Abstract
Endogenous tumor suppression provides a barrier against oncogenesis, but the molecular requirements of this process are not well understood. Here, we show that the dual specificity phosphatase PTEN, a gene almost universally altered in human tumors, silences the expression of survivin, an essential regulator of cell division and apoptosis in cancer. This pathway is independent of p53, involves active repression of survivin gene transcription, and is mediated by direct occupancy of the survivin promoter by FOXO1 and FOXO3a factors. Conditional deletion of PTEN in the mouse prostate causes deregulated induction of survivin before full-blown transformation in vivo, whereas expression of survivin and PTEN is inversely correlated in cancer patients. Therefore, silencing the survivin gene is an essential requirement of endogenous PTEN tumor suppression.
Introduction
The dual specificity phosphatase PTEN (Phosphatase and Tensin homologue deleted from chromosome Ten) functions as a nodal regulator of multiple signaling pathways that control cell proliferation, cell survival, and cell size (1). By removing the D3 phosphate from the lipid second messenger phosphatidylinositol triphosphate, PTEN shuts off growth factor receptor stimulation, and interrupts downstream signaling mediated by Akt and PDK1 kinases (2). This pathway provides a broad network of tumor suppression, not only by antagonizing cell proliferation and promoting apoptosis, but also by maintaining chromosomal integrity (3), and directly cooperating with p53-dependent responses (4). In this context, the PTEN pathway is almost universally disabled in human cancer (1), often involving deletion of the PTEN gene in common malignancies of prostate, brain, breast, and colon (5). Although Forkhead transcription factors of the FOXO subfamily have been shown to contribute to PTEN signaling (6), their downstream target genes potentially cooperating in tumor suppression have not been conclusively identified (7).
Survivin is a unique member of the Inhibitor of Apoptosis (IAP) gene family with essential roles in mitosis, the cellular stress response, and inhibition of cell death (8). These properties are exploited in cancer, where survivin is universally overexpressed, and promotes unfavorable outcome (8). Although several signaling pathways, including Akt (9), have been associated with elevated survivin levels in cancer, the interplay between oncogenic mechanisms and tumor suppression networks in controlling survivin gene transcription has remained largely elusive.
In this study, we investigated the effect of the PTEN pathway on the expression of the survivin gene.
Materials and Methods
Cells and antibodies
Breast adenocarcinoma MCF-7 and MDA-MB-231, and prostate adenocarcinoma PC3 cells were obtained from the American Type Culture Collection. p53+/+ or p53−/− colorectal cancer HCT116 cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD). MCF-7 cells stably expressing survivin (MCF-7 SVV) were described previously (10). MDA-MB-231 cells stably transfected with the proximal 830nt of the mouse survivin promoter fused to GFP (ms-830-GFP) were as described (11). A lentiviral plasmid encoding control pLKO or PTEN-directed short hairpin RNA (Open Biosystems) was transfected into 293T packaging cells together with pCMVΔ8.9 and pMDG to generate lentiviruses. MCF-7 cells were infected with control or PTEN-directed lentivirus, and positive clones were selected in 1 µg/mL Puromycin-containing (Sigma) medium. Antibodies to PTEN, phospho-AKT (S473), cleaved caspase 3 (Cell Signaling), XIAP, GFP (BD Biosciences), FOXO1, FOXO3a, 14-3-3β (Santa Cruz Biotechnology), RNA Polymerase II, IgG (Active Motif), survivin (Novus Biologicals), or β-actin (Sigma) were used.
RNA and protein analysis
Total RNA (Qiagen) was reverse-transcribed (Invitrogen), and amplified with primers for survivin, 5′-GCATGGGTGCCCCGACGTTG- 3′ ( forward) and 5′-GCTCCGGCCAGAGGCCTCAA-3′ (reverse); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-ACGGATTTGGTCGTATTGGGCG- 3′ ( forward) and 5′-CTCCTGGAAGATGGTGATGG- 3′ (reverse); FOXO1, 5′-AAGAGCGTGCCCTACTTCAA-3′ ( forward) and 5′-AGGCCATTTGGAAAACTGTG-3′ (reverse); and FOXO3a, 5′-GCAAGCACAGAGTTGGATGA- 3′ ( forward) and 5′-CTGGCGTAGGGAGTTCAGAG- 3′ (reverse). Protein expression was analyzed by Western blotting.
Transfections
β-Galactosidase–normalized survivin promoter luciferase activity (pLuc-3000, pLuc-1430, pLuc-649, and pLuc-441) was quantified as described (12). Cells were transfected with SMARTpool siRNA directed to PTEN, FOXO1 or FOXO3a (Dharmacon), or nontargeted siRNA (10). LY294002 (Calbiochem) was used as a PI3 kinase inhibitor. MDA-MB-231 cells stably transfected with ms-830-GFP were transduced with adenoviruses encoding FOXO variants at 30to 50 multiplicity of infection for 8 h, and harvested after 24 to 48 h.
Analysis of apoptosis
Cells transfected with pEGFP or PTEN-GFP were analyzed after 24 h by multiparametric flow cytometry using PE-Annexin V and 7-AAD staining (BD Biosciences; ref. 10), or, alternatively, for DNA content by propidium iodide staining and flow cytometry.
Chromatin immunoprecipitation
Colorectal adenocarcinoma HCT116 cells (4.7 × 107) were fixed in 1% formaldehyde for 10min at 22°C, lysed, and nuclear fractions were isolated before DNA shearing ( fragment size of 200–600 nt) by sonication. Samples were incubated with protein G magnetic beads (Active Motif) and various antibodies for 5 h at 4°C. The beads were precipitated, reverse cross-linked, treated with proteinase K, and DNA was amplified by PCR using primers for regions in the human survivin promoter containing putative FOXO binding sites (−1428 nt): 5′-TGAGCTGAGATCATGCCACT- 3′ ( forward), and 5′-CTGGTGCCTCCACTGTCTTT-3′ (reverse), or devoid of FOXO binding sites (−2269 nt): 5′-TTGTTCCTTTCCTCCCTCCTGAG- 3′ ( forward), and 5′-GTCAACTGGATTTGATAACTGCA-3′ (reverse). Primers to amplify FOXO binding sites in the p27Kip1 promoter (13), or RNA polymerase II binding sites in the GAPDH promoter, were used as control.
Oncomine analysis
Oncomine data were reviewed for microarray analysis of differential gene expression in cancer versus matched normal tissues. Raw gene expression data were extracted for both PTEN and survivin from the same studies, and comparative analysis was performed using GraphPad Software (Prism 4.0). A P value was calculated using Spearman rank correlation test.
Histology
Five-micrometer sections cut from prostate tissues of 20to 24-wk-old prostate-specific PTEN conditional knockout mice (14) were stained with antibodies to survivin or control IgG by immunohistochemistry, as described (11).
Results and Discussion
PTEN regulation of survivin gene expression
Transfection of breast adenocarcinoma MCF-7 cells with PTEN-directed siRNA efficiently suppressed PTEN expression, by Western blotting (Fig. 1A). This was associated with increased levels of endogenous survivin, compared with cultures transfected with nontargeted siRNA (Fig. 1A). XIAP, another IAP family protein stabilized by survivin (8), was also increased in MCF-7 cells after PTEN knockdown, whereas the levels of 14-3-3β were unchanged (Fig. 1A). siRNA silencing of PTEN indistinguishably increased survivin expression in p53+/+ or p53−/− HCT116 cells, indicating that p53 was not required for this response (ref. 4; Fig. 1A). Consistent with a transcriptional mechanism, PTEN knockdown resulted in a 2-fold increase in survivin mRNA, by reverse transcription-PCR (RT-PCR; Fig. 1B), and increased survivin promoter activity, by luciferase reporter assay (Fig. 1C). Accordingly, MCF-7 cells transfected with the proximal 830nt of the survivin promoter fused to GFP (ms-830-GFP) exhibited increased number of GFP-expressing cells after PTEN silencing, by fluorescence microscopy (Fig. 1D). In contrast, PTEN siRNA did not affect the expression of control pEGFP, and a nontargeted siRNA was ineffective (Fig. 1D).
Figure 1.
PTEN regulation of survivin gene expression. A, the indicated cell types were transfected with control (ctrl) or PTEN-directed siRNA, and analyzed by Western blotting. *, nonspecific. B, siRNA-transfected HCT116 cells were analyzed by RT-PCR. Numbers correspond to densitometric quantification of mRNA bands. C, PTEN siRNA–silenced HCT116 cells were transfected with a survivin promoter luciferase construct (pLuc-441), and analyzed for luciferase activity. RLU, relative luciferase units. *, P = 0.025. D, HCT116 cells expressing pEGFP or ms-830-GFP were transfected with the indicated siRNA, and analyzed by fluorescence microscopy. Right, quantification of GFP-expressing cells. **, P = 0.0061. Columns, mean of replicates from a representative experiment of at least two independent determinations; bars, SE (C and D).
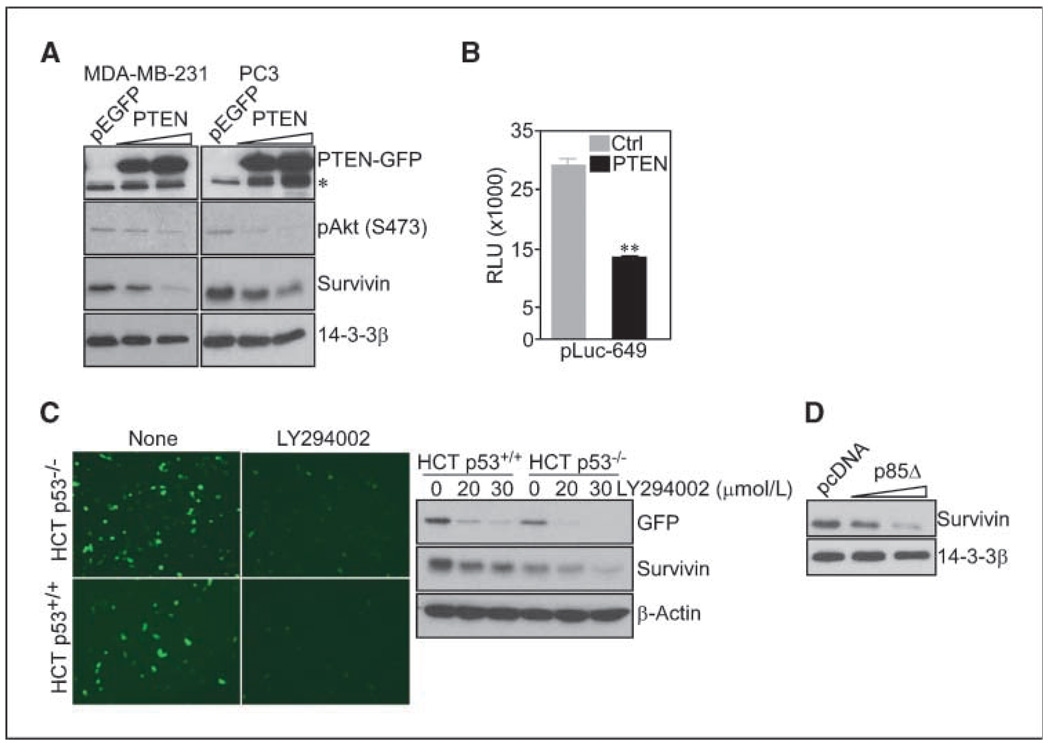
In complementary experiments, transfection of a PTEN cDNA in PTEN-null breast adenocarcinoma MDA-MB-231 or prostate adenocarcinoma PC3 cells resulted in nearly complete suppression of Akt phosphorylation on Ser473 (Fig. 2A). This was associated with concentration-dependent reduction in endogenous survivin levels, by Western blotting (Fig. 2A), and repression of survivin gene transcription, by luciferase promoter assay (Fig. 2B). Similarly, downstream targeting of the PI3 kinase pathway with the pharmacologic inhibitor LY294002, or after transfection of a dominant negative (DN) mutant of the p85 regulatory subunit (p85Δ), suppressed survivin promoter–directed GFP expression in HCT116 cells (Fig. 2C), and reduced endogenous survivin protein levels in MCF-7 cells (Fig. 2C and D), respectively.
Figure 2.
Transcriptional repression of survivin by PTEN. A, the indicated cell types were transfected with pEGFP or increasing concentrations of PTEN cDNA, and analyzed by Western blotting. *,nonspecific. B, HCT116 cells expressing pEGFP or PTEN cDNA were transfected with a survivin promoter luciferase construct (pLuc-649), and analyzed for luciferase activity. **, P = 0.0048. Columns, mean of replicates of a representative experiment of at least two independent determinations; bars, SE. C, HCT116 cells were transfected with ms-830-GFP, treated with vehicle or LY294002 (30 µmol/L), and analyzed by fluorescence microscopy. Right, Western blotting. D, MCF-7 cells were transfected with pcDNA or PI3 kinase p85Δ DN mutant and analyzed by Western blotting.
FOXO silencing of survivin gene transcription
The FOXO family of Forkhead transcription factors functions as potential effectors of PTEN signaling (7). Accordingly, transduction of MDA-MB-231 cells with an adenovirus encoding a FOXO1 DN mutant increased endogenous survivin levels, compared with cultures transduced with wild-type (WT) FOXO1 (Fig. 3A). Conversely, a constitutively active FOXO1 construct (ADA) slightly reduced survivin expression in MDA-MB-231 cells (Fig. 3A). In control experiments, expression of FOXO1 ADA induced spontaneous apoptosis in transduced cultures, whereas FOXO1 WT or DN mutant had no effect, by DNA content analysis and flow cytometry (Fig. 3A). In complementary experiments, siRNA silencing of FOXO1 or FOXO3a (Fig. 3B) increased the expression of endogenous survivin in HCT116 cells, by Western blotting (Fig. 3B), and enhanced survivin promoter luciferase activity (Fig. 3C), compared with nontargeted siRNA. When analyzed in chromatin immunoprecipitation studies, FOXO1 and FOXO3a physically associated with a segment of the proximal survivin promoter (−1428 nt) containing putative FOXO binding sites, whereas an upstream region (−2269 nt) in the survivin gene was ineffective (Fig. 3D, left). In control experiments, FOXO1 and FOXO3a also bound to the p27Kip1 promoter, which is regulated via FOXO-dependent transcription (13) but not to an unrelated promoter, GAPDH (Fig. 3D, left). Finally, stable short hairpin RNA silencing of PTEN in MCF-7 cells resulted in increased survivin expression (data not shown), and reduced formation of FOXO3a complexes with the survivin promoter, whereas FOXO1 interactions were less prominently affected (Fig. 3D, right).
Figure 3.
FOXO regulation of survivin gene transcription. A, MDA-MB-231 cells stably expressing ms-830-GFP were transduced with the indicated FOXO1 adenoviruses (pAd), and analyzed by Western blotting. ADA, constitutively active. Right, DNA content analysis of transduced cells. The percentage of cells with sub-G1 DNA content is indicated. B, HCT116 cells transfected with control, FOXO1-directed (left), or FOXO3a-directed (right) siRNA were analyzed by RT-PCR (top) or Western blotting (bottom). C, HCT116 cells silenced for FOXO1 or FOXO3a were transfected with survivin promoter luciferase constructs (pLuc-1430 or pLuc-3000) and analyzed for luciferase activity. Columns, mean of replicates of a representative experiment of at least two independent determinations; bars, SE. D, nuclear extracts of HCT116 cells (left) or MCF-7 cells stably expressing control pLKO or PTEN-directed short hairpin RNA (right) were immunoprecipitated (IP) with the indicated antibodies, and the immune complexes were amplified with primers corresponding to survivin, p27Kip1, or GAPDH promoter sequences. Top, position of putative FOXO sites in the survivin promoter.
Survivin modulation of PTEN tumor suppression
Transfection of a PTEN cDNA in MCF-7 cells minimally affected cell viability, whereas the combination of PTEN expression plus the broad cell death stimulus, staurosporine, caused approximately a 2-fold increase in apoptosis, by multiparametric flow cytometry (Fig. 4A). Stable expression of survivin in MCF-7 SVV cells reversed apoptosis induced by expression of PTEN plus staurosporine to background levels of untreated cells (Fig. 4A), and reduced the extent of effector caspase 3 cleavage, by Western blotting (Fig. 4B). When analyzed in vivo, the expression of survivin and PTEN was inversely correlated in published microarray data sets of patients with glioblastoma (P < 0.0001; ref. 15), or colon cancer (P = 0.0017; Fig. 4C; ref. 16). In addition, conditional deletion of PTEN in the mouse prostate resulted in a dramatic increase in survivin expression at the earliest stages of prostatic tumorigenesis, i.e., atypical hyperplasia, which persisted in more advanced disease phases of carcinoma in situ and invasive carcinoma, in vivo (Fig. 4D).
Figure 4.
Regulation of PTEN tumor suppression by survivin. A, MCF-7 or MCF-7 SVV cells were transfected with pEGFP or PTEN with our without staurosporine (1 µmol/L), and analyzed by multiparametric flow cytometry. The percentage of cells in each quadrant is indicated. B, transfected MCF-7 or MCF-7 SVV cells were analyzed by Western blotting. C, microarray data sets of patients with glioblastoma (77) or colon cancer (81) were examined for expression of PTEN or survivin by linear regression analysis. D, prostate tissues from 20- to 24-wk-old prostate-specific PTEN conditional knockout mice were stained with IgG or antibodies to survivin. Arrow, area of local invasion. Magnification, ×400.
In summary, we have shown that PTEN-mediated tumor suppression (1) involves acute silencing of the survivin gene, and this response is mediated by binding of FOXO1 and FOXO3a factors to the proximal survivin promoter (6, 7). This mechanism may contribute to the differential expression of survivin in many disparate types of cancer (8), given that defects in the PTEN pathway, or deregulated Akt signaling, occur in nearly every human tumor (1). Similar to PTEN (this study), other pivotal tumor suppressors, including p53 (17, 18), APC (19), or SIRT1 (20) have been shown to acutely silence the survivin gene, by different mechanisms. This suggests that maintaining low to undetectable levels of survivin is a general mechanism of an endogenous tumor suppression network, and provides a requisite to effectively antagonize neoplastic transformation.
Acknowledgments
Grant support: NIH grants CA78810, CA90917, HL54131 (D.C. Altieri), CA89720, and CA109874 (L.R. Languino).
We thank Drs. Bert Vogelstein for HCT116 cells, Domenico Accili for FOXO1 adenoviral constructs, Alex Toker for p85Δ cDNA, and Alonzo Ross for PTEN-GFP cDNA.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 2.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen WH, Balajee AS, Wang J, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 4.Li AG, Piluso LG, Cai X, Wei G, Sellers WR, Liu X. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol Cell. 2006;23:575–587. doi: 10.1016/j.molcel.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 7.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 8.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Yang L, Yang J, et al. Transforming growth factor β induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Res. 2008;68:3152–3160. doi: 10.1158/0008-5472.CAN-07-5348. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 11.Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Res. 2006;66:3392–3395. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- 12.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344(Pt 2):305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Wang Z, Kong D, Li R, Sarkar SH, Sarkar FH. Regulation of Akt/FOXO3a/GSK-3β/AR signaling network by isoflavone in prostate cancer cells. J Biol Chem. 2008;283:27707–27716. doi: 10.1074/jbc.M802759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Gao J, Lei Q, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 15.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Ki DH, Jeung HC, Park CH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 18.Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 19.Zhang T, Otevrel T, Gao Z, Ehrlich SM, Fields JZ, Boman BM. Evidence that APC regulates survivin expression: a possible mechanism contributing to the stem cell origin of colon cancer. Cancer Res. 2001;61:8664–8667. [PubMed] [Google Scholar]
- 20.Wang RH, Zheng Y, Kim HS, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008;32:11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]