 years) sex (patients with cancer, 29 males and 11 females; control subjects, 19 males and 22 females), or smoking history (active/former/never: patients with cancer, 22/18/0; control subjects, 20/16/5).
Undifferentiated NSCLC or inadequate tissue to characterize histology.
Phage Libraries and Biopanning
One T7 phage NSCLC cDNA library was purchased (Novagen, Madison, WI) and a second was constructed from the adenocarcinoma cell line NCI-1650, using Novagen OrientExpress cDNA synthesis and cloning systems (17). The libraries were biopanned with pooled plasma from five patients with NSCLC (stages 2–4; histology “NSCLC”) and normal healthy donors, to enrich the population of phage-expressed proteins recognized by tumor-associated antibodies as previously described (17). Briefly, the phage-displayed library was affinity selected by incubation with protein G–agarose beads coated with antibodies from pooled normal sera (250 μl of pooled normal sera, diluted 1:20, at 4°C overnight) to remove non–tumor-specific proteins. Unbound phages were separated from phages bound to antibodies in normal plasma by centrifugation. The supernatant was then biopanned against protein G–agarose beads coated with pooled patient plasma (4°C overnight) and separated from unbound phages by centrifugation. The bound/reactive phages were eluted with 1% sodium dodecyl sulfate and centrifugation. The phages were amplified in Escherichia coli BLT5615 (GIBCO-BRL, Grand Island, NY) in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside and carbenicillin (50 μg/ml) until lysis. Amplified phage-containing lysates were collected and subjected to three additional sequential rounds of biopan enrichment. Phage-containing lysates from the fourth biopan were amplified, and individual phage clones were isolated and then incorporated into protein arrays as described below.
Array Construction and High-throughput Screening
Phage lysates from the fourth round of biopanning were amplified and grown on LB–agar plates covered with 6% agarose for isolating individual phage. A colony-picking robot (QPixII; Genetix, New Milton, UK) was used to pick 4,000 individual colonies (2,000 per library). The picked phages were reamplified in 96-well plates and then 5-nl samples of clear lysate from each well were robotically spotted in duplicate on FAST slides (Schleicher & Schuell BioScience, Keene, NH), using an Affymetrix 417 Arrayer (Affymetrix, Santa Clara, CA).
Five individual NSCLC patient plasma samples not used in the biopan were used to identify immunogenic phage-displayed proteins from the screening slides. Rabbit anti-T7 primary antibody (Jackson ImmunoResearch, West Grove, PA) was used to detect T7 capsid proteins as a control for phage amount. Both preabsorbed plasma (plasma:bacterial lysate, 1:30) samples and anti-T7 antibodies were diluted 1:3,000 with 1× Tris-buffered saline (TBS) plus 0.1% Tween 20 (TBST) and incubated with the screening slides for 1 hour at room temperature. Slides were washed and then probed with Cy5-labeled anti-human and Cy3-labeled anti-rabbit secondary antibodies (Jackson ImmunoResearch; each antibody diluted 1:4,000 in 1× TBST) together for 1 h at room temperature. Slides were washed again and then scanned with an Affymetrix 428 scanner. Images were analyzed with GenePix 5.0 software (Axon Instruments/Molecular Devices, Union City, CA). Phages bearing a Cy5:Cy3 signal ratio greater than 2 standard deviations from a linear regression were selected as candidates for use on a “diagnostic chip.”
Diagnostic Chip Design and Antibody Measurement
Two hundred and twelve immunoreactive phages identified by high-throughput screening (described above), plus 120 “empty” T7 phages, were combined, reamplified, and spotted in duplicate onto FAST slides as single diagnostic chips. Replicate chips were used to assay 40 NSCLC plasma samples, according to the protocol described above for screening. The median Cy5 signal was normalized to the median Cy3 signal (Cy5:Cy3 signal ratio) as the measurement of human antibody against a unique phage-expressed protein. To compensate for chip-to-chip variability, measurements were further normalized by subtracting background reactivity of plasma against empty T7 phage proteins and dividing by the median of the T7 signal [(CY5:Cy3 of phage – Cy5:Cy3 of T7)/Cy5:Cy3 of T7].
Statistical Analysis
Normalized Cy5:Cy3 for each of the 212 phage-expressed proteins was independently analyzed for statistically significant differences between the patient group and normal group by t test, using JMP statistical software (SAS, Inc., Cary, NC). Candidate phage markers were chosen if p < 0.01 and checked for redundancy by polymerase chain reaction amplification using commercially available T7 phage vector primers and protocol (Novagen) as previously described (17). Redundant clones were eliminated.
Using a panel of nonredundant phage-expressed proteins, logistic regression analysis was performed to predict the probability that a sample was from a patient with NSCLC. All 81 samples were divided into two groups. The first group, consisting of 21 normal and 20 patient plasma samples, randomly chosen, was used as a training set to build up classifiers that were able to distinguish patient from normal samples on the basis of an individual marker or a combination of markers. The second group, consisting of 20 patient and 20 normal samples, was used to validate the prediction rate of classifiers derived with the training group. Receiver operating characteristic curves were generated to compare the predictive sensitivity and specificity with different markers, and the area under the curve. The classifiers were further examined by leave-one-out cross-validation. Smoking and stage of disease were also analyzed and compared.
Sequence Identification of Phage-expressed Proteins
When possible, phage identity was made on the basis of significant nucleotide and translated nucleotide matches (bit score, e value, and percent sequence match) with a single gene in the GenBank database, using BLASTN and BLASTX search engines.
RESULTS
High-throughput Screening
To assess the efficiency of high-throughput fluorescent array screening of phage-expressed proteins, using antibodies in plasma from patients with NSCLC, two T7 phage NSCLC libraries were biopanned with NSCLC patient and normal plasma to enrich the population of immunogenic clones, using an approach similar to that described in previous work (17). Four thousand phages from the fourth biopan were then spotted on membrane-coated array slides and screened with five individual NSCLC patient plasma samples not used in the biopan to identify immunogenic phages. Linear regression of the Cy5:Cy3 signal revealed 212 individual phages with signal ratios greater than 2 standard deviations from the average, which were chosen as candidates for a “diagnostic chip.” An example of linear Cy5:Cy3 regression used for phage selection from a screening chip is shown in Figure 1.
Figure 1.
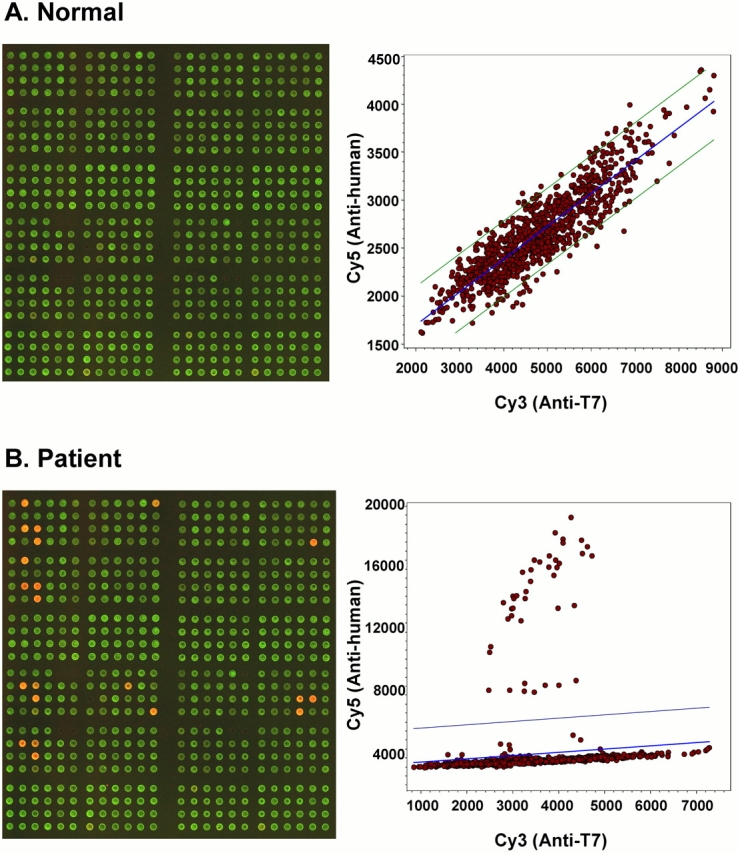
High-throughput screening of marker candidate phages. Biopanned phage clones were spotted on microarray slides and tested with NSCLC plasma samples. The partial array images show reactivity patterns for (A) normal and (B) patient samples, and the corresponding scatter plots show different distributions of reactive phage clones. The computer-generated regression line and standard deviation lines on the scatter plots assist in identifying candidate marker phages.
Data Processing and Choice of Candidate Markers
Forty NSCLC, 36 high-risk control, and 5 nonsmoking control samples not used in the biopan or in the high-throughput screening described above were assayed with a diagnostic chip containing 212 candidate phage-expressed proteins. Each specific signal (Cy5) was normalized for number of phage particles by the Cy3 (anti-T7 capsid) signal. Chip-to-chip variability, a suspected function of variable total IgG concentration in individual plasma samples, was normalized relative to the signal from empty T7 (Figure 2). A Student t test of normalized signal from 40 patients and 41 normal subjects afforded a statistical cutoff (p < 0.01) that suggested relative predictive value of each candidate marker. Of the 212 candidates, 17 met this stringent cutoff (p = 0.00003 to p = 0.01). Redundancy within this group was assessed by polymerase chain reaction and sequence analysis revealing several duplicate and triplicate clones. When redundant clones were eliminated a set of seven phage-expressed proteins was available for further analysis.
Figure 2.
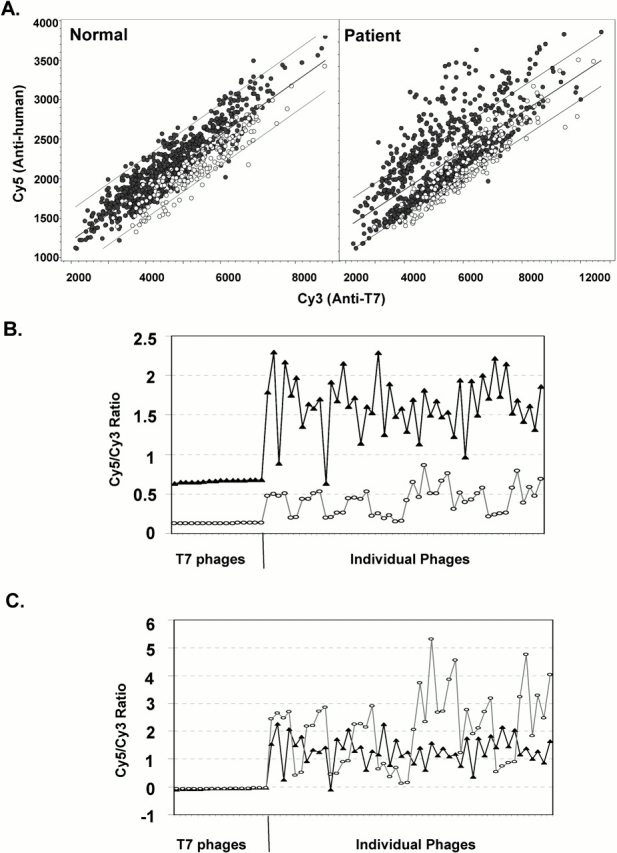
Normalization of array data among chips. “Empty” T7 phage proteins were spotted as a standard control on the diagnostic chips. Open circles on the scatter plots in (A) show the T7 phages on two different chips. (B) Patterns of Cy5:Cy3 signal ratio distribution before normalization; (C) distribution after normalization to T7 proteins. (B and C) Triangles, normal subjects; circles, patients.
Sequence Analysis of Phage-expressed Proteins
The 17 phages that were chosen for their putative predictive value, using the t test and p < 0.01, were sequenced to identify redundancy, which revealed seven unique sequences. Although the identity of the phage-expressed proteins is not critical for use in a diagnostic assay, the sequences were compared with those obtained in a previous study that used a different (independent) screening methodology (17) and also were compared with the GenBank database to obtain possible identity. Nucleotide sequences obtained from these seven clones showed homology to GAGE7, EEF1A, PMS2L15, NOPP140, paxillin, SEC15L2, and BAC clone RP11-499F19. The first four were identified in previous work as immunoreactive with patient plasma, using qualitative histochemical detection of plaque lifted colonies probed with patient plasma (17). Of these seven proteins, EEF1A (eukaryotic translation elongation factor 1A), a core component of the protein synthesis machinery, and GAGE, a cancer testis antigen, are overexpressed in some lung cancers (18–22). Paxillin is a focal adhesion protein that regulates cell adhesion and migration. Aberrant expression and anomalous activity have been associated with an aggressive metastatic phenotypic in some malignancies including lung cancer (23–28). PMS2L is a DNA mismatch repair–related protein, but no mutation has yet been identified in cancer (29). Similarly, SEC15L2, an intracellular trafficking protein and NOPP140, a nucleolar protein involved in regulation of transcriptional activity, do not have known malignant association (30, 31). The physiologic function of these three proteins, however, suggests each could have a role in the malignant phenotype. The BAC clone has, at present, no known associated protein function.
Statistical Modeling and Assay Prediction Accuracy
To develop classifiers using the unique seven phage-expressed proteins for a better predictive rate, we divided the total 81 samples randomly into two groups: one was used for training purposes and the other for validation. Logistic regression was used to calculate the sensitivity and specificity for predictive accuracy, using individual phage-expressed proteins as well as a combination of multiple phage-expressed markers. Results show that five phage markers had significant ability to distinguish patient samples from normal control subjects in the training set. The area under the receiver operating characteristic curve for each individually ranged from 0.79 to 0.86. Combinations of these five markers achieved a promising prediction rate (area under the receiver operating characteristic curve, 0.98), with 95% sensitivity and 85% specificity (Table 2). Using this statistical model to test the validation group consisting of 20 control normal samples and 20 NSCLC samples achieved a sensitivity of 90% and a specificity of 95% (Table 2). It is worth noting that, because these five markers were chosen for their individual predictive value (p < 0.01) using all 81 patient and control samples, this predictive accuracy may be biased upward. In our training set of 20 patient and 21 control samples we found that these same five markers retained p values less than 0.01 and would have been chosen for possible inclusion in our models, and thus the sensitivity of 90% and specificity of 95% for our training data are unbiased.
TABLE 2.
AREAS UNDER RECEIVER OPERATING CHARACTERISTICS CURVES AND PREDICTIVE ACCURACY
| Training Set*
|
Validation Set†
|
||||
|---|---|---|---|---|---|
| Phage Clone | AUC | Specificity (%) | Sensitivity (%) | Specificity (%) | Sensitivity (%) |
| 1864 | 0.857 | 75 | 81 | 65 | 85 |
| 1896 | 0.857 | 70 | 86 | 70 | 75 |
| 1919 | 0.824 | 75 | 81 | 70 | 90 |
| 1761 | 0.798 | 70 | 81 | 70 | 85 |
| 1747 | 0.864 | 70 | 86 | 70 | 80 |
| Five combined | 0.983 | 85 | 95 | 95 | 90 |
Definition of abbreviation: AUC = area under the receiver operating characteristic curve.
Training set consisted of 21 normal samples and 20 samples from patients with NSCLC.
Validation set consisted of 20 normal samples and 20 samples from patients with NSCLC.
To further examine the association of the classifiers with diagnostic sensitivity and specificity, we performed class prediction using leave-one-out cross-validation on all 81 chips. Using all 81 chips to estimate predictive ability results in upwardly biased estimates. The leave-one-out method removes the chips one at a time and uses the remaining 80 chips to classify the one left out. These classifications are used to compute sensitivity and specificity to correct for the fact that all 81 chips are being used. The results showed that sensitivity and specificity were 90 and 87%, respectively, with 81 samples, and the overall diagnostic accuracy was 89% (Table 3). Also, when using all 81 chips, the corresponding clone ID, gene name, and p value were as follows: 1864, GAGE7, p = 9.1 × 10−9; 1896, BAC clone RP11-499F19, p = 3.5 × 10−8; 1919, SEC15L2, p = 1.2 × 10−6; 1761, PMS2L15, p = 5.2 × 10−7; and 1747, EEF1A, p = 5.9 × 10−7. All five markers passed a Bonferroni correction of 0.001/262 = 3.8 × 10−6, making the probability of one or more of them being false positive less than 0.001.
TABLE 3.
LEAVE-ONE-OUT VALIDATION*
| Phage Clone | Specificity (%) | Sensitivity (%) | Diagnostic Accuracy† (%) |
|---|---|---|---|
| 1864 | 70 | 82.9 | 76.5 |
| 1896 | 70 | 82.9 | 75.3 |
| 1919 | 70 | 82.9 | 76.5 |
| 1761 | 60 | 82.9 | 71.6 |
| 1747 | 72.5 | 82.9 | 77.8 |
| Five combined | 87.5 | 90.2 | 88.9 |
Leave-one-out validation: one sample was removed from the testing set containing a total of 81 samples and a classifier was generated to predict the status (normal or patient) of the removed samples, using the rest of the samples. This procedure was repeated for all samples.
Diagnostic accuracy = (number of true positive + number of true negative)/total number of samples.
We were unable to accurately distinguish stage or histologic type of tumor in this sample set, probably because of sample size.
DISCUSSION
Serum tumor markers have the potential of being incorporated into diagnostic and therapeutic practice to improve historically dismal outcomes in NSCLC (2). Potential uses include early detection or screening, differentiation of benign from malignant disease, differentiating histologies, defining stages and responses to therapy, identifying recurrences, and defining prognosis. These goals have generated considerable interest in identifying predictive tumor markers (1–3, 32–34). Although a number of NSCLC tumor markers are measurable and a combination of available NSCLC markers can enhance diagnostic value, limited sensitivity and specificity of these markers preclude their widespread clinical use (2, 3, 35–38). In context, tumor-associated antibodies may expand the range of available NSCLC markers (14–17). Consistent with the knowledge that an antibody response to a single protein is unlikely to be a universal marker (10, 11, 36), we have been exploring methodology for efficiently identifying and measuring multiple tumor-associated antibodies. This article describes a technique for enriching and screening phage-expressed tumor-associated proteins that are, in turn, used in array fashion to measure multiple antibodies simultaneously. We biopan-enriched the immunogenic phage content of two NSCLC T7 libraries, used fluorescent microarrays to screen 4,000 clones and sequentially refined a list of potential markers to generate a diagnostic array capable of distinguishing patients with NSCLC from normal control subjects.
High-throughput screening identified 212 candidate markers that were included in a diagnostic chip for further testing. Each phage clone was then individually analyzed for its potential to discriminate 40 patients with NSCLC from 41 normal subjects. Only the most promising markers, those that were identified at a high level and frequency in patients with cancer to make them significantly different from normal subjects by Student t test, made the final cut. The t test provided a stringent cutoff, although other methods could have been applied to increase the number of markers to be assessed. The list size was further reduced by finding that several of the 17 phages were redundant clones. Notably, the fact that four of seven unique markers identified by this high-throughput approach were also identified by a qualitative histochemical screening method previously described (17) not only validates the high-throughput screening methodology, but also provides some validation of the markers themselves, because different sets of patient samples were used for both biopanning and screening in the independent experiments.
The classifier using five combined phage markers has given us good predictive accuracy using both the training and validation method and the leave-one-out method. Data were assessed for the relative importance of smoking, stage, and histology, although numbers in each group were too small to draw definitive conclusions. For reasons of assay development, we tested predominantly advanced-stage disease as these individuals are theoretically more likely to express high levels and greater variety of antibodies. In this population, compared with high-risk control subjects, we obtained diagnostic accuracy of 88.9%. This exceeds the currently reported prediction values of the clinically available markers tissue polypeptide antigen, 80%; CA19-9, 62%; carcinoembryonic antigen, 73%; squamous cell carcinoma antigen, 62%; and neuron-specific enolase, 63% (38). Additional validation of these data should include samples from a broader range of normal control subjects who are carefully matched for age and smoking history, and also include individuals with a variety of benign lung diseases common in the high-risk population, including chronic obstructive pulmonary disease and granulomatous lung disease. We have also not assayed plasmas from other malignancies. Knowing that several of the tumor-associated antibodies we have identified in NSCLC plasma samples have been described in other malignancies and that several more of the tumor-associated proteins we describe in NSCLC are known to be expressed by other cancers (17, 39), NSCLC specificity will have to be independently evaluated.
From the standpoint of application, our most immediate interest is to develop a predictive assay that can assist in early diagnosis of lung cancer. This interest is driven by the fact that only 25% of new cases of lung cancer are diagnosed at an early stage, when curative surgical resection is still possible (1, 40). An appropriately sensitive and specific assay could help address some concerns surrounding the use of low-dose spiral chest computed tomography (CT) as a stand-alone test for lung cancer screening. Specifically, low-dose CT screening is costly, is characterized by high sensitivity but just 64% specificity, and screening criteria of age and smoking history have limited ability to refine the target population (40–42). Moreover, the routine identification of indeterminate pulmonary nodules during radiographic imaging frequently leads to expensive workup and often potentially harmful intervention, including major surgery (40–42). Antibody profiling could enhance screening efforts by defining a population with high probability of disease in which screening is most warranted, by assisting in the differentiation of benign from malignant nodules detected on screening CT scans and potentially by detecting occult lung cancer at a time before a nodule can be seen on CT scan. Testing with early-stage cancers and matched control subjects will be the immediate focus of subsequent studies. Investigations will then determine the ability of antibody profiling to distinguish benign from malignant disease in a population presenting with indeterminate radiographic pulmonary nodules. Linking biomarker analysis to radiographic screening trials will provide necessary validation for these two related applications and, in addition, will determine whether antibody profiling can detect lung cancer before it is radiographically apparent. Other applications such as staging and prognosis are also compelling and will similarly require independent sample analysis.
Relevant to the preceding discussion, the markers described in this article were selected for their predictive value, using the available patient and control blood samples. If on further validation we see a decrease in sensitivity or specificity, or if the defined marker set does not have adequate diagnostic accuracy for a specific application (e.g., early detection), inclusion of additional markers or selection of alternative marker sets may help achieve appropriate predictive accuracy. If necessary, additional patient samples can be used to screen available or newly constructed tumor libraries to expand the number and range of existing markers, although we did not find this necessary to generate a panel of markers capable of distinguishing our patient from control samples.
Importantly, we have not exhaustively screened these libraries for all possible markers and have likely not yet identified some significantly predictive circulating tumor-associated antibodies. Specifically, we have chosen markers using only two NSCLC tumor libraries and antibodies found in five advanced-stage NSCLC plasma samples that will not be representative of the full spectrum of disease. Further, phage-expressed proteins, derived from a cDNA tumor library expressed within a prokaryotic T7 bacteriophage system, would not be expected to include the wide variety of proteins with posttranslational modifications typical of a mammalian system and often anomalous in human tumors. As such, this approach is not going to be useful for generating a comprehensive proteomic profile, although the intrinsic ability of this approach to potentially identify a variety of aberrantly expressed tumor-associated proteins is attractive.
In summary, this article describes efficient methodology for screening, identifying, and measuring multiple circulating antibodies as markers of disease. Although we have not generated a comprehensive panel of antibodies and associated proteins we have obtained an inventory of markers that can distinguish NSCLC patient samples from control subject samples. These data support the rationale for further validation, development, and application of this approach for management of lung cancer.
Acknowledgments
The authors thank Dr. Xiaoju Wang for helpful discussion.
Supported by a National Institutes of Health grant (R01, CA10032-01), the Veteran's Administration Merit Review Program, and the Kentucky Lung Cancer Association.
Originally Published in Press as DOI: 10.1164/rccm.200505-830OC on August 18, 2005
Conflict of Interest Statement: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Hoffman PC, Mauer Am, Vokes EE. Lung cancer. Lancet 2000;5:479–485. [DOI] [PubMed] [Google Scholar]
- 2.Niklinski J, Furman M. Clinical tumour markers in lung cancer. Eur J Cancer Prev 1995;4:129–138. [DOI] [PubMed] [Google Scholar]
- 3.Ferrigno D, Buccheri G, Biggi A. Serum tumour markers in lung cancer: history, biology and clinical applications. Eur Respir J 1994;7:186–197. [DOI] [PubMed] [Google Scholar]
- 4.Gure AO, Altorki NK, Stockert E, Scanlan MJ, Old LJ, Chen YT. Human lung cancer antigens recognized by autologous antibodies: definition of a novel cDNA derived from the tumor suppressor gene locus on chromosome 3p21.3. Cancer Res 1998;58:1034–1041. [PubMed] [Google Scholar]
- 5.Stockert E, Jager E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med 1998;187:1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tureci O, Sahin U, Zwick C, Neumann F, Pfreundschuh M. Exploitation of the antibody repertoire of cancer patients for the identification of human tumor antigens. Hybridoma 1999;18:23–28. [DOI] [PubMed] [Google Scholar]
- 7.Jager D, Stockert E, Gure AO, Scanlan MJ, Karbach J, Jager E, Knuth A, Old LJ, Chen YT. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res 2001;61:2055–2061. [PubMed] [Google Scholar]
- 8.Stevanovic S. Identification of tumour-associated T-cell epitopes for vaccine development. Nat Rev Cancer 2002;2:514. [DOI] [PubMed] [Google Scholar]
- 9.Diesinger I, Bauer C, Brass N, Schaefers HJ, Comtesse N, Sybrecht G, Meese E. Toward a more complete recognition of immunoreactive antigens in squamous cell lung carcinoma. Int J Cancer 2002;102:372–378. [DOI] [PubMed] [Google Scholar]
- 10.Abu-Shakra M, Buskila D, Ehrenfeld M, Conrad K, Shoenfeld Y. Cancer and autoimmunity: autoimmune and rheumatic features in patients with malignancies. Ann Rheum Dis 2001;60:433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheever MA, Disis ML, Bernhard H, Gralow JR, Hand SL, Huseby ES, Qin HL, Takahashi M, Chen W. Immunity to oncogenic proteins. Immunol Rev 1995;145:33–59. [DOI] [PubMed] [Google Scholar]
- 12.Sioud M, Hansen MH. Profiling the immune response in patients with breast cancer by phage-displayed cDNA libraries. Eur J Immunol 2001;31:716–725. [DOI] [PubMed] [Google Scholar]
- 13.Hanash S. Disease proteomics. Nature 2003;422:226–232. [DOI] [PubMed] [Google Scholar]
- 14.Hanash S. Harnessing immunity for cancer marker discovery. Nat Biotechnol 2003;21:37–38. [DOI] [PubMed] [Google Scholar]
- 15.Brichory FM, Misek DE, Yim AM, Krause MC, Giordano TJ, Beer DG, Hanash SM. An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proc Natl Acad Sci USA 2001;98:9824–9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brichory F, Beer D, LeNaour F, Giordano T, Hanash S. Proteomics-based identification of protein gene product 9.5 as a tumor antigen that induces a humoral immune response in lung cancer. Cancer Res 2001;61:7908–7912. [PubMed] [Google Scholar]
- 17.Zhong L, Peng X, Hidalgo GE, Doherty DE, Stromberg AJ, Hirschowitz EA. Identification of circulating antibodies to tumor-associated proteins for combined use as markers of non–small cell lung cancer. Proteomics 2004;4:1216–1225. [DOI] [PubMed] [Google Scholar]
- 18.Brasseur F, Chomez P, Van den Eynde B, Boon T, van der Bruggen P. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res 1999;59:3157–3165. [PubMed] [Google Scholar]
- 19.Melloni G, Ferreri AJ, Russo V, Gattinoni L, Arrigoni G, Ceresoli GL, Zannini P, Traversari C. Prognostic significance of cancer-testis gene expression in resected non–small cell lung cancer patients. Oncol Rep 2004;12:145–151. [PubMed] [Google Scholar]
- 20.Lamberti A, Caraglia M, Longo O, Marra M, Abbruzzese A, Arcari P. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids 2004;26:443–448. [DOI] [PubMed] [Google Scholar]
- 21.Joseph P, O'Kernick CM, Othumpangat S, Lei YX, Yuan BZ, Ong TM. Expression profile of eukaryotic translation factors in human cancer tissues and cell lines. Mol Carcinog 2004;40:171–179. [DOI] [PubMed] [Google Scholar]
- 22.Grant AG, Flomen RM, Tizard ML, Grant DA. Differential screening of a human pancreatic adenocarcinoma lambda gt11 expression library has identified increased transcription of elongation factor EF-1 α in tumour cells. Int J Cancer 1992;50:740–745. [DOI] [PubMed] [Google Scholar]
- 23.Chen HY, Shen CH, Tsai YT, Lin FC, Huang YP, Chen RH. Brk activates rac1 and promotes cell migration and invasion by phosphorylating paxillin. Mol Cell Biol 2004;24:10558–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowe DL, Ohannessian A. Recruitment of focal adhesion kinase and paxillin to β1 integrin promotes cancer cell migration via mitogen activated protein kinase activation. BMC Cancer 2004;4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo W, Li H. Relationship of the expression of CD44v6 and paxillin to the prognosis of non–small cell lung carcinoma. Sichuan Da Xue Xue Bao Yi Xue Ban 2003;34:484–485. [PubMed] [Google Scholar]
- 26.Nagata M, Fujita H, Ida H, Hoshina H, Inoue T, Seki Y, Ohnishi M, Ohyama T, Shingaki S, Kaji M, et al. Identification of potential biomarkers of lymph node metastasis in oral squamous cell carcinoma by cDNA microarray analysis. Int J Cancer 2003;106:683–689. [DOI] [PubMed] [Google Scholar]
- 27.Azuma K, Tanaka M, Uekita T, Inoue S, Yokota J, Ouchi Y, Sakai R. Tyrosine phosphorylation of paxillin affects the metastatic potential of human osteosarcoma. Oncogene 2005;24:4754–4764. [DOI] [PubMed] [Google Scholar]
- 28.Li HG, Xie DR, Shen XM, Li HH, Zeng H, Zeng YJ. Clinicopathological significance of expression of paxillin, syndecan-1 and EMMPRIN in hepatocellular carcinoma. World J Gastroenterol 2005;11:1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicolaides NC, Carter KC, Shell BK, Papadopoulos N, Vogelstein B, Kinzler KW. Genomic organization of the human PMS2 gene family. Genomics 1995;30:195–206. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XM, Ellis S, Sriratana A, Mitchell CA, Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J Biol Chem 2004;279:43027–43034. [DOI] [PubMed] [Google Scholar]
- 31.Miau LH, Chang CJ, Tsai WH, Lee SC. Identification and characterization of a nucleolar phosphoprotein, Nopp140, as a transcription factor. Mol Cell Biol 1997;17:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foa P, Fornier M, Miceli R, Seregni E, Santambrogio L, Nosotti M, Cataldo I, Sala M, Caldiera S, Bombardieri E. Tumour markers CEA, NSE, SCC, TPA and CYFRA 21.1 in resectable non–small cell lung cancer. Anticancer Res 1999;19:3613–3618. [PubMed] [Google Scholar]
- 33.Buccheri G, Ferrigno D. Lung tumor markers of cytokeratin origin: an overview. Lung Cancer 2001;34:S65–S69. [DOI] [PubMed] [Google Scholar]
- 34.Ebert W, Muley T, Drings P. Does the assessment of serum markers in patients with lung cancer aid in the clinical decision making process? Anticancer Res 1996;16:2161–2168. [PubMed] [Google Scholar]
- 35.Buccheri G, Violante B, Sartoris AM, Ferrigno D, Curcio A. Clinical value of a multiple biomarker assay in patients with bronchogenic carcinoma. Cancer 1986;57:2389–2396. [DOI] [PubMed] [Google Scholar]
- 36.Lombardi C, Tassi GF, Pizzocolo G, Donato F. Clinical significance of a multiple biomarker assay in patients with lung cancer: a study with logistic regression analysis. Chest 1990;97:639–644. [DOI] [PubMed] [Google Scholar]
- 37.Ando S, Kimura H, Iwai N, Shima M, Ando M, Kuriyama T. Optimal combination of seven tumour markers in prediction of advanced stage at first examination of patients with non–small cell lung cancer. Anticancer Res 2001;21:3085–3092. [PubMed] [Google Scholar]
- 38.Mizushima Y, Tsuji H, Izumi S, Hirata H, Kin Y, Kawasaki A, Matsui S, Yano S. Clinical evaluation of five tumor marker assays in patients with lung cancer. Anticancer Res 1991;11:91–96. [PubMed] [Google Scholar]
- 39.Pei J, Balsara BR, Li W, Litwin S, Gabrielson E, Feder M, Jen J, Testa JR. Genomic imbalances in human lung adenocarcinomas and squamous cell carcinomas. Genes Chromosomes Cancer 2001;31:282–287. [DOI] [PubMed] [Google Scholar]
- 40.Bach PB, Kelley MJ, Tate RC, McCrory DC. Screening for lung cancer: a review of the current literature. Chest 2003;123:72S–82S. [DOI] [PubMed] [Google Scholar]
- 41.MacRedmond R, Logan PM, Lee M, Kenny D, Foley C, Costello RW. Screening for lung cancer using low-dose CT scanning. Thorax 2004;59:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McWilliams A, Mayo J, MacDonald S, leRiche JC, Palcic B, Szabo E, Lam S. Lung cancer screening: a different paradigm. Am J Respir Crit Care Med 2003;168:1143–1144. [DOI] [PubMed] [Google Scholar]


