Abstract
Restoration of functionally intact chromatin structure following DNA damage processing is crucial for maintaining genetic and epigenetic information in human cells. Here, we show the UV-induced uH2A foci formation in cells lacking XPC, DDB2, CSA or CSB, but not in cells lacking XPA, XPG or XPF indicating that uH2A incorporation relied on successful damage repair occurring through either GGR or TCR sub-pathway. In contrast, XPA, XPG or XPF were not required for formation of γH2AXfoci in asynchronous cells. Notably, the H2A ubiquitin ligase Ring1B, a component of Polycomb repressor complex 1, did not localize at DNA damage sites. However, histone chaperone CAF-1 showed distinct localization to the damage sites. Knockdown of CAF-1 p60 abolished CAF-1 as well as uH2A foci formation. CAF-1 p150 was found to associate with NER factors TFIIH, RPA p70 and PCNA in chromatin. These data demonstrate that successful NER of genomic lesions and prompt CAF-1-mediated chromatin restoration link uH2A incorporation at the sites of damage repair within chromatin.
Keywords: DNA damage, DNA repair, Nucleotide excision repair, Ubiquitinated histone H2A, CAF-1, Chromatin assembly
1. Introduction
DNA damage response, including damage-induced checkpoint signaling and DNA repair pathways, enables cells to overcome genotoxicity and maintain genomic integrity. A wide variety of helix-distorting DNA lesions including ultraviolet light (UV)-induced cyclobutane pyrimidine dimers (CPD) and 6-4 pyrimidine-pyrimidone photoproducts (6-4PP) are eliminated by nucleotide excision repair (NER) [1]. Defects in NER are associated with several rare autosomal recessive genetic disorders, e.g., Xeroderma pigmentosum (XP) and Cockayne syndrome (CS). Seven XP proteins, corresponding to XP complementary group A to G, have been identified; whereas two CS proteins, CSA and CSB, have been discovered. These components constitute two distinct NER sub-pathways: global genomic repair (GGR), which operates throughout the genome, and transcription-coupled repair (TCR), which processes the damage within transcribed DNA strands of transcriptionally active genes. Biochemically, NER includes damage recognition, dual incision, and gap-filling DNA synthesis steps [2,3]. In GGR, DNA lesions are recognized by concerted action of UV-damaged DNA-binding protein (DDB) and XPC-hHR23B protein complexes [4,5]. Transcription factor II H (TFIIH) protein complex, containing XPB and XPD and other components, is recruited by XPC complex to open the DNA helix around the damage site [6–8]. In TCR, lesions are detected by stalling of RNA polymerase II in coordination with recognition of stalled transcription by XPG, CSB and TFIIH [9,10]. Other NER factors, such as XPA and RPA are believed to join the TFIIH-containing repair complex to verify the nature of DNA structure alteration [11]. Endonucleases XPF-ERCC1 and XPG are responsible for dual incision and removal of the damage containing oligonucleotide of ~24–32 nt [8]. Subsequent gap-filling DNA synthesis is performed by concerted action of pol δ or pol ε and the cofactors PCNA, RF-C as well as RPA.
In eukaryotic cells, the NER machinery operates on lesions situated within chromatin and the folding of DNA within histones into nucleosome and higher order chromosomal structure [12] poses a crucial structural barrier for lesion repair. An “access-repair-restore” model was proposed to delineate how repair machinery operates on chromatin-embedded substrates [13,14]. During access stage, nucleosome organization is transiently disrupted to expose the chromatin-embedded lesions to the repair machinery. The rearrangement or alteration of chromatin may occur through different mechanisms, e.g., post-translational histone modifications, chromatin remodelling and disruption of nucleosomal structure due to removal of histones [15,16]. Restoration of chromatin structure after DNA repair involves histones chaperones, and, among them, chromatin assembly factor 1 (CAF-1) has been suggested to play a pivotal role in chromatin assembly after DNA replication and repair [17–22].CAF-1 consists of three subunits,p150, p60 and p48. During DNA replication,CAF-1complex binds to newly synthesized histone H3 and H4 and deposits the histone tetramers onto replicating DNA to form chromatin precursor in a PCNA-dependent manner [17]. The replicated precursor then serves as the template for deposition of either old or new histone H2A and H2B [20,23]. Similar to its role in chromatin assembly during DNA replication, CAF-1 is also presumed to couple chromatin assembly to NER by acting at the sites of damage and repair [20–22]. How chromatin restoration is actually achieved in vivo, nevertheless, remains unclear. Chromatin restoration does not simply recycle histones, but new histones and histones with distinct post-translational modification are incorporated into restored chromatin. For example, new histone H3.1, deposited during DNA replication, is incorporated into chromatin as a marker for the sites of NER of UV-induced DNA damage [24]. Furthermore, it was recently found that restoration of the chromatin following double-strand break repair is driven by acetylated H3lysine 56 and signals completion of repair [25,26]. The chromatin restoration and histone marks are believed to present the critical trigger for checkpoint recovery, extending the “histone code” hypothesis [27] to nucleosome assembly. It is possible that multiple histone modifications are instrumental for DNA repair related chromatin assembly by histone chaperones, and, some of these histone modifications play a regulatory role in resuming cell cycle after completion of DNA repair.
Our present work has focused on the recruitment of ubiquitinated histone H2A (uH2A) during NER of UV-induced lesions in human cells, to address the post-repair chromatin restoration in relation to histone modifications within damaged chromatin in vivo. The uH2A, comprising about 5–15% of available H2A, is the most abundant histone modification in higher eukaryotic cells [28]. Establishing the function of this histone modification has remained elusive. Recent identification of enzymes involved in histone H2A ubiquitination and deubiquitination has revealedcertain functional aspects of this modification in gene silencing, transcriptional regulation and cell cycle progression [29–31]. Monoubiquitination of nucleosome histone H2A at lysine 119 by Polycomb repressor complex 1 (PRC1), an E3 ubiquitin ligase complex, regulates polycomb silencing [29]. In mouse embryonic cells, PRC1 protein Ring1B is required to maintain global uH2A level. Moreover, both Ring1A and Ring1B have an overlapping function in maintaining uH2A on inactive X chromosome in differentiated cells in female mammals [30]. In connection with NER, histone H2A is suggested to be one of the substrates of Cul4A-DDB ubiquitin ligase [32]. The Cul4A-DDB ubiquitin ligase contains DDB heterodimeric complex of DDB1 and DDB2, whose function is lost due to amutation in DDB2 gene representing XP complementary group E [33]. It has been proposed that Cul4A-DDB ubiquitin ligase also ubiquitinates histone H3 and H4 in facilitating NER [34]. We have previously demonstrated that Cul4A-DDBubiquitin ligase is recruited to the sites of DNA damage through its damage recognition via DDB[35,36]. Interestingly, a recent study demonstrated UV damage induced monoubiquitination of histone H2A in the vicinity of DNA lesions [37]. The monoubiquitination of H2A was shown to be specifically dependent on functional GGR pathway and required Ring1B. Furthermore, the UV-induced ubiquitination of H2A was dependent on the DNA damage signaling kinase ATR, and therefore, it forms part of the cellular response to UV damage. We now report on the recruitment of uH2A to sites of DNA damage as a post-excision repair event, in which transiently disrupted chromatin is restored through repair synthesis-coupled chromatin assembly. We show that formation of uH2A foci do not involve pre-incision events mediated by Cul4A-DDB ubiquitin ligase, but require successful NER through either GGR or TCR sub-pathway. These pathways, however, are not required for formation of γH2AXfoci, which is a signature of DNA damage signaling by ATM or ATR kinase, suggesting different cellular responses are involved in uH2A foci formation. We further demonstrate that Ring1B does not localize to DNA damage, and propose a model foruH2A incorporation through chromatin restoration by histone chaperone CAF-1, rather than Ring1B-mediated local monoubiquitination of histone H2A.
2. Materials and methods
2.1. Cell culture
HeLa cells stably expressing N-terminal FLAG-HA-DDB2 (HeLa-DDB2) were provided by Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute, Boston, MA). HeLa-DDB2 cells stably expressing V5-His epitope-tagged XPC (HeLa-DDB2-XPC) or CBP (calmodulin-binding peptide) and SBP (streptavidin-binding peptide) epitope-tagged ubiquitin were established and selected with G418, sub-cloned further by single cell dilution. XP and CS fibroblasts, including GM04429 (XP-A, XP12BE), GM15983 (XPC, XP4PA-SV-EB), GM02096 (XP-C, XP1MI), GM02415 (XP-E, XP2RO), GM04313 (XP-F, XP2YO), GM03021 (XPG, XP2BI), GM0296 (CS-A) and GM01098 (CS-B), were obtained from the Coriell Cell Repository. The cell lines were grown in DMEM or MEM supplemented with 10%fetal calf serum, antibiotics and without or with 500 µg/ml G418 at 37 °C, in a humidified atmosphere of 5% CO2.
2.2. Antibodies
Primary rabbit anti-XPC and anti-CPD antibodies were raised and characterized in our laboratory as previously described [35,38]. Anti-HA (clone 12CA5), anti-uH2A (clone E6C5) and anti-γH2AX (SER139) monoclonal antibodies were purchased from Roche Diagnostics (Indianapolis, IN) and Upstate Biotechnology (Lake placid, NY), respectively. Monoclonal CAF-1 p150 antibody (ab7655), CAF-p60 antibody ab8133 and clone p60-24 were from Abcam Inc. (Cambridge, MA) and MBL International (Woburn, MA). Other primary antibodies were monoclonal anti-Sug-1 and anti-Rad23B from BD Biosciences (San Jose, CA), anti-XPA (12F5) from NeoMarkers (Fremont, CA), rabbit polyclonal antibody against S1 subunit of proteasome 19S from Affinity BioReagents (Golden, CO), rabbit polyclonal anti-XPB (S-19), anti-XPA(FL-273) and anti-XPF (H-300), goat anti-PCNA (C-20), monoclonal anti-PCNA (PC-10), anti-XPG (8H7) and anti-p62 (G-10) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA), rabbit polyclonal anti-CBP epitope antibody from Upstate Biotechnology, as well as goat anti-hHR23A from Rockland Inc. (Gilbertsville, PA). Fluorescent-conjugated antibodies were from Santa Cruz Biotechnology.
2.3. UV irradiation
The cells at desired stages of growth were washed twice with phosphate-buffered saline (PBS). The UV-C light (254 nm) was delivered at a dose rate of 0.5 J/m2/sec as measured by a Kettering model 65 radiometer (Cole-Palmer, Vernon Hills, IL). For local UV irradiation, the cells, grown on glass coverslips, were washed with PBS and an isopore polycarbonate filter (Millipore, Bedford, MA) with a pore size of 3, 5, or 8 µm diameter, was placed on top of the cell monolayer. The coverslips were irradiated and the cells were processed immediately, or maintained in a suitable medium for the desired period and processed thereafter.
2.4. Immunofluorescence
The immunofluorescence staining was conducted according to the method established in our laboratory [39]. Briefly, the micro-pore UV irradiated cells were washed twice with cold PBS and then fixed with 2% paraformaldehyde in 0.5% Triton X-100 at 4 °C for 30 min. For immunofluorescent detection of uH2A, the cells were successively rinsed with PBS, washed twice with CSK buffer (100 mM NaCl, 300 mM Sucrose, 10 mM PIPES pH 7.0, 3 mM MgCl2), permeabilized with CSK buffer plus 0.5% Triton X-100 for 8 min at room temperature and then fixed with 2% paraformaldehyde in PBS. For DNA denaturation, the cells were incubated in 2N HCl for 5min at 37°C. After cell fixation, the coverslips were rinsed three times with PBS and blocked with 20% normal goat serum (NGS) in washing buffer (0.1% Triton X-100/PBS) at room temperature for 30 min. Primary rabbit anti-XPC, anti-CPD, anti-S1 and mouse monoclonal anti-HA, anti-uH2A, anti-Sug1, and anti-hHR23 B antibodies (1:50 to 1:1000 dilution) as well as fluorescent (FITC or Texas Red)-conjugated secondary antibodies (1:200 to 1:1000 dilution) were all prepared in washing buffer containing 5% NGS and layered on the coverslips for 1 h at room temperature. Following each antibody incubation step, the cells were washed with 0.1% Tween-20 in PBS four times for 5 min each. Fluorescence images were obtained with a Nikon Fluorescence Microscope E80i (Nikon, Tokyo, Japan). The digital images were then captured with a cooled CCD camera and processed with SPOT analysis software (Diagnostic Instruments, Sterling Heights, MI).
2.5. Co-immunoprecipitation, cross-linking and chromatin immunoprecipitation
The HeLa-DDB2-XPC cells grown to ~70% confluence were left untreated or UV-irradiated at 20 J/m2 and incubated further for h in fresh medium. For regular co-immunoprecipitation, the cells were washed twice with PBS then lysed in RIPA buffer (50 mM Tris–HCl [pH, 7.5], 150 mM NaCl, 5mM EDTA, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS) containing a protease inhibitor cocktail. For in vivo cross-linking, the cells were synchronized in G0 phase by maintaining in DMEM media with 0.2% fetal calf serum for minimum of 5 days. The serum-starved cells were UV irradiated and incubated further for 1 h in fresh medium. The cells were then washed twice with PBS and fixed with 1% formaldehyde in PBS at room temperature for 10 min, followed by addition of glycine to a final concentration of 125 mM and incubating for 5 min. The chromatin solution was made by sonication of cell lysates in RIPA buffer on ice to break the DNA to ~500 bp fragments. The whole cell extracts or chromatin solutions containing ~2 mg protein were precleared with protein A/G agarose beads then incubated with ~2 µg specific antibodies in RIPA buffer at 4 °C overnight, followed by addition of 25 µl protein A/G agarose beads and incubation for another 3 h. The matrix beads were washed 4 times with RIPA buffer and the bound proteins were eluted with 50 µl of Laemmli SDS sample buffer. The immunoprecipitates were boiled for 5 min or incubated at 95 °C for 1 h to reverse cross-links prior to electrophoresis.
2.6. RNA interference
Small interference RNA oligonucleotides were obtained from Dharmacon (Lafayette, CO). The sequences targeting human CAF-1 p60 gene are 286AATGATAACAAGGAGCCGGAG306 and 1577AAATTCAGTCAGAGACGCCTG1597 [40]. The later was chosen through pilot experiments and nontarget siRNA was used as a control. The siRNA transfection experiments were carried out using Lipofectamine transfection reagent according to manufacturer’s instruction. Briefly, the Lipofectamine 2000 and OPTI-MEM medium (Invitrogen, Carlsbad, CA) were mixed for 5 min and then incubated with siRNA for 20 min at room temperature. After addition of proper amount of 10%-FBS/DMEM medium to the mixture, the siRNA-Lipofectamine mix was then applied to the cell culture. For immunofluorescence staining experiments, the siRNA transfection was performed for 48 h with the cells grown on coverslips in 60-mm dishes.
3. Results
3.1. Accumulation of ubiquitin conjugates at DNA damage sites
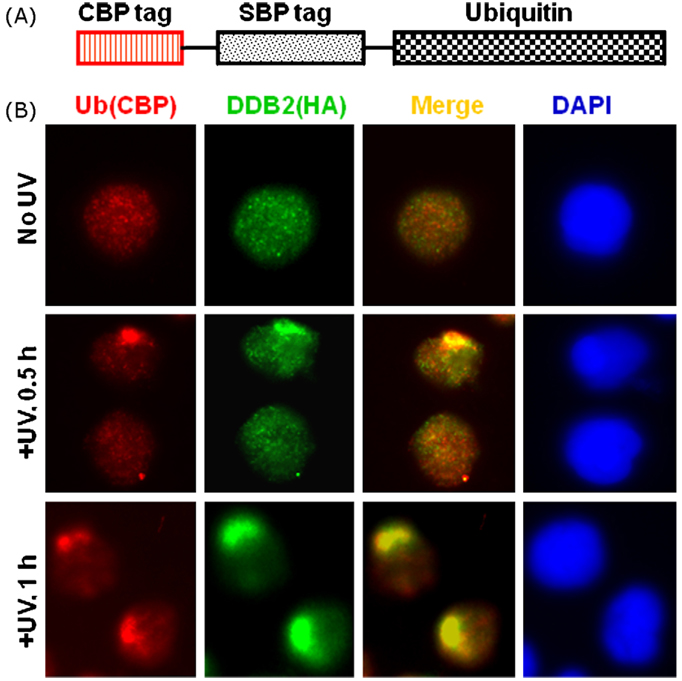
We have previously demonstrated that Cul4A-DDB ubiquitin ligase is recruited to local DNA damage sites immediately after UV irradiation [35]. In search of the ligase substrates other than NER factors XPC and DDB2, CBP and SBP-tagged ubiquitin (Fig. 1A) were stably transfected in HeLa-DDB2 cells expressing HA and FLAG-tagged DDB2. We first tested the competence of epitope-tagged ubiquitin for conjugating function in transfected cells. High molecular weight ubiquitin conjugates were detected by Western blot analysis of extracts from cells treated with 10 µM proteasome inhibitor MG 132, indicating that the epitope-tagged ubiquitin is functional and can appropriately conjugate with target proteins (data not shown). The epitope-tagged ubiquitin expressing cells were then examined by immunofluorescence for the accumulation of DDB2 and epitope-tagged ubiquitin at damaged sites introduced by local UV irradiation through a 5 µm micropore polycarbonate filter. As shown in Fig. 1B, both Triton X-100-resistant fraction of epitope-tagged ubiquitin as well as DDB2 are uniformly distributed within the nuclei of unirradiated cells. Local UV irradiation provoked an accumulation of DDB2 (HA, green) and ubiquitin (CBP, red) to the irradiated subnuclear spots at 0.5 and 1 h post-irradiation (Fig. 1B). The ubiquitin clearly co-localized with DDB2 foci, indicating that the ubiquitin conjugates were readily targeted to the DNA damage sites. The data provided an initial impetus for the speculation that some NER factors or proteins related to UV damage response are either ubiquitinated at the damaged site or ubiquitinated proteins within cells are promptly mobilized to DNA damage following UV irradiation.
Fig. 1.
Immunofluorescent detection of ubiquitin after micropore UV irradiation in cells expressing epitope-tagged ubiquitin. (A) Diagramrepresents CBP (calmodulin binding peptide)-and SBP (streptavidin binding peptide)-tagged ubiquitin stably transfected in HeLa-DDB2 cells, which also express HA and FLAG-tagged UV-damaged DNA binding protein 2 (DDB2). (B) Immunofluorescent detection of ubiquitin by immunostaining of CBP (red) after local UV irradiation at 100 J/m2 delivered through a polycarbonate micropore (5 µm) filter. The accumulation of DDB2 at damage spots were determined by immunostaining of HA tags (green). The merged images show co-localization of ubiquitin and DDB2. Nuclei of cells were illustrated with DAPI staining (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2. uH2A localizes to DNA damage sites and persists in chromatin
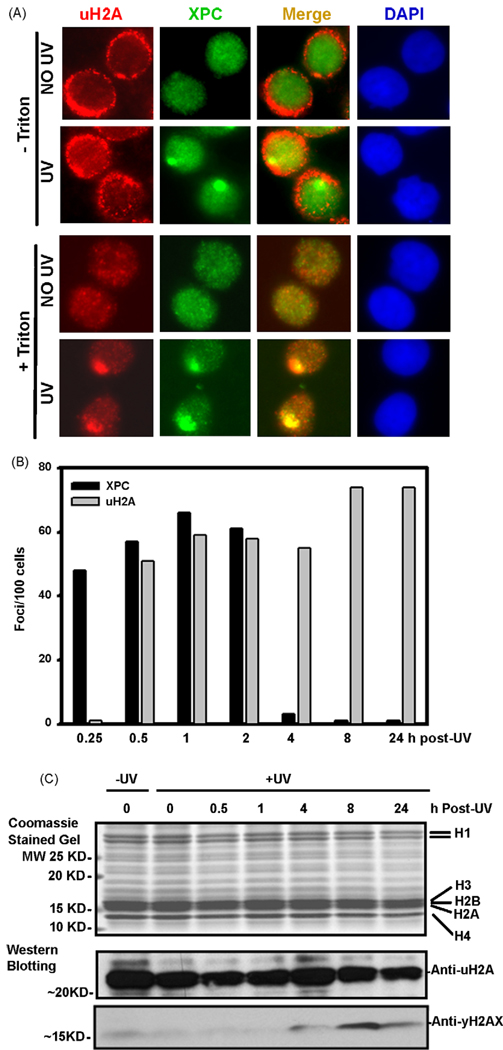
It has been suggested that histone H2A is a substrate of Cul4A-DDB ubiquitin ligase, and DNA damage triggers NER-dependent monoubiquitination of histone H2A [32,37]. We, therefore, examined the cellular distribution of uH2A, accumulation of uH2A at DNA damage sites and dynamics of uH2A foci formation upon UV irradiation. Fig. 2 shows that unless cells were pretreated with 0.5% Triton X-100 before fixation, uH2A could be detected both in cytoplasm and nuclei. Highly intense fluorescent signal (red) from cytoplasmic uH2A indicated that considerable amount of uH2A resides in the cytoplasm. On the contrary, XPC (green) resides exclusively within the nuclei (Fig. 2A). As shown earlier, local UV irradiation triggered an instant accumulation of XPC within damaged sites indicating an initiation of NER process. Although uH2A appeared to co-localize with XPC foci, the results were mostly equivocal. Nevertheless, treatment of cells with 0.5% Triton X-100 depleted the cytoplasmic and unbound nuclear uH2A to allow distinct visualization of chromatin-associated uH2A and its co-localization with XPC foci. Thus, uH2A also spatially tracks to the DNA damage sites as observed with other NER factors. Nonetheless, appearance of XPC and uH2A foci exhibits a clear temporal difference. For example, XPC foci appeared readily and could be seen in almost all damaged cells within 15 min and completely disappeared at 4 h after local UV irradiation (Fig. 2B). On the other hand, the uH2A foci did not form until 30 min and persisted for at least 24 h post-irradiation. Notably, however, majority (~90%) of uH2A foci co-localized with XPC foci within 2 h after UV irradiation and once formed do not disappear from the lesion sites. Relatively rapid loss of XPC foci could be attributed to fast repair of 6-4 photoproducts. These results provide evidence that formation of uH2A foci is not directly associated with damage recognition and recruitment of XPC. More importantly, uH2A shown to co-localize with XPC is incorporated in chromatin, which makes uH2A resistant to Triton X-100 extraction.
Fig. 2.
uH2A forms Triton X-100-resistant foci at DNA damage spot after micropore UV irradiation. (A) HeLa-DDB2 cells were locally UV irradiated and incubated further for 1 h. The cells were pre-treated with CSK buffer containing 0.5% Triton X-100 and then fixed or directly fixed with 2% paraformaldehyde in presence of 0.5% Triton X-100 in PBS. The uH2A (red) and XPC (green) proteins were visualized by indirect immunofluorescence with specific antibodies. (B) uH2A and XPC proteins were detected by indirect immunofluorescence at different time points of post-UV incubation. The XPC and uH2A foci were counted from 3 to 5 fields and average number of foci per 100 cells were plotted to show dynamics of appearance and disappearance of both XPC and uH2A foci. (C) Acid extracts were made from 20 J/m2 UV irradiated HeLa-DDB2 cells at indicated time after global UV irradiation. The proteins were separated by 16% acrylamide gel, stained with Coomassie bright blue (upper panel) or immunoblotted and analyzed for uH2A and γH2A (lower two panels). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
We also determined the cellular level of unmodified and modified proteins using acid-extracted histones from unirradiated and UV irradiated HeLa cells (Fig. 2C). UV irradiation had no significant effect on the composition of various types of histones as determined by their Coomassie blue staining. Also, uH2A level did not exhibit an appreciable change for a period of 24 h after global UV irradiation. The intensification of uH2A at the damage sites while maintaining the constant levels in irradiated cells argues in favor of recruitment of pre-existing uH2A. By contrast, phosphorylated form of histone variant H2AX (γH2AX), amarker of strand break formation, demonstrated a clear UV-dependent induction that peaked at about 8 h post-irradiation. These distinct patterns of uH2A and γH2AX following cellular irradiation purport to a separate basis of their origin.
3.3. Recruitment of uH2A requires either GGR or TCR sub-pathway but not functional DDB2 protein
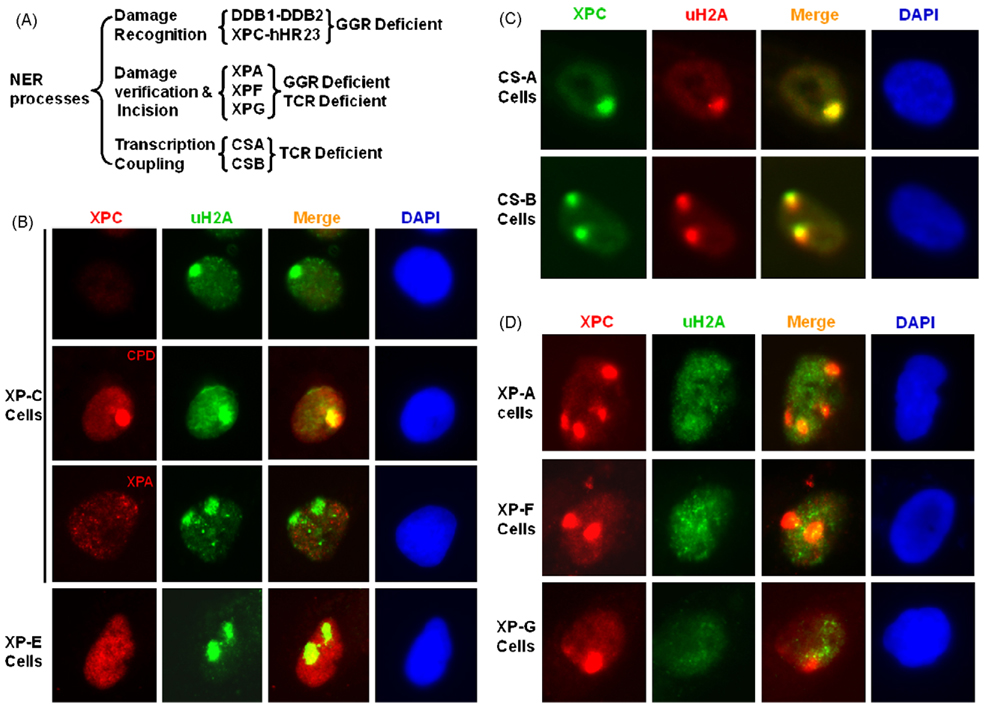
To determine the relationship between NER and formation of uH2A foci, we examined the localization of uH2A to DNA damage in various cells representing XP and CS disorders, which by virtue of the lack of a specific repair function are known to be defective in GGR, TCR or NER (Fig. 3A). Thus, any cell lacking a GGR-specific protein, i.e., XPC or a TCR-specific protein, i.e., CSA will be considered partially repair-proficient as it can still conduct repair of a significant portion of lesions by the complementary sub-pathway. While any cells lacking a factor like XPA, which is common downstream protein for both GGR and TCR sub-pathways, will be considered repair-deficient and unable to exhibit any degree of NER. Fig. 3B shows that XP-C fibroblasts exhibit clear formation of uH2A foci. It should be mentioned that in XP-CGM15983 cells, harboring a truncated XPC protein, no recruitment of XPC or XPA was detectable at the lesion sites (Fig. 3B). Yet, the uH2A foci in these cells are shown to distinctly co-localize with DNA damage sites revealed by their staining with CPD specific antibodies. The formation of uH2A foci in XP-C cells was also confirmed in XP-C GM02096 cells which harbor a Pro218His point mutation (data not shown). In XP-E cells which are defective in DDB activity, XPC protein although present cannot be recruited to the damage sites. Nevertheless, the ability to form uH2A foci is uncompromised in GGR-deficient XPE cells (Fig. 3B). These observations offer two critical conclusions. First, the recruitment of uH2A to damage does not depend on absolute presence of XPC. So, GGR, which requires functional XPC, is not the only trigger to form uH2A foci. Second, DDB activity is not needed for the appearance of uH2A foci. This is important because it strongly suggests that ubiquitinated form of H2A, seen at DNA damage site, is not the result of Cul4A-DDB ubiquitin ligase activity.
Fig. 3.
Formation of local uH2A foci is dependent on functional NER sub-pathway, GGR or TCR. (A) A schematic depiction of functionality of NER factors in different repair processes. (B) uH2A, XPC and XPA recruitment was visualized 1 h after micropore UV irradiation in fibroblasts deficient in GGR (XP-C and -E). UV-induced CPD were visualized in XP-C cells to show DNA damage spots which received micropore UV irradiation at a dose of 100 J/m2. (C) uH2A foci and XPC recruitment in fibroblasts deficient in TCR (CS-A and -B). (D) uH2A foci and XPC recruitment in fibroblasts deficient in NER (XP-A, -F and -G).
Next, we tested the premise that uH2A is recruited to the damage sites in cells that are proficient in repair by virtue of any one of the two functional NER sub-pathways. For this, we determined the formation of uH2A foci in different TCR-deficient albeit GGR-proficient CS-A and CS-B cell lines. CS cells are UV sensitive and defective in recovery of RNA synthesis after UV irradiation and are normal for repair of lesions in the overall genome [41]. Our data clearly show that uH2A foci formation in CS cells was analogous to GGR-deficient cells and that TCR defect did not compromise the recruitment of uH2A foci in CS-A or -B cells (Fig. 4C). Obviously, active TCR in cells is not the only repair-related stimuli triggering the formation of uH2A foci. More importantly, the results provide evidence that formation of uH2A foci is not a consequence of stalled transcription machinery which is known to initiate TCR.
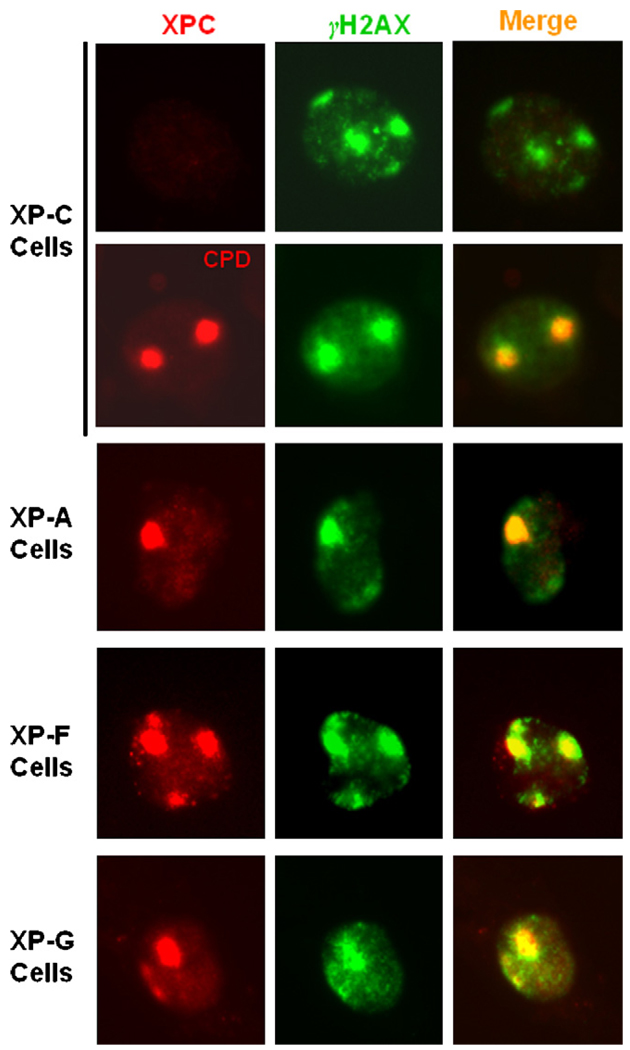
Fig. 4.
Formation of γH2A foci at DNA damage and repair spots is independent of functional NER pathways. γH2A foci and XPC recruitment were visualized 1 h after micropore UV irradiation in NER-deficient XP-A, -F and -G fibroblasts. UV-induced CPD were visualized in XP-C cells to indicate DNA damage spots upon micropore UV irradiation at a dose of 100 J/m2.
Finally, we examined the formation of uH2A foci in NER-deficient XP-A, -F and -G cells. These cells lack factors that are critical for the excision of DNA damage. For instance, XPF and XPG cells are deficient in nucleases that incise the damaged strand 3′ and 5′ of the lesion, respectively. Since binding of XPC to damage occurs prior to the action of these repair factors, XPC localization to damaged foci was unaffected in all three NER-deficient cells (Fig. 3D).None of these cells, however, showed any accumulation of uH2A. In agreement with previous observation [37], this data reaffirms that formation of uH2A foci is not directly associated with damage recognition by XPC and, that it occurs at a later stage of NER. Given that uH2A foci persist at damage sites for considerably long time, we reckon that recruitment of uH2Afoci is a post-incision event. Taken together, the results of repair-proficient and -deficient cells help conclude that formation of uH2A foci requires functional NER through either GGR or TCR sub-pathways.
UV irradiation of cells has also been shown to induce γH2AX foci, a hallmark of strand breaks induced by agents like ionizing radiation. We assessed the appearance of γH2AX following UV irradiation of various repair-proficient and -deficient cells used in studying the formation of uH2A. Recruitment of XPC or CPD (in XPC cells) was again used to mark the damage induction at locally irradiated sites in different cells. Contrary to the results of uH2A recruitment, γH2AX foci were distinctly apparent in TCR-proficient XP-C as well as repair-deficient XP-A, -F and -G cells (Fig. 4). Moreover, γH2AX exhibited unambiguous co-localization with XPC or CPD containing sites. It should be mentioned that γH2AX foci were visualized in ~15% of the irradiated XP cells. This response is expected as the experiments were performed with asynchronous cells in which the γH2AX foci are generated through UV-induced replication stress [42,43]. This result indicated that formation of γH2AX foci in asynchronous cells is independent of NER, suggesting that γH2AX and uH2A foci formation occurs by two different mechanisms.
3.4. uH2A at DNA damage sites fails to recruit proteasome
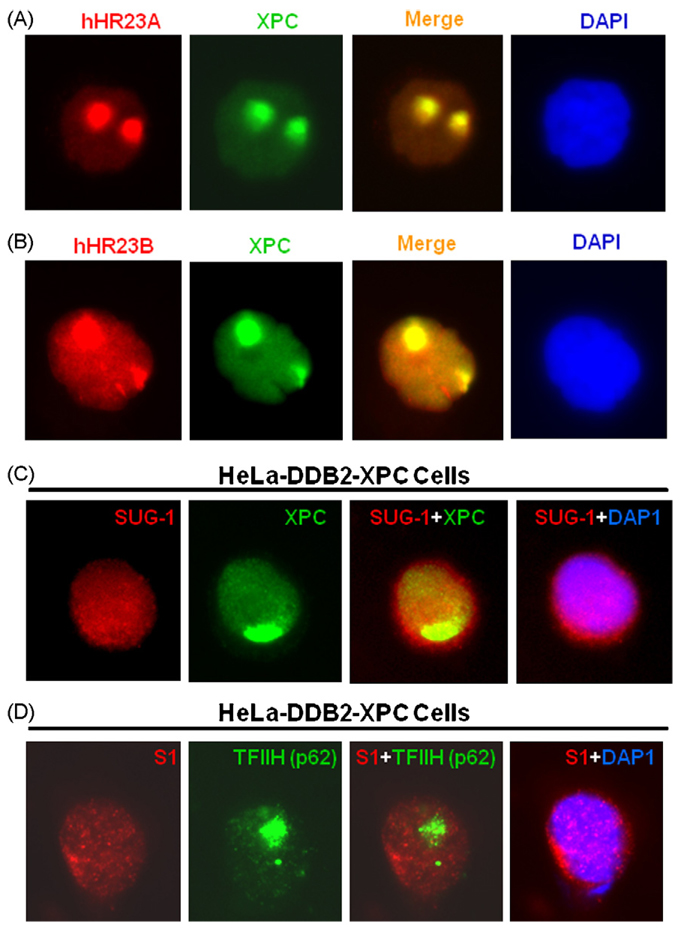
Although proteasome recognizes polyubiquitin conjugated proteins and targets them for proteolysis [44], it also regulates NER independent of the proteolytic function [45,46]. Moreover, it is well established that hHR23B/A, present in complex with XPC, aids in recognizing DNA damage and protecting XPC from proteolysis [47]. To discern the role of uH2A, recruited to damage sites, we determined the possible recruitment of proteasome to DNA damage sites via ubiquitin or Ubl moieties of uH2A and hHR23B/A, respectively. Local UV irradiation revealed the focal accumulation of both hHR23A and hHR23B (Fig. 5A and B) and co-localization with damage targeted XPC. The 19S proteasomal component, Sug-1, was shown to uniformly distribute in both cytoplasm and nucleus of HeLa cells. Nevertheless, there was no recruitment of proteasome to DNA damage as revealed by the failure of foci formation and co-localization with XPC under various local irradiation conditions (Fig. 5C). The failure to recruit proteasome was confirmed with another proteasome component, S1, in HeLa cells (Fig. 5D). Upon local UV irradiation, S1 protein exhibited a uniform distribution in the cytoplasm as well as nuclei. Although local UV-induced TFIIH component p62 foci were easily seen, S1-specific foci formation could not be observed. Consistent with the notion that proteasome is not recruited to DNA damage sites [37], the combined data clearly argued against the possibility of uH2A or hHR23B/A playing any role in recruiting 19S proteasome to the DNA damage sites.
Fig. 5.
Formation of uH2A foci and hHR23A/B at damage spots fails to recruit proteasome. (A) Recruitment of hHR23A to damage spots and its co-localization with XPC protein are visualized 1 h after micropore UV irradiation by indirect immunofluorescence. (B) Localization of hHR23B to damage spots. (C) Proteasomal components SUG-1 and S1 do not accumulate at damage spots. The proteasomal SUG-1 is visualized by indirect immunofluorescence (red) in HeLa-DDB2-XPC cells in which XPC localization (green) is provoked by local UV irradiation. (D) Cellular distribution of proteasomal S1 (red) and TFIIH component p62 (green) are examined in Hela-DDB2-XPC cells upon micropore UV irradiation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.5. Ring1B component of PRC1 E3 ubiquitin ligase complex does not localize at DNA damage sites
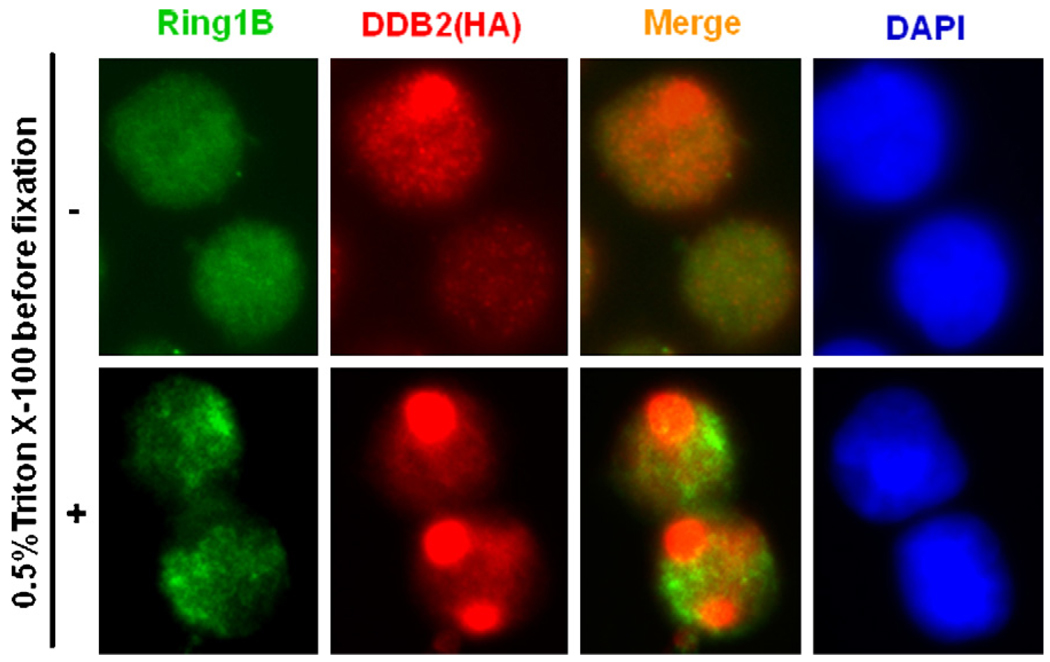
It has been demonstrated that PRC1 is the dominant E3 ubiquitin ligase for monoubiquitination of histone H2A [29]. Interestingly recent study has reported that the Ring1B component of the PRC1 complex is required for UV-induced H2A ubiquitination [37]. Therefore, to test whether the site specific uH2A accumulation results from local de novo ubiquitination, we detected the presence of Ring1B and its potential recruitment to local DNA damage sites. Our data showed that Ring1B uniformly distributed within the nuclei of both unirradiated as well as locally irradiated cells. Moreover, pretreatment with 0.5% Triton X-100 revealed a distinct signal intensification of the insoluble Ring1B presumed to exist in tight association with heterochromatin (Fig. 6). Under both these conditions Ring1B exhibited no specific affinity for the DNA damage amply present in the damage-marked subnuclear areas. On the other hand, both conditions revealed an unambiguous localization of control DDB2 protein to the corresponding damage sites that failed to recruit Ring1B. These results do not cohere with the reported UV-induced H2A ubiquitination by Ring1B ligase complex [37]. The data, per se, does not support the hypothesis that UV irradiation induces local de novo H2A ubiquitination through PRC1 E3 ubiquitin ligase. Nevertheless, it is possible that Ring1B transiently recruited to damage sites does not accumulate at levels sufficient for immunofluorescent detection.
Fig. 6.
Ring1B component of PRC1 E3 ubiquitin ligase complex does not localize at DNA damage sites. HeLa-DDB2 cells were locally UV irradiated through a polycarbonate micropore (5 µm) filter. After 1 h incubation in fresh medium, the cells were pre-treated with CSK buffer containing 0.5% Triton X-100 and then fixed or directly fixed with 2% paraformaldehyde in presence of 0.5% Triton X-100 in PBS. The Ring1B (green) and DDB2 (HA, red) proteins were visualized by indirect immunofluorescence with specific antibodies. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
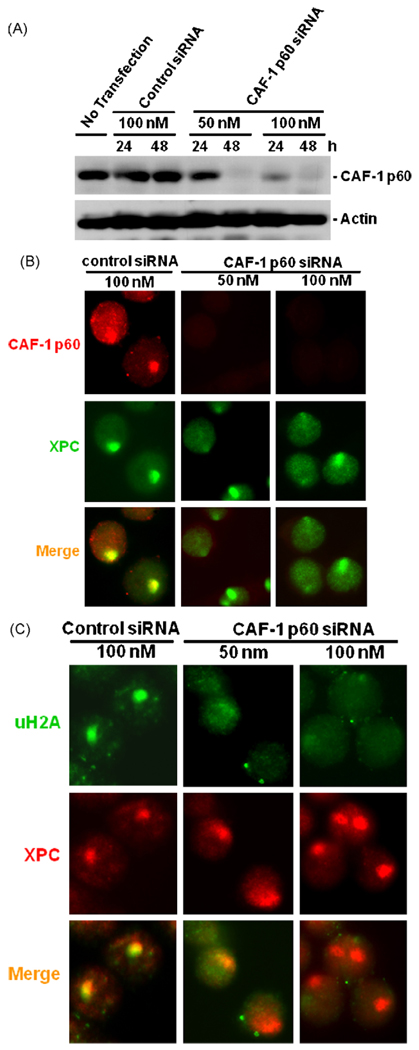
3.6. CAF-1 p60 is necessary for recruiting uH2A to DNA repair sites
Since uH2A recruitment is dependent on successful excision and most likely a post-repair event, we wanted to determine whether this recruitment involves the active participation of histone chaperone, CAF-1. Thus, we targeted the p60 subunit of CAF-1 protein for knockdown by cellular transfection with siRNA and subsequent analysis of DNA damage specific factor recruitment. Western blotting showed that transfection of siRNA specifically targeting CAF-1 exhibited a siRNA concentration and time dependent reduction in the expressed protein. Treatment for 48 h with 50 and 100 nM siRNA quantitatively depleted the cells of CAF-1 (Fig. 7A). These conditions were then used for assessing the recruitment of various NER factors. The depletion of CAF-1 was first confirmed by immunofluorescent detection of the protein in control and CAF-1 siRNA transfected cells (Fig. 7B). More importantly, CAF-1 in cells was shown to get recruited to locally damaged DNA sites. Besides, CAF-1 foci were observed in ~60% cells that also exhibited co-localized XPC foci. Depletion of CAF-1 had no impact on the recruitment of XPC that is known to track to DNA damage in the initial phase of NER. However, the knockdown of CAF-1 in cells abolished the recruitment of uH2A, as uH2A foci were seen to co-localize with less than 2% of XPC foci (Fig. 7C). These results demonstrate that histone chaperone CAF-1 is intimately associated with the late recruitment of uH2A following the successful repair of lesions through NER.
Fig. 7.
Knockdown of CAF-1 p60 prevents the formation of uH2A foci at DNA repair sites. (A) HeLa-DDB2-XPC cells were transiently transfected for indicated time period with control siRNA or CAF-1 siRNA targeting p60 subunit of CAF-1. The cellular level of CAF-1 after siRNA transfection was determined by Western blotting. (B) CAF-1 and XPC proteins were visualized in control or CAF-1 siRNA transfected HeLa-DDB2-XPC cells 1 h after local UV irradiation at a dose of 100 J/m2. (C) uH2A and XPC was visualized in control or CAF-1 siRNA transfected HeLa-DDB2-XPC cells by indirect immunofluorescence.
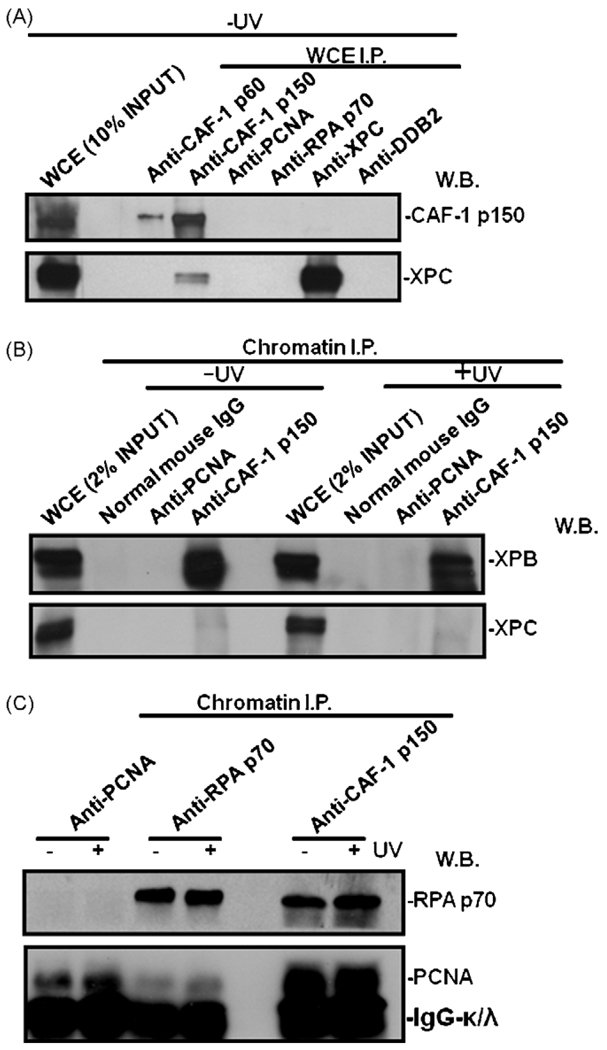
3.7. CAF-1 p150 interacts with RPA and PCNA in chromatin
Previous studies have shown that replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin [48]. DNA synthesis in vitro, however, is not required for bidirectional chromatin assembly from the site of NER [21]. We wanted to determine the basis of CAF-1 recruitment to DNA damage and its interaction with various NER component(s). It is believed that NER machinery does not exist in a pre-assembled state but individual components are sequentially brought to the DNA damage sites within chromatin [4].With this in mind, we first explored the protein–protein interaction between CAF-1 p150 with NER factors through immunoprecipitation using protein extracts from unirradiated cells. Fig. 8A shows that CAF-1 p150 did not interact with DDB2, XPC, PCNA and RPA p70 as it was absent in immunoprecipitates pulled down with cognate factor-specific antibodies. A weak signal of XPC was found in anti-CAF-1 p150 immunoprecipitates indicating a possible interaction. However, this could not be confirmed as XPC was absent in anti-CAF-1 p60 immunoprecipitates and CAF-1 p150 was not recovered in anti-XPC immunoprecipitates. We next tested the interaction between CAF-1 p150 and NER factors through chromatin immunoprecipitation using formaldehyde cross-linked chromatin obtained from serum-starved cells (Fig. 8B and C). The serum starvation would minimize active DNA replication and the cell would exhibit less protein–protein interaction related to replication. No interaction of CAF-1 was observed with NER factors except XPB. In this case, XPB component of TFIIH was found in anti-CAF-1 p150 immunoprecipitates in both unirradiated and UV irradiated cells (Fig. 8B). Interestingly, RPA p70 and PCNA were also present in anti-CAF-1 p150 immunoprecipitates regardless of UV irradiation of cells. These results confirm the interaction between PCNA and CAF-1 p150 and revealed that CAF-1 binds DNA synthesis machinery through multiple protein–protein interactions. The interactions, however, are not induced by UV irradiation and therefore, are not repair-specific, signifying the dual role of CAF- 1 in DNA replication and DNA repair. The interaction between CAF-1 and XPB (TFIIH) may be relevant to both transcription and DNA repair. Nevertheless, RPA is involved in both pre-incision and post-incision steps of NER [49], the interactions between CAF-1 and PCNA as well as CAF-1 and RPA may explain the coupling of chromatin assembly to new DNA synthesis, including repair synthesis.
Fig. 8.
CAF-1 p150 interacts with RPA and PCNA in chromatin. (A) Whole cell extracts from HeLa-DDB2-XPC cells were made in RIPA buffer and immunoprecipitated with indicated antibodies. The immunoprecipitates were Western blotted and analyzed for the presence of CAF-1 p150, XPC or XPG. (B) Immunoprecipitation was performed using soluble chromatin made from unirradiated or UV-irradiated, formaldehyde-cross-linked HeLa-DDB2-XPC cells as described in Section 2. Presence of NER factors XPC, TFIIH (XPB), XPF and XPG in immunoprecipitates was detected by factor-specific antibodies. (C) Examination of interactions between PCNA, RPA p70 and CAF-1 p150 by chromatin immunoprecipitation and Western blotting.
4. Discussion
In eukaryotic cells, repair of DNA lesions buried within chromosome involves transient chromatin disruption to allow damage processing and subsequent chromatin restoration. Chromatin dynamics during DNA repair plays an important role in reestablishing epigenetic status of a repaired chromatin segment or marking the site of once damaged chromatin with newly incorporated structural histones. Here, we demonstrate the UV-induced accumulation of uH2A at damage sites as part of the chromatin restoration process. We have shown that Triton X-100-resistant uH2A foci are formed at sites of DNA damage in a process that is functionally dependent on NER. We have provided evidence that uH2A foci formation does not occur through the action of Cul4ADDB ubiquitin ligase which is known to directly ubiquitinate NER factors, DDB and XPC, at the damage sites. On the other hand, we show that uH2A foci formation is functionally dependent on the presence of histone chaperone CAF-1. These results, together with recently published observations of histone 3.1 incorporation at DNA repair sites [24], point to a new phenomenon that chromatin restoration accompanying repair of DNA damage must provoke critical changes in epigenetic information in which newly incorporated structural histones present historical marks of the DNA repair event.
A recent study indicated that DNA damage triggers NER-dependent monoubiquitination of histone H2A in a manner requiring XPC function [37]. Our observations, however, argue that DDB2 or XPC-mediated GGR is not the only stimuli triggering the accumulation of uH2A. It can also be accomplished following TCR of DNA lesions. The discrepancy could be attributed to different UV doses used for local irradiation in these two studies. The presented data also clearly favor the placement of uH2A as a post-incision event during chromatin restoration steps. This notion of uH2A recruitment to damage sites, as a post-incision event, is supported by several lines of evidence: (i) Local UV-induced uH2A foci at damage sites were resistant to Triton X-100 and the uH2A within foci persisted long after completion of repair of 6-4PP, suggesting that uH2A is incorporated in chromatin. The endurance of uH2A foci is critical, because XPC and DDB were already released at 4 h time point (Fig. 2B). (ii) Although formation of uH2A foci was functionally dependent on NER, it did not solely rely on either GGR or TCR sub-pathway. (iii) In asynchronous cells, functional XPA, XPF and XPG were required for the formation of uH2A but not of γH2A foci (Fig. 4). This difference between UV-induced uH2A and γH2A foci formation is noteworthy because γH2AX is considered as a marker for the presence of strand breaks in chromatin. Although UV-induced H2AX phosphorylation has been shown to occur in non-replicating G0-arrested cells undergoing NER [50], it is most likely activated through transient gaps introduced by damage excision [51]. For example, gap-filling step of NER is highly inefficient in non-replicating G0-arrested cells due to the lack of crucial replication factors. Thus, perturbed repair synthesis leads to unfilled gaps that would present strand ends as triggers to initiate H2AX phosphorylation in an ATR-dependent manner. Nevertheless, we were able to show the appearance of γH2AX foci in asynchronous repair-deficient XP cells that are unable to reach the incision stage and form any excision-mediated gaps. Therefore, in this situation H2AX phosphorylation would occur due to UV-induced replication stress which is established to be an main trigger for γH2AX foci formation [42,43]. In contrast, however, neither UV-induced replication stress nor perturbed gap-filling repair could account for the formation of uH2A foci, as it is functionally dependent on NER within asynchronous cells. (iv) Lastly, our data clearly show that the formation of uH2A foci is dependent on participation of histone chaperone CAF-1 (Fig. 7). This fully coheres with previous reports indicating that functional NER pathway, specifically the dual incision, is required for recruiting CAF-1 and PCNA to DNA damage site [22]. Thus, it is reasoned that uH2A is incorporated into repaired chromatin as a post-repair processing event.
Several DNA repair factors, including XPC, DDB2 and PCNA, are subjected to ubiquitination triggered by UV-induced DNA damage [52]. Whether UV exposure of cell also induces de novo ubiquitination of histone H2A remains controversial. It is well established that uH2A is maintained at abundant levels in mammalian cells within the nucleus as well as cytoplasm. Thus, it is available for incorporation during chromatin assembly following DNA replication and repair. Interestingly, one previous study showed a dramatic loss of cellular uH2A immediately after UV irradiation and its total restoration by 2 h [32]. Another study, monitoring the mobility GFP-tagged ubiquitin in the nucleus of live cells, showed that global UV exposure induced a massive histone H2A ubiquitination [37]. Our results (Fig. 2C) as well as other studies [34] failed to detect any noticeable change in global level of cellular uH2A. Moreover, our rigorous attempts also failed to demonstrate the recruitment Ring1B component of PRC1 E3 ubiquitin ligase complex to damage sites (Fig. 6). Therefore, our results are consistent with the idea incorporation of uH2A at repair sites from the pre-existing cellular uH2A pool. This uH2A pool is indeed maintained by the PRC1 proteins, Ring1A and Ring1B [29,30]. Nonetheless, it remains possible that UV-induced ubiquitination of histone H2A occurs at relatively low level at the damage sites and consequently does not cause a discernable effect on total cellular level of uH2A. In such case, the putative ubiquitin ligase will have to act on newly assembled chromatin or work cooperatively with CAF-1 to incorporate uH2A into chromatin at the site of damage. In any event, CAF-1-mediated new chromatin assembly would be required for local histone H2A ubiquitination or spread the uH2A around the restored chromatin initiated by lesion repair. In support of such a scenario, CAF-1 was shownto initiate and propagate chromatin assembly in vitro several hundred base pairs distant from a lesion site [21].
Our demonstration of uH2A incorporation during NER suggests that what occurs at UV-induced damage sites is fundamentally different than RNF8-mediated histone H2A and γH2AX ubiquitination at DSB, despite that they are conceptually similar [53–55]. First, RNF8 and E2 ubiquitin-conjugating enzyme UBC13 are known form lysine63-ubiquitination chain; second, the ubiquitination dependent on MDC1, whose BRCT domain binds γH2AX. Therefore, RNF8 integrates protein phosphorylation and ubiquitination signaling and plays a critical role in the cellular response to DSB. The NER-dependent uH2A incorporation during chromatin restoration, on the other hand, is consistent with recently reported CAF-1 dependent deposition of newly synthesized histone H3.1 in chromatin [24]. This report has raised an important question of how epigenetic information is preserved or revised after DNA damage repair, given that both post-translational modifications and histone variants contribute to epigenetic marking. Moreover, in relation regulation of epigenetic information, it has been shown that CAF- plays a prominent role in heterochromatin organization in embryonic cells [56], heterochromatin silencing in yeast [57,58] and maintaining transcriptional gene silencing in mammalian cells [59]. We anticipate that CAF-1-mediated deposition of histone H3.1 and uH2A into chromatin following damage repair are analogous events and both can be viewed as imprints of repaired chromatin. Other histone modifications can also be envisaged at the sites of NER, existing at least temporarily. These imprints would allow repaired chromatin to distinctively stand out from the undisturbed neighboring chromatin region and represent a memory of damage repair. A critical question to ask here is what this memory means to subsequent life of the cell and how it impacts other cellular pathways, e.g., damage signaling and transcription recovery. Future studies in this realm will have to address unanswered questions of how the complex chromatin structure is modulated during different DNA-templated cellular processes without the loss of epigenetic information needed to preserve the genomic integrity, stability and function.
Acknowledgements
The work carried out by Q.Z.,G.W., M.E.,A.R. and A.A.W. was supported by Public Health Service Grants ES2388 and ES12991 from National Institute of Environmental Health Sciences and CA93413 from National Cancer Institute. We thank Haroon Haque for excellent technical assistance and Qi-en Wang, The Ohio State University, for advice on immunofluorescence experimentation. We are grateful to Yoshihiro Nakatani, Harvard Medical School, for providing HeLa-DDB2 cells.
Abbreviations
- 6-4PP
(6-4) pyrimidine-pyrimidone photoproducts
- CAF-1
chromatin assembly factor 1
- CPD
cyclobutane pyrimidine dimers
- CS
Cockayne syndrome
- DDB
damaged DNA-binding protein
- γH2AX
phosphorylated histone H2AX
- GGR
global genomic repair
- NER
nucleotide excision repair
- siRNA
small interference RNA
- TCR
transcription-coupled repair
- Ub
ubiquitin
- uH2A
ubiquitinated histone H2A
- PRC1
Polycomb repressor complex 1
- UV
ultraviolet light
- XP
Xeroderma pigmentosum
Footnotes
Conflict of interest
None.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.Petit C, Sancar A. Nucleotide excision repair: from E. coli to man. Biochimie. 1999;81:15–25. doi: 10.1016/s0300-9084(99)80034-0. [DOI] [PubMed] [Google Scholar]
- 3.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 4.Volker M, Mone MJ, Karmakar P, Van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, Van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 8;2001:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 5.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 6.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The Xeroderma pigmentosum group C proteincomplex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 7.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;16:6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans E, Fellows J, Coffer A, Wood RD. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 1997;16:625–638. doi: 10.1093/emboj/16.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 10.Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: insights for transcription-coupled repair and Cockayne Syndrome. Mol. Cell. 2005;20:187–198. doi: 10.1016/j.molcel.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 13.Smerdon MJ. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- 14.Green CM, Almouzni G. When repair meets chromatin. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolffe AP, Guschin D. Chromatin structural features and targets that regulate transcription. J. Struct. Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 16.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 18.Verreault A. De novo nucleosome assembly: newpieces in an old puzzle. Genes Dev. 2000;14:1430–1438. [PubMed] [Google Scholar]
- 19.Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillard PH, Martini EMD, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 21.Gaillard PH, Moggs JG, Roche DM, Quivy JP, Becker PB, Wood RD, Almouzni G. Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J. 1997;16:6281–6289. doi: 10.1093/emboj/16.20.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith S, Fellows J. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 28.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 29.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 30.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 32.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2588–2593. doi: 10.1073/pnas.0511160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response toDNA damage. Mol. Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 35.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cull in 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, Wani AA. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006;66:8590–8597. doi: 10.1158/0008-5472.CAN-06-1115. [DOI] [PubMed] [Google Scholar]
- 37.Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de WH, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, Hoeijmakers JH, Vermeulen W, Dantuma NP. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wani AA, D’Ambrosio SM, Alvi NK. Quantitation of pyrimidine dimers by immunoslot blot following sublethal UV-irradiation of human cells. Photochem. Photobiol. 1987;46:477–482. doi: 10.1111/j.1751-1097.1987.tb04798.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang QE, Zhu Q, Wani MA, Wani G, Chen J, Wani AA. Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Repair. 2003;2:483–499. doi: 10.1016/s1568-7864(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 40.Nabatiyan A, Krude T. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell Biol. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayne LV, Lehman AR. Failure of RNA synthesis to recover after UV irradiation:an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- 42.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent mannerin response to replicational stress. J. Biol. Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 43.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated andRad3-related (ATR) activation requires replication stress. J. Biol. Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 44.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillette TG, Huang W, Russell SJ, Reed SH, Johnston SA, Friedberg EC. The 19S complex of the proteasome regulates nucleotide excision repair in yeast. Genes Dev. 2001;15:1528–1539. doi: 10.1101/gad.869601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- 47.Ng JM, Vermeulen W, Van der Horst GT, Bergnik S, Sugasawa K, Vrieling H, Hoeijmakers JH. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 49.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 51.Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- 52.O’Connell BC, Harper JW. Ubiquitin proteasome system(UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 2007;19:206–214. doi: 10.1016/j.ceb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 56.Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
- 59.Tchenio T, Casella JF, Heidmann T. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol. Cell Biol. 2001;21:1953–1961. doi: 10.1128/MCB.21.6.1953-1961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]