Abstract
Rationale: Multiple infections with different strains of Mycobacterium tuberculosis may occur in settings where the infection pressure is high. The relevance of mixed infections for the patient, clinician, and control program remains unclear. Objectives: This study aimed to describe reinfection and mixed infection as underlying mechanisms of changing drug-susceptibility patterns in serial sputum cultures. Methods: Serial M. tuberculosis sputum cultures from patients diagnosed with multi-drug-resistant (MDR) tuberculosis were evaluated by phenotypic drug-susceptibility testing and mutation detection methods. Genotypic analysis was done by IS6110 DNA fingerprinting and a novel strain-specific polymerase chain reaction amplification method. Measurements and Main Results: DNA fingerprinting analysis of serial sputum cultures from 48 patients with MDR tuberculosis attributed 10 cases to reinfection and 1 case to mixed infection. In contrast, strain-specific polymerase chain reaction amplification analysis in 9 of the 11 cases demonstrated mixed infection in 5 cases, reinfection in 3 cases, and laboratory contamination in 1 case. Analysis of clinical data suggests that first-line therapy can select for a resistant subpopulation, whereas poor adherence or second-line therapy resulted in the reemergence of the drug-susceptible subpopulations. Conclusions: We have shown that, in some patients with MDR tuberculosis, mixed infection may be responsible for observations attributed to reinfection by DNA fingerprinting. We conclude that treatment and adherence determines which strain is dominant. We hypothesize that treatment with second-line drugs may lead to reemergence of the drug-susceptible strain in patients with mixed infection.
Keywords: drug resistance, mixed infections, Mycobacterium tuberculosis, reinfection
Traditionally, infection by Mycobacterium tuberculosis was assumed to be caused by a single strain, and recurrences were believed to be due to reactivation of the strain that caused the first episode (1). Infection with multiple M. tuberculosis strains within one patient before, during, or after successful treatment was rarely considered. The development of DNA fingerprinting methods to differentiate M. tuberculosis strains (2) has made it possible to document reinfection with a genetically different M. tuberculosis strain in patients with recurrent tuberculosis who were resident in high or low tuberculosis incidence settings, and who were either immunocompetent or immunosuppressed (3–11).
Evidence of mixed infection in a single host at a single point in time, suggesting reinfection before or during disease, was first observed using the phage typing method (12, 13) and was later confirmed using DNA fingerprint analysis (8, 14–19). The infrequency of observing mixed infections probably reflects the insensitivity of the DNA fingerprinting method (16). It has recently been shown, using a highly specific polymerase chain reaction (PCR)–based genotyping method, that patients with tuberculosis in a high-incidence setting often have different M. tuberculosis strains in the same sputum specimen (20).
It has been suggested that mixed infections may be of concern for tuberculosis control because such infections could influence the diagnosis of drug resistance if a patient is infected with both a sensitive and a resistant strain (21). Furthermore, it is possible that undetected drug-resistant strains may emerge under the pressure of antibiotic treatment (21).
This study aimed to describe whether reinfections and mixed infections can be underlying mechanisms of drug-susceptibility variation in serial M. tuberculosis sputum cultures collected from patients diagnosed with multi-drug-resistant (MDR) tuberculosis (defined as bacillary resistance to at least isoniazid and rifampin). We show that the type of information available (clinical, microbiological, DNA fingerprinting, or PCR results) influences the interpretation of the observed variation in drug-susceptibility patterns. We demonstrate how the selective pressure of the antibiotic therapy determines which strain is dominant in the sputum culture. This work was presented at the South African Society for Biochemistry and Molecular Biology conference held in Stellenbosch, South Africa, in 2005 (31).
METHODS
Study Population
Clinical data and sputum specimens were collected as part of an ongoing, prospective study in Cape Town, South Africa (22). The incidence of new smear and/or culture-positive tuberculosis in the study communities (population, ∼ 35,000) was, on average, 313 per 100,000 per year (1993–1998) (22). A database search was performed to identify patients with MDR tuberculosis and who had at least two serial sputum cultures of M. tuberculosis. Only data from patients fulfilling these criteria were included in the analysis. This study was approved by the Ethics Review Board at Stellenbosch University.
Drug-Susceptibility Testing
Drug-susceptibility testing was done by the National Health Laboratory Service using the indirect proportion method on Löwenstein-Jensen medium containing critical concentrations of 0.2 μg/ml isoniazid and 30 μg/ml rifampin. Drug-susceptibility testing for other drugs was done for sputum cultures resistant to isoniazid and/or rifampin.
Mutations conferring resistance to isoniazid, rifampin, streptomycin, and ethambutol were determined by DNA sequencing or PCR dot-blot hybridization (23).
Genotyping by Molecular Techniques
M. tuberculosis strains present in serial sputum cultures were genotyped by the IS6110 DNA fingerprinting method (2). In addition, each sputum culture was assessed by strain-specific PCR amplification (20), using a universal forward primer (complementary to an internal sequence of the IS6110 element) in combination with the respective reverse primer (complementary to the strain-specific IS6110 insertion junction; Table 1). Strain-specific primers were designed after sequencing of the IS6110 insertion junctions from different strains identified in serial sputum cultures by DNA fingerprinting (see the online supplement). PCR reactions were performed in a total volume of 25 μl, according to previously described reaction conditions (20). PCR-amplified products were electrophoretically fractionated in 2.0% agarose and visualized by staining with ethidium bromide. Sensitivity and specificity of each primer set were determined by amplification of pure DNA from a panel of genetically unrelated and related strains (20), and were shown to be 100% (95% confidence interval, 85–100%) when compared with the gold standard of IS6110 DNA fingerprinting. Using the described amplification conditions (20), underlying strains could be detected at a molar ratio of 1:125.
TABLE 1.
Primer sequences
| Primer Name | Sequence | IS6110 Insertion Point |
|---|---|---|
| Universal forward | TTC AAC CAT CGC CGC CTC TAC | |
| Strain a reverse | GAG CGC GCC GAA GGC GGC CAT GAA C | 2602188 |
| Strain b reverse | GGC CAA ATC CAG CAC CAC GGT GAA C | 3096762 |
| Strain c reverse | GCG CCA ATG AAG CCA GCA ACG CCG T | 3379767 |
| Strain d reverse | CAC CCT CTA CTC TGC GCT TTG | 3127922 |
| Strain e reverse | TTG CTT TGA GGC GAC TTC C | 3125705 |
| Strain f reverse | GCG CGT GTC CCGA TGT GAG GTG GT | 1989058 |
| Strain g reverse | TCA GCC CGC CGC GAC TGT ATG AAC C | 2627510 |
| Strain h reverse | CAG GAC AAA GGT CGG CAA CCT GAA CC | 1996100 |
| Internal control forward | GAG CAG CAG TGG AAT TTC GC | |
| Internal control reverse | TCC CAG TGA CGT TGC CTT C |
To minimize laboratory cross-contamination, sputum cultures from each patient were PCR-amplified on separate days, and each procedure (preparation of the PCR reaction mixes, the addition of the DNA, the PCR amplification, and the electrophoretic fractionation) was conducted in physically separated rooms. Primers complementary to the M. tuberculosis gene Rv3875 were included in each amplification reaction as a positive amplification control. Negative controls (water) were included to detect reagent contamination.
Reagent contamination could not be detected because all negative controls were negative on amplification. The inability to detect specific strains was not due to the presence of PCR inhibitors because positive controls produced amplification products in all sputum cultures. This PCR method was highly consistent as amplification of each sputum culture gave identical products on repeated amplification.
Definitions of Reinfection and Mixed Infection
Reinfection was confirmed, according to the gold standard (5), when analysis of serial sputum cultures from a single patient showed the appearance of a genetically different strain (in two or more cultures) during the course of disease. If the genetically different strain was only identified in a single sputum culture, reinfection was considered as probable. Mixed infection was confirmed when two genetically distinct strains were present in the initial sputum culture as well as in at least one subsequent sputum culture.
Statistical Methods
Fisher's exact test was used to identify significant differences between patient groups.
RESULTS
Study Population
During the period from January 1993 to December 1998, 1,023 patients resident in the epidemiologic field site in Cape Town, South Africa, were culture-positive for M. tuberculosis (22). Sputum cultures from 768 of these patients were available for further analysis. Cultures from the remaining 255 patients were contaminated, were lost, or failed to produce usable DNA fingerprints. Phenotypic drug-susceptibility testing classified 48 of the 768 patients as having an episode of MDR tuberculosis.
Genotyping by DNA Fingerprinting
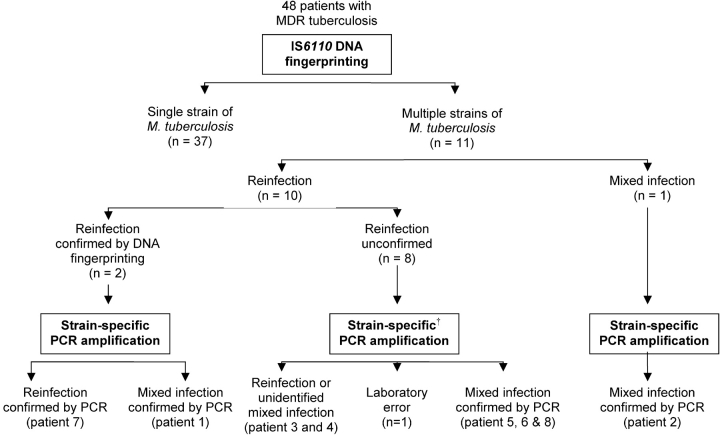
DNA fingerprinting showed a single strain in serial sputum cultures from 37 of the 48 patients with MDR tuberculosis. Serial sputum cultures from the remaining 11 patients showed the presence of genetically distinct strains (Figure 1). Among these, two different strains were isolated from one patient (Patient 2) on the day of diagnosis, as well as from subsequent sputum cultures, demonstrating mixed infection (Figure 2). In the remaining 10 patients, DNA fingerprinting data suggested reinfection. Reinfection could be confirmed by using fingerprint methods in two patients (HIV-uninfected Patient 1 [Figure 2] and HIV-infected Patient 7 [Table 2]), and was suggested (probable reinfection) in the remaining eight patients, for whom confirmatory sputum cultures for both genetically distinct M. tuberculosis strains were not available.
Figure 1.
Schematic diagram showing the grouping of patients according to DNA fingerprinting and strain-specific polymerase chain reaction (PCR) amplification methods. †Sputum culture from two patients could not be tested. MDR = multi-drug-resistant.
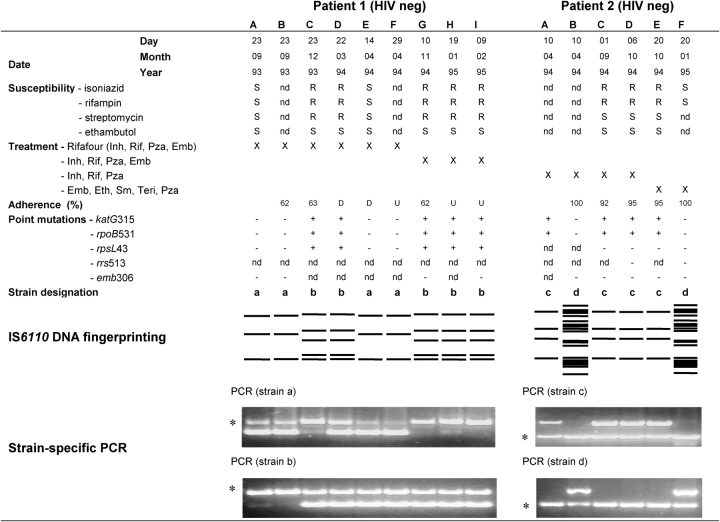
Figure 2.
Phenotypic and genotypic characterization of sputum cultures from Patients 1 and 2. Serial M. tuberculosis sputum cultures were obtained from patients diagnosed with MDR tuberculosis. Phenotypic culture-based drug-susceptibility testing was performed by the direct proportion method. Treatment regimen implemented at each visit is indicated, whereas adherence was measured for the period between each visit. Mutations conferring resistance were detected by DNA sequencing or PCR dot blot (23). All sputum cultures were genotyped by IS6110 DNA fingerprinting (2), and the strain(s) present was randomly assigned an alphabetic designation according to its strain family classification. Presence of multiple strains in each sputum culture was determined using strain-specific PCR amplification (20). D = default (stopped therapy for a period of > 2 months); Emb = ethambutol; Eth = ethionamide; Inat = isoniazid and thiacetazone; Inh = isoniazid; Kana = kanamycin; nd = not determined; neg = negative; Oflox = ofloxacin; pos = positive; Pza = pyrazinamide; R = drug resistant; Rif = rifampin; S = drug sensitive; Sm = streptomycin; Teri = terizidone; thia = thiacetazone; U = unknown; + = mutation present; − = mutation absent; *internal positive PCR control (Rv3875).
TABLE 2.
Summary of the phenotypic and genotypic characteristics of sputum cultures from patients 3 to 8
| Patient 3 (HIV neg)
|
Patient 4 (HIV neg)
|
|||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | A | B | C | D | E | F | G | H | ||||||||||||||
| Day | U | 25 | 20 | 16 | 29 | 05 | 09 | 31 | 29 | 04 | 15 | 05 | 14 | U | 14 | 09 | 20 | 21 | 28 | 05 | 08 | |||||||||||||
| Month | 11 | 05 | 07 | 08 | 09 | 10 | 11 | 01 | 02 | 04 | 10 | 02 | 08 | 11 | 04 | 06 | 06 | 07 | 07 | 01 | 06 | |||||||||||||
| Year | 94 | 95 | 95 | 95 | 95 | 95 | 95 | 96 | 96 | 96 | 96 | 98 | 98 | 93 | 94 | 94 | 94 | 94 | 94 | 95 | 95 | |||||||||||||
| Susceptibility | ||||||||||||||||||||||||||||||||||
| Isoniazid | nd | R | R | R | R | R | R | S | R | R | S | R | R | R | R | S | R | R | S | R | R | |||||||||||||
| Rifampin | nd | S | R | R | R | R | R | S | R | R | S | R | R | R | R | S | R | R | S | R | R | |||||||||||||
| Treatment | ||||||||||||||||||||||||||||||||||
| Rifafour (Inh, Rif, Pza, Emb) | U | X | X | X | X | X | ||||||||||||||||||||||||||||
| Rifater (Inh, Rif, Pza) | X | |||||||||||||||||||||||||||||||||
| Emb, Eth, Inat, Kana | X | X | X | X | X | X | X | |||||||||||||||||||||||||||
| Emb, Eth, Inat, Kana, Teri | X | |||||||||||||||||||||||||||||||||
| Eth, Inat, Kana, Teri, | X | X | X | X | ||||||||||||||||||||||||||||||
| Emb, Eth Inat, | X | |||||||||||||||||||||||||||||||||
| Emb, Eth, Inat, Teri, Pza | X | |||||||||||||||||||||||||||||||||
| Adherence, % | U | 87 | 79 | 90 | 90 | 95 | 99 | 71 | 79 | 53 | 49 | D | U | 83 | 80 | 82 | 100 | 100 | 68 | 57 | ||||||||||||||
| Point mutations | ||||||||||||||||||||||||||||||||||
| katG315 | nd | + | + | + | + | + | + | + | + | + | − | + | + | nd | + | − | + | + | + | + | + | |||||||||||||
| rpoB531 | nd | + | − | + | + | + | + | + | ||||||||||||||||||||||||||
| rpoB516 | nd | + | + | + | + | + | + | + | + | + | − | + | + | |||||||||||||||||||||
| Strain designation (DNA fingerprinting) |
nd | b | b | b | b | b | b | b | b | b | e | b | b | nd | d | f | d | d | d | d | d | |||||||||||||
| Strain population present (PCR) |
nd | b | b | b | b | b | b + e | b | b | b + e | b + e | b | b | nd | d + f | f | d | d + f | d | d + f | d | |||||||||||||
| Patient 5 (HIV nd)
|
Patient 6 (HIV nd)
|
Patient 7 (HIV pos)
|
Patient 8 (HIV nd)
|
|||||||||||||||||||||||||||||||
| A | B | C | D | A | B | C | A | B | C | D | E | F | A | B | C | D | ||||||||||||||||||
| Day | 03 | 26 | 05 | 05 | 17 | 23 | 10 | 01 | 02 | 07 | 15 | 15 | 21 | 19 | 24 | 24 | 05 | |||||||||||||||||
| Month | 06 | 11 | 01 | 01 | 06 | 09 | 11 | 07 | 07 | 12 | 12 | 12 | 12 | 02 | 02 | 02 | 03 | |||||||||||||||||
| Year | 93 | 93 | 95 | 95 | 93 | 93 | 93 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 98 | 99 | |||||||||||||||||
| Susceptibility | ||||||||||||||||||||||||||||||||||
| Isoniazid | S | nd | R | R | R | R | R | nd | nd | S | R | R | R | nd | R | R | R | |||||||||||||||||
| Rifampin | S | nd | R | R | R | R | R | nd | nd | S | R | R | R | nd | R | R | R | |||||||||||||||||
| Treatment | ||||||||||||||||||||||||||||||||||
| Rifater (Inh, Rif, Pza) | X | X | X | X | ||||||||||||||||||||||||||||||
| Emb, Eth, Kana, Inat, Oflox, Sm, Teri, Pza |
X | X | X | |||||||||||||||||||||||||||||||
| Rifinah (Inh, Rif) | X | X | X | X | X | |||||||||||||||||||||||||||||
| Emb, Pyrifin (Inh, Rif, Pza) | X | X | ||||||||||||||||||||||||||||||||
| Emb, Eth, Sm, Inat, Pza | X | |||||||||||||||||||||||||||||||||
| Adherence, % | U | 78 | 78 | 97 | U | 95 | 85 | 85 | 100 | 38 | D | |||||||||||||||||||||||
| Point mutations | ||||||||||||||||||||||||||||||||||
| KatG315 | − | + | + | + | − | + | + | − | − | + | + | + | + | + | + | + | − | |||||||||||||||||
| rpoB531 | − | + | + | + | − | + | + | − | − | + | + | + | + | + | + | + | − | |||||||||||||||||
| Strain designation (DNA fingerprinting) |
g | d | d | d | h | b | b | e | e | d | d | d | d | c | c | c | e | |||||||||||||||||
| Strain population present (PCR) |
g + d | g + d | g + d | g + d | b + h | b | b + h | e | e | d + e | d + e | d + e | d + e | c | c + e | c | c + e | |||||||||||||||||
Definition of abbreviations: D = default (stopped therapy for a period of > 2 months); Emb = ethambutol; Eth = ethionamide; Inat = isoniazid and thiacetazone; Inh = isoniazid; Kana = kanamycin; nd = not determined; neg = negative; Oflox = ofloxacin; PCR = polymerase chain reaction; pos = positive; Pza = pyrazinamide; R = drug resistant; Rif = rifampin; S = drug sensitive; Sm = streptomycin; Teri = terizidone; U = unknown; + = mutation present; − = mutation absent.
For complete dataset, see online supplement (Figure E1).
Genotyping by PCR Amplification
According to the strain-specific PCR amplification method results, only 3 of the 11 patients with genetically distinct strains present on DNA fingerprint analysis were classified as confirmed or probable reinfection (Figure 1). Reinfection with an MDR strain during treatment with first-line antituberculous drugs was confirmed in Patient 7, an HIV-infected patient (Table 2). Reinfection could not be distinguished from mixed infection in the remaining two patients (Table 2, Patients 3 and 4) because their initial sputum culture was not available for genotypic analysis.
Five cases were classified as mixed infection based on analysis of the strain-specific PCR amplification results. The diagnosis of mixed infection on the basis of the DNA fingerprinting results in Patient 2 was confirmed by the PCR method (Figures 1 and 2). In addition, four patients classified as probable reinfection by DNA fingerprinting were reclassified as cases of mixed infection, because two genetically distinct strains could be identified by PCR in both the initial and subsequent sputum cultures (Figure 2 and Table 2; Patients 1, 5, 6, and 8).
In one patient classified as probable reinfection by the DNA fingerprinting method, the presence of a second strain could not be confirmed by PCR amplification in more than one sputum culture (data not shown), suggesting laboratory error (Figure 1).
Strain-specific primers were not available for the analysis of serial sputum cultures from the remaining two patients. Similarly, strain-specific primer sets were also not available for the 37 patients with MDR tuberculosis with genetically identical strains in serial sputum cultures, because the development of primers was dependent on the identification of different M. tuberculosis strains in serial sputum cultures.
Clinical and Demographic Characteristics of Patients with Mixed Infection and Reinfection
No difference could be found between patients infected with either a single or two genetically distinct strains, when comparing sex, age, type of tuberculosis, and treatment outcome (Table 3).
TABLE 3.
Characteristics of patients diagnosed with multi-drug-resistant tuberculosis between 1993 and 1998
| Single DNA Fingerprint, % (n = 37) |
Multiple DNA Fingerprints, %* (n = 10) |
p Value | |
|---|---|---|---|
| Male | 51 | 50 | 1.00 |
| Mean age, yr | 30.8 | 36.8 | 0.22 |
| Disease classification | |||
| Pulmonary TB | 95 | 100 | 1.00 |
| Primary TB | 5 | 0 | |
| Treatment history | |||
| New | 51 | 40 | 0.72 |
| Retreatment | 49 | 60 | |
| HIV status | |||
| Negative | 46 | 50 | 0.08 |
| Positive | 0 | 10 | |
| Unknown | 54 | 40 | |
| Smear at diagnosis | |||
| Positive | 54 | 80 | 0.26 |
| Negative | 35 | 20 | |
| Unknown | 11 | 0 | |
| Outcome | |||
| Culture negative | 59 | 60 | 0.68 |
| Culture positive | 22 | 10 | |
| Death | 14 | 20 | |
| Unknown | 5 | 10 |
Definition of abbreviation: TB = tuberculosis.
One patient for whom the sputum culture was identified as laboratory error by strain-specific polymerase chain reaction method was omitted from this group.
Mechanisms for Emergence and Reemergence of Strains in M. tuberculosis Sputum Cultures of Mixed Infection Cases
To explore for an association between treatment regimen, adherence, and changing strain populations during therapy in patients with MDR tuberculosis, the genotype data were compared with the drug-susceptibility patterns, the treatment regimen, and the adherence records (Figure 2 and Table 2).
Emergence of a drug-resistant strain on treatment with first-line antituberculosis drugs.
Analysis of the drug-susceptibility results of the sputum cultures of Patient 1 (Figure 2, Patient 1, lane A through D) shows fully susceptible initial sputum cultures followed by the appearance of MDR tuberculosis. Without additional information of genotypic data, this would have been interpreted as acquisition of drug resistance in the same strain. Conversely, according to the analysis of the DNA fingerprinting data, this patient would have been classified as confirmed reinfection with a genetically distinct MDR strain during therapy (Figure 2, Patient 1; compare lanes A and B to lanes C and D). In contrast to this, the PCR-based method showed that both the drug-susceptible and the MDR strains were present at diagnosis (Figure 2, Patient 1, lane A), classifying the patient as a case of mixed infection. These findings suggest that antibiotic pressure with a standard first-line regimen may have led to reduced growth of the drug-susceptible strain population, and selection and subsequent culture dominance of the previously undetected genetically distinct MDR strain population. Similar results were observed for Patient 2 (Figure 2), as well as Patients 5 and 6 (Table 2).
Treatment interruption and reemergence of a drug-susceptible strain in the patient with MDR tuberculosis.
Analysis of the drug-susceptibility results of the subsequent sputum cultures from Patient 1 shows the reemergence of drug-susceptible sputum cultures (Figure 2, Patient 1, lanes E and F) followed again by MDR tuberculosis (Figure 2, Patient 1, lanes G to I). Without additional information, the presence of the drug-susceptible strain would have been interpreted as inaccuracy of the phenotypic drug-susceptibility tests. Analysis of DNA fingerprinting data, however, confirms the presence of the drug-susceptible strain (the strain the patient was initially dually infected with), thereby refuting the hypothesis of inaccuracy of the phenotypic drug-susceptibility tests. The PCR-based method showed that both the drug-susceptible and the MDR strain were present throughout the 17-month treatment period. These results have demonstrated that in this patient with MDR tuberculosis, the underlying drug-susceptible strain reemerged and became the dominant strain population in the sputum cultures after partial or complete removal of the antibiotic pressure through poor adherence or default (Figure 2, Patient 1, lane D). The data for Patients 3 and 8 showed a similar pattern, with the underlying drug-susceptible strain reemerging after a period of poor treatment adherence or default (Table 2, Patient 3, lane J and K, and Patient 8, lane C and D).
Treatment with second-line drugs and reemergence of a drug-susceptible strain in a patient with MDR tuberculosis.
Analysis of the drug-susceptibility results of the sputum cultures from Patient 2 shows the presence of drug-susceptible and drug-resistant strains at time of diagnosis (Figure 2, Patient 2, lanes A and B). Without additional information, this would have been interpreted as inaccuracy of the phenotypic drug-susceptibility tests. Analysis of DNA fingerprinting data and PCR-based data confirmed the presence of both a drug-susceptible and a drug-resistant strain at diagnosis, indicating mixed infection. Three months after starting second-line treatment, the drug-susceptibility tests demonstrate the presence of a fully susceptible sputum culture (Figure 2, Patient 2, lane F), which was confirmed by DNA fingerprinting and PCR-based genotyping methods. Together, these results suggest that, in this patient, the lowered antibiotic pressure was not due to poor adherence (adherence was 92 to 100% throughout treatment) but rather due to treatment with less effective second-line drugs. A similar result was observed for Patient 4 (Table 2).
The analysis of the combined data (clinical, phenotypic drug susceptibility test, mutation detection analysis, IS6110 DNA fingerprinting analysis, and strain-specific PCR amplification) allowed most observations to be resolved; however, some results remained discordant. In all of these instances, the phenotypic drug-susceptibility test data were in conflict with the results of mutation analysis. In three cases, the culture-based phenotyping method failed to detect the presence of the drug-resistant strain, despite this strain being overrepresented in these cultures (Table 2, Patients 3, lanes B and H, 4, lane F, and 7, lane C). This result demonstrates poor-quality drug-susceptibility testing. In the remaining two cases, the mutation analysis failed to detect the presence of the drug-resistant strain, probably as a result of the preferential amplification of the overrepresented genomic locus of the drug-susceptible strain in comparison to the underrepresented genomic locus in the underlying drug-resistant strain (Table 2, Patients 6, lane A, and 8, lane D).
DISCUSSION
The use of the standard IS6110 DNA fingerprinting method has provided insight into the relative importance of recent infection and reactivation (24, 25) and has established reinfection as a mechanism leading to the recurrence of tuberculosis (3, 5, 9). DNA fingerprinting has also been used to gain insight into the mechanisms resulting in changing drug-susceptibility patterns during the course of disease (17, 26, 27). These studies have demonstrated that reinfection, before or during therapy, can be a mechanism leading to the development of drug resistance. Using DNA fingerprinting as the gold standard, our study supports reinfection as a mechanism leading to changing drug-susceptibility patterns.
Our PCR-based strain-typing method, challenged, in certain instances, the validity of the interpretation based on the DNA fingerprinting data. The only patient with confirmed reinfection by both methods was coinfected with HIV and therefore was probably unable to resist reinfection with a drug-resistant strain despite receiving therapy for the drug-susceptible tuberculosis (4). The PCR-based strain-typing method contested the DNA fingerprinting classification of reinfection in certain cases by demonstrating the presence of undetected drug-resistant strains (not detected by drug-susceptibility testing or DNA fingerprinting) in the initial sputum culture. It is unlikely that these results reflect cross-contamination because the strains causing mixed infection were found to be present in multiple sputum cultures taken from these patients on different occasions. Only one isolate from one patient was classified as cross-contamination. This level of cross-contamination is similar to the 3.8% previously described for this laboratory (19).
We present new data to support three mechanisms whereby mixed infections can lead to changing drug-susceptibility patterns during therapy. We suggest first that, during the initial treatment period, the first-line antibiotics reduced the drug-susceptible strain population, while allowing the drug-resistant strain population to grow, thereby converting the patient from an apparently drug-susceptible tuberculosis case to an MDR tuberculosis case. We propose that this represents a mechanism of selection through antibiotic pressure.
Second, when the antibiotic pressure was removed by poor adherence or default, the underlying drug-susceptible strain population reemerged as the dominant population. This suggests a mechanism of selection in the absence of antibiotic pressure. The reason for the observed “overgrowth” by the drug-susceptible population remains unknown. We propose that this could reflect a difference in the level of “fitness” between the drug-susceptible and drug-resistant populations present in these patients. Similar observations were made when drug-susceptible and drug-resistant populations were cultured in vitro and in macrophage cell lines (28, 29). The reemergence of the drug-susceptible population demonstrates that the initial period of therapy in these patients was insufficient to enable complete sterilization. This supports the need to ensure adherence over the full course of therapy.
Third, we observed the reemergence of the underlying drug-susceptible strain population when the antibiotic pressure was changed by the introduction of second-line therapy. It is well known that second-line antibiotics have lower bactericidal activities when compared with first-line antibiotics and therefore we propose a mechanism of selection due to a reduced antibiotic pressure. This study supports the suggestion by Post and coworkers (30) that the treatment of patients with MDR tuberculosis may require antibiotics which target both drug-susceptible and drug-resistant subpopulations.
Comparison of the clinical parameters between patients infected with a single strain or multiple strains did not identify significant differences. However, because this study was not designed to detect a difference between these patient groups and precludes the period of disease before diagnosis, we cannot make definite conclusions on the influence of mixed infection on the disease presentation, disease progression, or treatment outcome. Most important, this study demonstrates the inability of routine culture-based drug-susceptibility methods to identify the presence of underlying drug-resistant strains, which is of concern for the patient and their contacts. Diagnostic delays could prolong the implementation of an appropriate treatment regimen and thereby extend the window of opportunity for transmission. Furthermore, inappropriate therapy during the period in which drug-susceptibility testing is being done could enhance the risk of the drug-resistant strain acquiring additional resistance mutations.
We acknowledge that the conclusions drawn from this study are limited by the small number of patients, although, with this study, we did not aim to provide a measure of the frequency of these processes but rather to demonstrate the mechanisms and the fact that they are able to occur in an epidemic setting. It is, however, warranted that our hypothesis generated on the basis of a limited number of observations be confirmed in different geographic settings.
In conclusion, this study describes the first molecular analysis of the population structure of drug-resistant and drug-susceptible M. tuberculosis strains in serial sputum cultures during the course of MDR tuberculosis. We have demonstrated that mixed infection is an important mechanism underlying changing drug-susceptibility patterns in a high-incidence region. Drug-susceptibility patterns change through the presence or absence of antibiotic pressure, which determines the dominant growth of the coinfecting strain. The inability to accurately determine resistance patterns in cases of mixed infection may exacerbate delays in the diagnosis of drug-resistant tuberculosis, which could have implications for the individual patient and the spread of drug-resistant strains.
Supplementary Material
Acknowledgments
The authors thank the National Health Laboratory Service for providing access to specimens, Mrs. A. Huysamen for her excellent technical support and the preparation of cultures, and Mrs. E. Hanekom-Keet for DNA sequencing. They thank Dr. I. Toms, Department of Health, City of Cape Town, for permission to do research in the clinics, and they are indebted to the residents of the epidemiologic field site.
Supported by GlaxoSmithKline Action TB Program for funding the collection of clinical and demographic data, sputum culturing, and DNA fingerprinting; the Harry Crossley Foundation and the National Research Foundation (project 2054201 and the DST/NRF Centre of Excellence for Biomedical TB Research) for funding the development of the polymerase chain reaction–based strain-typing method; and the National Institutes of Health (R21 A155800-01) and the Wellcome Trust (DDS PC3145) for funding the identification of genotypic mechanisms conferring drug resistance.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Conflict of Interest Statement: A.v.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.C.V. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.R. is paid a salary from a research grant sponsored by GlaxoSmithKline (GSK) and does not have a conflict of interest. R.J. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.D.v.d.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. N.B. received £500 in 2003 for speaking at a conference where one session was sponsored by GSK. In 2004, she was reimbursed for travel to give a talk at a “Diseases of the Developing World” day held at GSK, United Kingdom. She received £111,150 in 2000 and £150,000/year for 2001–2003 from the GSK Action TB Program as research grants for developing and maintaining an epidemiologic field site and for doing a study aimed at identifying surrogate markers for response to treatment in patients with tuberculosis. From 2004–2005, funding (£72,000/year) is received from GSK for maintaining the epidemiology field site. N.C.G.v.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.D.v.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript R.M.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Stead WW. Pathogenesis of a first episode of chronic pulmonary tuberculosis in man: recrudescence of residuals of the primary infection or exogenous reinfection? Am Rev Respir Dis 1967;95:729–745. [DOI] [PubMed] [Google Scholar]
- 2.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993;31:406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Rie A, Warren R, Richardson M, Victor TC, Gie RP, Enarson DA, Beyers N, van Helden PD. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med 1999;341:1174–1179. [DOI] [PubMed] [Google Scholar]
- 4.Small PM, Shafer RW, Hopewell PC, Singh SP, Murphy MJ, Desmond E, Sierra MF, Schoolnik GK. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in patients with advanced HIV infection. N Engl J Med 1993;328:1137–1144. [DOI] [PubMed] [Google Scholar]
- 5.Caminero JA, Pena MJ, Campos-Herrero MI, Rodriguez JC, Afonso O, Martin C, Pavon JM, Torres MJ, Burgos M, Cabrera P, et al. Exogenous reinfection with tuberculosis on a European island with a moderate incidence of disease. Am J Respir Crit Care Med 2001;163:717–720. [DOI] [PubMed] [Google Scholar]
- 6.Garcia de Viedima D, Marin M, Henangomez S, Diaz M, Ruiz Serrano MJ, Alcala L. Tuberculosis recurrences: reinfection plays a role in a population whose clinical/epidemiological characteristics do not favor reinfection. Arch Intern Med 2002;162:1873–1879. [DOI] [PubMed] [Google Scholar]
- 7.Horn DL, Hewlett D Jr, Haas WH, Butler WR, Alfalla C, Tan E, Levine A, Nayak A, Opal SM. Superinfection with rifampin-isoniazid-streptomycin-ethambutol (RISE)-resistant tuberculosis in three patients with AIDS: confirmation by polymerase chain reaction fingerprinting. Ann Intern Med 1994;121:115–116. [DOI] [PubMed] [Google Scholar]
- 8.Niemann S, Richter E, Rusch-Gerdes S, Schlaak M, Greinert U. Double infection with a resistant and a multidrug-resistant strain of Mycobacterium tuberculosis. Emerg Infect Dis 2000;6:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnenberg P, Murray J, Glynn JR, Shearer S, Kambashi B, Godfrey-Faussett P. HIV-1 and recurrence, relapse, and reinfection of tuberculosis after cure: a cohort study in South African mineworkers. Lancet 2001;358:1687–1693. [DOI] [PubMed] [Google Scholar]
- 10.Jasmer RM, Bozeman L, Schwartzman K, Cave MD, Saukkonen JJ, Metchock B, Khan A, Burman WJ, and the Tuberculosis Trials Consortium. Recurrent tuberculosis in the United States and Canada: relapse or reinfection? Am J Respir Crit Care Med 2004;170:1360–1366. [DOI] [PubMed] [Google Scholar]
- 11.Verver S, Warren RM, Beyers N, Richardson M, van der Spuy GD, Borgdorff MW, Enarson DA, Behr MA, van Helden PD. Rate of reinfection TB after successful treatment is higher than the rate of new TB. Am J Respir Crit Care Med 2005;171:1430–1435. [DOI] [PubMed] [Google Scholar]
- 12.Mankiewicz E, Liivak M. Phage types of Mycobacterium tuberculosis in cultures isolated from Eskimo patients. Am Rev Respir Dis 1975;111:307–312. [DOI] [PubMed] [Google Scholar]
- 13.Bates JH, Stead WW, Rado TA. Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am Rev Respir Dis 1976;114:353–358. [DOI] [PubMed] [Google Scholar]
- 14.Yeh RW, Hopewell PC, Daley CL. Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int J Tuberc Lung Dis 1999;3:537–539. [PubMed] [Google Scholar]
- 15.du Plessis DG, Warren R, Richardson M, Joubert JJ, van Helden PD. Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS6110 and other repetitive element-based DNA fingerprinting. Tuberculosis (Edinb) 2001;81:211–220. [DOI] [PubMed] [Google Scholar]
- 16.Richardson M, Carroll NM, Engelke E, van der Spuy GD, Salker F, Munch Z, Gie RP, Warren RM, Beyers N, van Helden PD. Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J Clin Microbiol 2002;40:2750–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braden CR, Morlock GP, Woodley CL, Johnson KR, Colombel AC, Cave MD, Yang ZH, Valway SE, Onorato IM, Crawford JT. Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin Infect Dis 2001;33:E42–E47. [DOI] [PubMed] [Google Scholar]
- 18.Pavlic M, Allerberger F, Dierich MP, Prodinger WM. Simultaneous infection with two drug-susceptible Mycobacterium tuberculosis strains in an immunocompetent host. J Clin Microbiol 1999;37:4156–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia de Viedma D, Marin M, Ruiz Serrano MJ, Alcala L, Bouza E. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J Infect Dis 2003;187:695–699. [DOI] [PubMed] [Google Scholar]
- 20.Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, van Helden PD. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med 2004;169:610–614. [DOI] [PubMed] [Google Scholar]
- 21.Behr MA. Tuberculosis due to multiple strains: a concern for the patient? A concern for tuberculosis control? Am J Respir Crit Care Med 2004;169:554–555. [DOI] [PubMed] [Google Scholar]
- 22.Verver S, Warren RM, Munch Z, Vynnycky E, van Helden PD, Richardson M, van der Spuy GD, Enarson DA, Borgdorff MW, Behr MA, et al. Transmission of tuberculosis in a high incidence urban community in South Africa. Int J Epidemiol 2004;33:351–357. [DOI] [PubMed] [Google Scholar]
- 23.Victor TC, Jordaan AM, van Rie A, van der Spuy GD, Richardson M, van Helden PD, Warren R. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuber Lung Dis 1999;79:343–348. [DOI] [PubMed] [Google Scholar]
- 24.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N Engl J Med 1994;330:1703–1709. [DOI] [PubMed] [Google Scholar]
- 25.Alland D, Kalkut GE, Moss AR, McAdam RA, Hahn JA, Bosworth W, Drucker E, Bloom BR. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med 1994;330:1710–1716. [DOI] [PubMed] [Google Scholar]
- 26.Shafer RW, Singh SP, Larkin C, Small PM. Exogenous reinfection with multidrug-resistant Mycobacterium tuberculosis in an immunocompetent patient. Tuber Lung Dis 1995;76:575–577. [DOI] [PubMed] [Google Scholar]
- 27.Kruuner A, Pehme L, Ghebremichael S, Koivula T, Hoffner SE, Mikelsaar M. Use of molecular techniques to distinguish between treatment failure and exogenous reinfection with Mycobacterium tuberculosis. Clin Infect Dis 2002;35:146–155. [DOI] [PubMed] [Google Scholar]
- 28.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2004;48:1289–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toungoussova OS, Caugant DA, Sandven P, Mariandyshev AO, Bjune G. Impact of drug resistance on fitness of Mycobacterium tuberculosis strains of the W-Beijing genotype. FEMS Immunol Med Microbiol 2004;42:281–290. [DOI] [PubMed] [Google Scholar]
- 30.Post FA, Willcox PA, Mathema B, Steyn LM, Shean K, Ramaswamy SV, Graviss EA, Shashkina E, Kreiswirth BN, Kaplan G. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis 2004;190:99–106. [DOI] [PubMed] [Google Scholar]
- 31.van Rie A, Victor T, Richardson M, Johnson R, van der Spuy G, Murray M, Beyers N, Gey van Pittius NC, van Helden PD, Warren RM. Mixed Mycobacterium tuberculosis infection: changing drug-resistance patterns through antibiotic selection. Molecules-R-US, the 19th meeting of the South African Society of Biochemistry and Molecular Biology, Stellenbosch, South Africa, 2005.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.