Abstract
Rationale: Tumor necrosis factor is a proinflammatory cytokine found in increased concentrations in asthmatic airways. The TNF-α (TNF) and lymphotoxin-α (LTA) genes belong to the TNF gene superfamily located within the human major histocompatibility complex on chromosome 6p in a region repeatedly linked to asthma. The TNF position –308 and LTA NcoI polymorphisms are believed to influence TNF transcription and secretion, respectively. Objectives: This study sought to determine whether polymorphisms in TNF or LTA, or in TNF-LTA haplotypes, are associated with asthma and asthma phenotypes. Methods: We genotyped the TNF –308 and LTA NcoI polymorphisms, and two other haplotype-tagging polymorphisms in the TNF and LTA genes, in 708 children with mild to moderate asthma enrolled in the Childhood Asthma Management Program and in their parents. Using an extension of the family-based association tests in the PBAT program, each polymorphism was tested for association with asthma, age at onset of asthma, and time series data on baseline FEV1 % predicted, postbronchodilator FEV1 % predicted, body mass index, and log of PC20. Measurements and Main Results: Although no associations were found for the individual single-nucleotide polymorphisms, the haplotype analysis found the LTA NcoI_G/LTA 4371T/TNF –308G/TNF 1078G haplotype to be associated with asthma and with all five phenotype groups. Conclusions: We conclude that it is unlikely that the TNF –308 or LTA NcoI polymorphisms influence asthma susceptibility individually, but that this haplotype of variants may be functional or may be in linkage disequilibrium with other functional single-nucleotide polymorphisms.
Keywords: asthma, haplotypes, lymphotoxin-α polymorphism, tumor necrosis factor
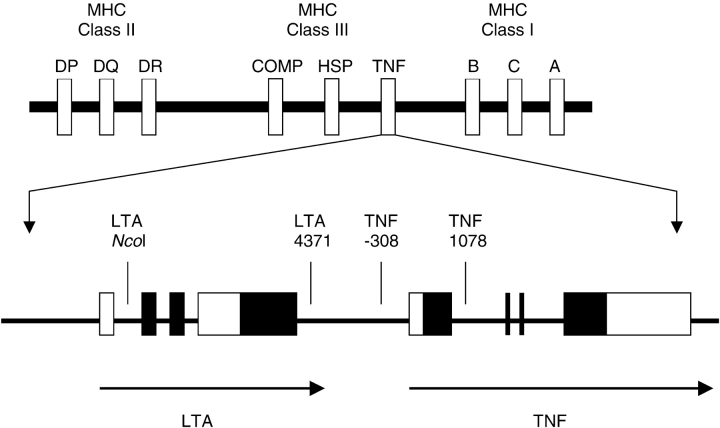
Tumor necrosis factor is a proinflammatory cytokine that is found in increased concentrations in asthmatic airways. The TNF-α (TNF) and lymphotoxin-α (LTA) genes are members of the TNF gene superfamily located within the human major histocompatibility complex on chromosome 6p in a region repeatedly linked to asthma (1, 2). As shown in Table 1, association between a position –308 guanine (G)-to-adenine (A) polymorphism in the TNF promoter and asthma has been tested in 18 published studies that included different age groups of individuals with asthma from a range of ethnic backgrounds, and an association between the A allele and asthma was reported in seven of these studies (3–9). In nine additional studies (10–18) listed in Table 1, however, authors reported no association between asthma and this single-nucleotide polymorphism (SNP), and two other studies showed an association between the wild-type allele and asthma (19, 20). The TNF –308A allele is of interest because it is associated with increased in vitro transcription of TNF and with increased TNF levels in stimulated human white blood cells (21, 22). As shown in Figure 1, the LTA gene, previously called the TNF-β gene, is located close upstream to the TNF gene. An NcoI polymorphism in the LTA gene has also been associated with asthma in children (19) and adults (5), although each study reported that a different allele was associated and 13 other studies (4, 7, 9, 11, 13, 14, 16, 17, 20, 23–26) showed no association (see Table 1).
TABLE 1.
Studies evaluating the association of tnf 308 and lta ncoi polymorphisms for asthma
| TNF –308*,‡
|
LTA NcoI‡
|
||||||
|---|---|---|---|---|---|---|---|
| First Author, Year | Reference | Population | Design | G | A | G | A |
| Moffatt, 1997 | 5 | White, 413 subjects from 88 families: 318 nonasthmatic and 92 with asthma (32 asthmatic parents and 60 children) |
Case-control | – | + | + | – |
| Albuquerque, 1998* | 19 | White, 74 children with atopic asthma and 50 unrelated control subjects | Case-control | + | – | – | + |
| Moffatt, 1999† | 16 | White, 1,004 subjects in 230 nuclear families; 179 were asthmatic (66 parents and 113 children) | Case-control | NS | NS | NS | NS |
| Li Kam Wa, 1999* | 4 | Mixed race, 556 subjects tested for bronchial hyperreactivity (246 hyperreactive); also sample of 60 extended families of an asthmatic proband | Case-control | – | + | NS | NS |
| Chagani, 1999 | 3 | White, 92 with mild–moderate asthma, 159 with fatal/near-fatal asthma, 43 nonasthmatic, 252 random control subjects |
Case-control | – | + | ||
| Trabetti, 1999 | 17 | White, 600 families with 131 children with atopic asthma | Sibling-pairs | NS | NS | NS | NS |
| Winchester, 2000 | 8 | White, 20 subjects with asthma identified by questionnaire for history of childhood asthma and 416 control subjects | Case-control | – | + | ||
| Louis, 2000 | 15 | White adults, 95 subjects with asthma and 98 control subjects | Case-control | NS | NS | ||
| Zhu, 2000 | 18 | White children, 12 with asthma and 269 nonasthmatic | Family based | NS | NS | ||
| Cardaba, 2001 | 23 | Spanish gypsies, 5 families with 87 people | Case-control | NS | NS | ||
| Izakovicova, 2001 | 25 | Czech, 243 white with atopy (167 had asthma) and 184 control subjects | Case-control | NS | NS | ||
| Immervol, 2001 | 24 | White, 97 families with 2 or more siblings with asthma | Family based | NS | NS | ||
| Lin, 2002 | 14 | Taiwanese, 80 subjects with high-IgE asthma and 69 nonasthmatics | Case-control | NS | NS | NS | NS |
| Noguchi, 2002 | 26 | Japanese, 144 atopic asthma families; transmission disequilibrium tests done | Family based | NS | NS | ||
| Witte, 2002 | 9 | White, 169 asthma cases and 170 control subjects | Case-control | – | + | NS | NS |
| Buckova, 2002 | 11 | Czech, 151 patients with atopic asthma and 155 control subjects | Case-control | NS | NS | NS | NS |
| Di Somma, 2003 | 12 | Italian, 70 patients with asthma and 169 without asthma from 51 nuclear families | Case-control | NS | NS | ||
| El Bahlawan, 2003 | 13 | Mixed race, children: 38 with asthma and 231 control subjects | Case-control | NS | NS | NS | NS |
| Sandford, 2004 | 6 | Chinese, 107 children with wheeze in the last 12 mo and 118 without wheeze | Case-control | – | + | ||
| Shin, 2004 | 20 | Korean, 550 with asthma and 171 control subjects | Case-control | + | – | NS | NS |
| Beghe, 2004 | 10 | 142 toluene diisocyanate–induced asthma cases and 50 asymptomatic exposed control subjects | Case-control | NS | NS | ||
| Wang, 2004 | 7 | Taiwanese children, 128 with atopic asthma, 51 with nonatopic asthma, 55 atopic control subjects, and 78 nonatopic control subjects | Case-control | – | + | NS | NS |
Definition of abbreviations: LTA = lymphotoxin-α; NS = not significant; TNF = tumor necrosis factor.
Positive association found in individuals homozygous for the allele.
The Moffatt and coworkers (1999) study (16) includes the same population as the Moffatt and Cookson (1997) population (5).
A significant (p ≤ 0.05) association, with the + allele increased and the – allele decreased in the subjects with asthma; NS, no significant differences found.
Figure 1.
Top: Schematic representation of the TNF genes on chromosome 6 in relation to other important innate immunity genes in the area. TNF includes lymphotoxin-α (LTA), tumor necrosis factor-α (TNF), and lymphotoxin-β (LTB). COMP = complement; HSP = heat shock 70-kD protein-1 and 90-kD protein-1; MHC = major histocompatibility complex (human leukocyte antigens DP, DQ, DR); SNP = single-nucleotide polymorphism. Bottom: TNF and LTA with SNP locations; the solid boxes denote exons and the open boxes denote the 5′ and 3′ untranslated regions. Not drawn to scale. (The top portion of this diagram was modified from Reference 39.)
Conflicting findings in previous studies could be explained by the different populations studied, by population stratification, or by small sample sizes in case-control study designs. The TNF –308 and LTA NcoI polymorphisms are in strong linkage disequilibrium (5, 19). Moffatt and colleagues (16) have advocated that haplotypes created by the TNF –308 and LTA NcoI polymorphisms and surrounding innate immunity genes in the major histocompatibility complex may explain the conflicting results. A Japanese study showed significant transmission disequilibrium with haplotypes in the TNF genes (26). Using a family-based design to control for population stratification in a large cohort of children with mild to moderate asthma, we tested the TNF –308 and LTA NcoI polymorphisms, and extended TNF/LTA haplotypes for association with asthma. Some of the results of these studies have been previously reported in the form of an abstract (27).
METHODS
Population
The Childhood Asthma Management Program (CAMP) was a multicenter, randomized, double-masked, placebo-controlled clinical trial evaluating the long-term effects of inhaled antiinflammatory medications in children with mild to moderate asthma (28). The diagnosis of asthma was based on methacholine hyperreactivity (PC20 no greater than 12.5 mg/ml) and the meeting of one or more of the following criteria for at least 6 months in the year before recruitment: (1) asthma symptoms two times or more per week; (2) at least two uses per week of an inhaled bronchodilator; and (3) daily asthma medication. See the online supplement for a description of spirometry, including methacholine challenge (29), and other testing methods.
Of the 1,041 children participating in the original CAMP study (29), DNA samples were obtained from 968 participating children and 1,518 of their parents. Complete family trios were available for 652 nuclear families that included 708 children. Some families had multiple children with asthma (49 families had two children and two families had three children). The characteristics of these children are shown in Table 2.
TABLE 2.
Characteristics of population
| CAMP Children with Asthma
|
||
|---|---|---|
| All (n = 708) | White Subjects Only (n = 432) | |
| Sex, % male | 59.4 | 58 |
| Age, yr | 8.07 (SD, 2.1) | 8.84 (SD, 2.1) |
| Ethnicity, % | ||
| Non-Hispanic white | 73.4 | 100 |
| African American | 9.7 | NA |
| Hispanic | 7.5 | NA |
| Other | 9.2 | NA |
| Baseline FEV1, % predicted | 95.1 (SD, 13) | 96.4 (SD, 14) |
| Age of asthma onset, yr | ||
| Mean | 3.07 | 3.03 |
| Median, IQR | 2.5, 1.0–4.5 | 2.5, 1.0–4.0 |
| BMI at first visit | ||
| Mean | 18.2 | 17.7 |
| Median, IQR | 16.6, 15.6–19.8 | 17.7, 15.6–19.1 |
| Log of serum IgE, total | 2.62 | 2.56 |
Definition of abbreviations: BMI = body mass index; CAMP = Childhood Asthma Management Program; IQR = interquartile range; NA = not applicable.
Choice of SNPs and Molecular Methods
Choice of SNPs and SNP locations are described in detail in the online supplement, including representation of the SNPs reported as being significantly overtransmitted to Japanese individuals with atopic asthma (26). Briefly, haplotypes were identified with the PHASE program (30) and this output was analyzed by the BEST program (31) to identify haplotype-tagging SNPs. As shown in Table 3, the four SNPs chosen for genotyping should have been able to distinguish all of the common (more than 7%) haplotypes identified in Europeans. SNP genotyping was performed with the SEQUENOM system (see the online supplement for a detailed description).
TABLE 3.
Lymphotoxin-α and tumor necrosis factor haplotypes and haplotype tagging single-nucleotide polymorphisms for europeans in the ceph sample
| Haplotype No. | LTA 559 | LTA 2202 | LTANcoI | LTA 2820 | LTA 3842 | LTA 4371 | LTA 4539 | TNF –308 | TNF 352 | TNF 1078 | Haplotype Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | T | A | A | A | A | T | C | G | G | G | 11 (0.24) | ||||||||
| 2* | A | C | A | C | A | C | A | G | G | G | 7 (0.15) | ||||||||
| 3 | A | C | G | A | A | T | C | G | G | G | 7 (0.15) | ||||||||
| 4 | T | C | G | A | A | T | C | A | G | G | 5 (0.11) | ||||||||
| 5 | T | A | A | A | A | T | C | G | G | A | 4 (0.09) | ||||||||
| 6* | T | C | A | A | A | C | C | G | A | G | 3 (0.07) | ||||||||
| 37 (0.80) | |||||||||||||||||||
| Alternative SNPs†
|
|||||||||||||||||||
| LTA 1829 | LTA 2132 | LTA 1071 | LTA 2619 | LTA 306 | TNF 4101 | TNF 1009 | LTA 4545* | ||||||||||||
| LTA 2490 | LTA 2847 | TNF 2642 | LTA 1828 | TNF 1440 | TNF 4765 | ||||||||||||||
| TNF 4013 | TNF 1893 | ||||||||||||||||||
| TNF 4040 | |||||||||||||||||||
| TNF 4366 | |||||||||||||||||||
Definition of abbreviations: CEPH = Centre d'Etude du Polymorphisme Humain; LTA = lymphotoxin-α; SNP = single-nucleotide polymorphism; TNF = tumor necrosis factor.
Included are all haplotypes at 5% or greater frequency in the European population. SNPs in boldface were genotyped. SNP numbering is as reported by SeattleSNPs, NHLBI Program for Genomic Applications, University of Washington–Fred Hutchinson Cancer Research Center (Seattle, WA), except for LTA 2374 = LTA NcoI and TNF 282 = TNF –308 (URL: http://pga.mbt.washington.edu; accessed July 2005).
Haplotypes 2 and 6 cannot be distinguished from each other. Haplotypes 1, 3, 4, and 5 can be identified, using the SNPs highlighted in boldface.
Alternative SNPs were in 100% linkage disequilibrium for all haplotypes reported, accounting for any haplotype that occurred in at least two individuals.
Statistical Analysis
The PedCheck program was used to assess the genotype data for pedigree errors (32). Hardy-Weinberg equilibrium was tested in the parental data for each locus, using the χ2 goodness of fit test in SAS (version 8.1; SAS Institute, Cary, NC).
The primary analysis was for association of individual SNPs and haplotypes with asthma, applying the PBAT (33) program to the entire sample for the SNP analysis and to white individuals for the haplotype analysis. In the PBAT program the recessive model was used because that model had optimal power, as has been shown in previous studies (34, 35). The secondary analysis was to test each individual SNP/haplotype for association with a time series of asthma-related quantitative phenotypes, using the PBAT program (36). The time series data were obtained during multiple visits (before, at, and after study entry) to children in the CAMP population (see the online supplement for details). We tested the age at onset of asthma and the following four-time series: body mass index (BMI), logarithm of the provocative concentration of methacholine causing a 20% fall in FEV1 from baseline (LNPC20), baseline FEV1 % predicted (Pre-FEV), and postbronchodilator FEV1 % predicted (Pos-FEV). The time series data were tested with the FBAT-PC statistic for repeated measurements (36) and the time-to-onset data were tested with FBAT-LOGRANK (37). Both tests are available in PBAT for SNP and haplotype analysis.
Pairwise linkage disequilibrium between each pair of SNP loci was evaluated by a maximum likelihood method to infer phases for dual heterozygotes, expressed as r2.
RESULTS
Genotyping Results and Haplotype Evaluation
Genotypes for families with pedigree check errors were set to zero. The number of PedCheck errors for each SNP were as follows: LTA NcoI (23), LTA 4371 (9), TNF –308 (3), and TNF 1078 (3). Genotyping success rates were 97% or greater for each SNP. Hardy-Weinberg equilibrium was confirmed for all loci in the parents (all p values were greater than 0.1). There was significant linkage disequilibrium between the TNF –308 and LTA NcoI SNPs (r2 = 0.424). The LTA NcoI SNP and the LTA 4371 SNP had an r2 value of 0.172, and the TNF 282 SNP and LTA 4371 SNP had an r2 value of 0.083. The r2 values for all other SNP pairs were less than 0.04.
There were five four-SNP haplotypes represented in the sample at 5% or greater frequency in any ethnic group. The common haplotypes represented in the sample were the same as those with 5% or greater frequency reported in Table 3 for the Centre d'Etude du Polymorphisme Humain European sample.
Family-based Association Analysis of the TNF SNPs and Haplotypes with Asthma
As shown in Table 4, the A allele for the TNF –308 SNP was not overtransmitted to children with asthma from their parents. In contrast to previous reports of an increased frequency of the A allele in the asthmatic cohorts, we found a trend toward undertransmission of the A allele in the entire cohort. using additive or recessive models. There was also a trend for the G allele of the LTA NcoI polymorphism to be overtransmitted, but the p value (less than 0.05) was not corrected for multiple comparisons.
TABLE 4.
Family-based association test of tumor necrosis factor single-nucleotide polymorphisms from parents to children with asthma in the camp cohort: recessive model
| Marker | Allele | Allele Frequency | Informative Families* | p Value |
|---|---|---|---|---|
| CAMP cohort | ||||
| LTA NcoI | A | 0.657 | 337 | 0.912 |
| G | 0.343 | 184 | 0.048 | |
| LTA 4371 | C | 0.208 | 83 | 0.257 |
| T | 0.792 | 303 | 0.585 | |
| TNF –308 | A | 0.141 | 48 | 0.744 |
| G | 0.859 | 246 | 0.068 | |
| TNF 1078 | A | 0.094 | 23 | 0.251 |
| G | 0.906 | 163 | 0.125 | |
| White subjects only | ||||
| LTA NcoI | A | 0.646 | 260 | 0.775 |
| G | 0.354 | 134 | 0.101 | |
| LTA 4371 | C | 0.206 | 62 | 0.676 |
| T | 0.794 | 195 | 0.382 | |
| TNF –308 | A | 0.128 | 38 | 0.855 |
| G | 0.872 | 195 | 0.252 | |
| TNF 1078 | A | 0.085 | 17 | 0.261 |
| G | 0.915 | 121 | 0.686 |
Definition of abbreviations: CAMP = Childhood Asthma Management Program; TNF = tumor necrosis factor.
Informative families = at least one parent heterozygous for the allele.
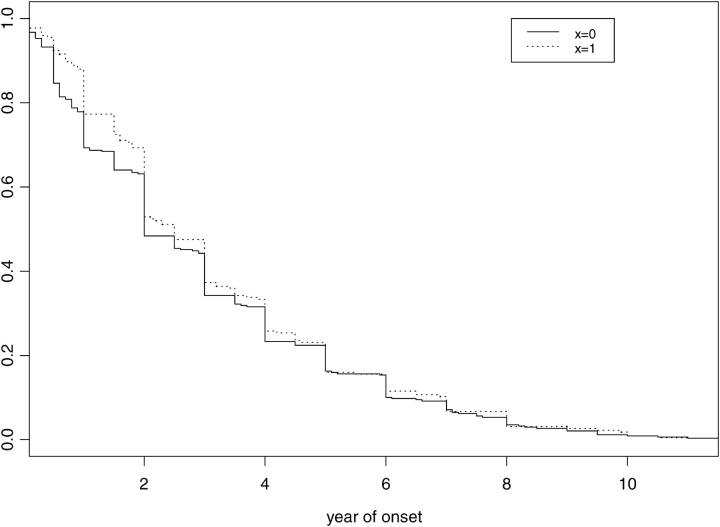
We restricted the extended haplotype analysis to white subjects. As shown in Table 5, there was an increase in transmission of the LTA NcoI_G/LTA 4371T/TNF –308G/TNF 1078G (GTGG) haplotype, using the recessive model (GTGG = 167 vs. other haplotypes = 146; relative risk, 2/0 copies GTGG = 3.1 and 1/0 copy GTGG = 2.75, p = 0.05). The p values shown in Table 5 are corrected for multiple comparisons. Table 5 also shows the results of the haplotype analysis for the GTGG haplotype for asthma-related phenotypes. The GTGG haplotype was associated with later age of onset of asthma based on the Kaplan-Meier plot (Figure 2). The GTGG haplotype was also associated with BMI, LNPC20, Pre-FEV, and Pos-FEV, using the time-series phenotype data.
TABLE 5.
| Phenotype | Test Statistic | p Value‡ | h |
|---|---|---|---|
| Asthma | HBAT | 0.05 | — |
| Later age of onset | HBAT-LOGRANK | 0.007 | — |
| BMI | HBAT-PC | 0.025 | 0.039 |
| LNPC20 | HBAT-PC | 0.014 | 0.037 |
| Pre-FEV | HBAT-PC | 0.019 | 0.062 |
| Pos-FEV | HBAT-PC | 0.044 | 0.054 |
Definition of abbreviations: h = percentage of phenotypic variance explained; BMI = body mass index; LNPC20 = logarithm of the provocative concentration of methacholine causing a 20% fall in FEV1 from baseline; Pre-FEV = baseline FEV1 % predicted; Pos-FEV = postbronchodilator FEV1 % predicted.
GTGG = LTA NcoI_G/LTA 4371_T/TNF –308_G/TNF 1078_G haplotype.
Restricted to white individuals in the Childhood Asthma Management Program.
p values are corrected for multiple comparisons.
Figure 2.
Kaplan-Meier curve of reported asthma age of onset by the presence (x = 1) or the absence (x = 0) of the LTA NcoI_G/LTA 4371T/TNF –308G/TNF 1078G haplotype. A statistically significant association for later age of asthma onset in carriers of the haplotype was found (p = 0.007).
DISCUSSION
Our data indicate that the TNF –308 and LTA NcoI polymorphisms do not influence asthma susceptibility individually, but that an extended haplotype in the TNF and LTA genes is associated with asthma susceptibility and with multiple common asthma phenotypes. In contrast to seven published case-control studies that showed an increased frequency of the TNF –308A allele in individuals with asthma (3–9), we instead found a strong trend toward an increased frequency of transmission from parents of the G allele to their child with asthma, consistent with other investigators in two case-control studies (19, 20). The TNF and LTA genes are located in the major histocompatibility complex on chromosome 6 between many innate immunity genes (see Figure 1). We believe that previous linkage with asthma in this region may be explained by extended haplotypes across the TNF and LTA genes.
Our study has many strengths. We used a family-based genetic study design including a large number of parent–child trios. Our population included more subjects with asthma than have been previously enrolled in other published studies evaluating SNPs in TNF and LTA genes (see Table 1). All children had mild to moderate asthma and extensive asthma phenotype data had been collected longitudinally (29). Genotyping success rates were high and we were able to infer haplotypes from family-based data at a high rate.
Our study is limited by the number of SNPs that were genotyped. Because our population was largely white, we chose SNPs to replicate findings in published reports and chose additional SNPs to ensure good representation of common haplotypes. We cannot rule out the possibility that linkage disequilibrium between polymorphisms in the asthmatic population may be different from those in the nonasthmatic population. This is unlikely, however, because our four-SNP haplotype results are consistent with those reported for the nonasthmatic European Centre d'Etude du Polymorphisme Humain sample. With the four-haplotype–tagging SNPs we chose, we were able to test the majority of common haplotypes across the TNF and LTA genes for association with asthma.
Noguchi and colleagues (26) reported an association between a TNF polymorphism and asthma susceptibility in a population of Japanese children with atopic asthma in 144 families. They reported increased transmission of the C allele of the TNF –857C/T SNP to children with asthma in 54 informative families. Further evaluation of haplotypes showed significantly increased transmission of the TNF –1031T/–857C (our SNPs LTA 4371T/TNF 1078G) haplotype in 74 informative families. Although we substituted the TNF 1078 SNP for their SNP labeled –857, these two SNPs are reported to be in 100% linkage disequilibrium in nonasthmatic white individuals. The TNF –308 SNP is not found in the Japanese population (26). Our results do not contradict the finding of Noguchi and colleagues (26), but our haplotypes are more extended. The TNF distal promoter haplotype they reported is broken into subhaplotypes in our mostly white sample, and we had a much larger population of individuals with asthma with many more informative families.
Whether the TNF –308 locus modulates the levels of tumor necrosis factor transcription is controversial. To resolve this controversy, Knight and colleagues (38) used chromatin immunoprecipitation and mass spectrometry (haplo-CHIP analysis) to identify differential protein–DNA binding in vivo associated with allelic variants of the TNF and LTA genes. Using Epstein-Barr virus–transformed B-cell lines, they evaluated RNA polymerase II loading and tumor necrosis factor mRNA expression. Their haplo-CHIP analysis of the –308 TNF polymorphism did not find differential expression of tumor necrosis factor by the two alleles of the SNP. They then extended their haplotypes to include SNPs across the LTA gene. They identified only three major haplotypes across these two genes. Two of the haplotypes had increased transmission of tumor necrosis factor (TNF –1031T_TNF–308A_LTA NcoIG and TNF –1031C_TNF –308G _LTA NcoIA) in comparison with the third haplotype (TNF –1031T_TNF –308G_LTA NcoIA). These haplotypes represent the following respective haplotypes in Table 3: haplotype 4, haplotype 2 or 6, and haplotype 1 or 5. Haplotype 3 (Table 3), which we found to be positively associated with asthma, was not represented in their analysis. Extrapolation from the study by Knight and colleagues (38) is limited because three common haplotypes found in asthmatic and nonasthmatic subjects are missing from their analysis. However, it is clear from their study that the TNF –308 SNP does not upregulate tumor necrosis factor levels and is not likely to be the important SNP as previously hypothesized. We were not able to test whether haplotype 3 (Table 3) had a regulatory effect on tumor necrosis factor levels.
In a large population of children with mild to moderate asthma, we have shown that the TNF –308 and LTA NcoI loci are not associated with asthma susceptibility or asthma phenotypes. Our results are consistent with the majority of other case-control and family-based studies shown in Table 1 that failed to find an association between asthma and these two loci. One haplotype across the TNF and LTA genes was found to be associated with asthma susceptibility and with asthma phenotypes in our cohort. Future studies of the association of polymorphisms in the TNF and LTA genes should evaluate extended haplotypes across the TNF and LTA genes and across other nearby genes in the major histocompatibility complex (5).
Supplementary Material
Acknowledgments
The authors thank all families for their enthusiastic participation in the CAMP Genetics Ancillary Study, supported by the National Heart, Lung, and Blood Institute (NO1-HR-16049). The authors also acknowledge the CAMP investigators and research team, supported by the NHLBI, for collection of CAMP Genetic Ancillary Study data. All work on data from the CAMP Genetics Ancillary Study was conducted at the Channing Laboratory, Brigham and Women's Hospital under appropriate CAMP policies and human subject protections.
Supported by National Institutes of Health (NIH) grants K23HL04278, U01 HL66795, PO1 HL67664, and P50 HL67664, and by MedImmune, Inc. A.G.R. is supported by NIH NHLBI K23 award HL04278.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Conflict of Interest Statement: A.G.R. received an unrestricted $10,000 education grant from MedImmune, Inc., to pay for genotyping costs. C.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. E.K.S. received grant support and honoraria from GlaxoSmithKline for a study of chronic obstructive pulmonary disease (COPD) genetics. He also received a $500 speaker fee from Wyeth for a talk on COPD genetics. R.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.T.W. received a grant for $900,065, Asthma Policy Modeling Study, from AstraZeneca from 1997 to 2003. He has been a coinvestigator on a grant from Boehringer-Ingelheim to investigate a COPD natural history model that began in 2003. He has received no funds for his involvement in this project. He has been an advisor to the TENOR Study for Genentech and has received $5,000 for 2003–2004. He received a grant from GlaxoWellcome for $500,000 for genomic equipment from 2000 to 2003. He was a consultant for Roche Pharmaceuticals in 2000 and received no financial remuneration for this consultancy.
References
- 1.Daniels SE, Bhattacharrya S, James A, Leaves NI, Young A, Hill MR, Faux JA, Ryan GF, le Souef PN, Lathrop GM, et al. A genome-wide search for quantitative trait loci underlying asthma. Nature 1996;383:247–250. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, et al. Genomewide screen and identification of gene–gene interactions for asthma-susceptibility loci in three US populations: collaborative study on the genetics of asthma. Am J Hum Genet 2001;68:1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chagani T, Pare PD, Zhu S, Weir TD, Bai TR, Behbehani NA, Fitzgerald JM, Sandford AJ. Prevalence of tumor necrosis factor-α and angiotensin converting enzyme polymorphisms in mild/moderate and fatal/near-fatal asthma. Am J Respir Crit Care Med 1999;160:278–282. [DOI] [PubMed] [Google Scholar]
- 4.Li Kam Wa TC, Mansur AH, Britton J, Williams G, Pavord I, Richards K, Campbell DA, Morton N, Holgate ST, Morrison JF. Association between –308 tumour necrosis factor promoter polymorphism and bronchial hyperreactivity in asthma. Clin Exp Allergy 1999;29:1204–1208. [DOI] [PubMed] [Google Scholar]
- 5.Moffatt MF, Cookson WO. Tumour necrosis factor haplotypes and asthma. Hum Mol Genet 1997;6:551–554. [DOI] [PubMed] [Google Scholar]
- 6.Sandford AJ, Chan HW, Wong GW, Lai CK, Chan-Yeung M. Candidate genetic polymorphisms for asthma in Chinese schoolchildren from Hong Kong. Int J Tuberc Lung Dis 2004;8:519–527. [PubMed] [Google Scholar]
- 7.Wang TN, Chen WY, Wang TH, Chen CJ, Huang LY, Ko YC. Gene–gene synergistic effect on atopic asthma: tumour necrosis factor-α-308 and lymphotoxin-α-NcoI in Taiwan's children. Clin Exp Allergy 2004;34:184–188. [DOI] [PubMed] [Google Scholar]
- 8.Winchester EC, Millwood IY, Rand L, Penny MA, Kessling AM. Association of the TNF-α-308 (G→A) polymorphism with self-reported history of childhood asthma. Hum Genet 2000;107:591–596. [DOI] [PubMed] [Google Scholar]
- 9.Witte JS, Palmer LJ, O'Connor RD, Hopkins PJ, Hall JM. Relation between tumour necrosis factor polymorphism TNFα-308 and risk of asthma. Eur J Hum Genet 2002;10:82–85. [DOI] [PubMed] [Google Scholar]
- 10.Beghe B, Padoan M, Moss CT, Barton SJ, Holloway JW, Holgate ST, Howell WM, Mapp CE. Lack of association of HLA class I genes and TNF α-308 polymorphism in toluene diisocyanate-induced asthma. Allergy 2004;59:61–64. [DOI] [PubMed] [Google Scholar]
- 11.Buckova D, Holla LI, Vasku A, Znojil V, Vacha J. Lack of association between atopic asthma and the tumor necrosis factor α-308 gene polymorphism in a Czech population. J Investig Allergol Clin Immunol 2002;12:192–197. [PubMed] [Google Scholar]
- 12.Di Somma C, Charron D, Deichmann K, Buono C, Ruffilli A. Atopic asthma and TNF-308 alleles: linkage disequilibrium and association analyses. Hum Immunol 2003;64:359–365. [DOI] [PubMed] [Google Scholar]
- 13.El Bahlawan L, Christensen M, Binaei S, Murphy C, Zhang Q, Quasney M. Lack of association between the tumor necrosis factor-α regulatory region genetic polymorphisms associated with elevated tumor necrosis factor-α levels and children with asthma. Chest 2003;123:374S–375S. [PubMed] [Google Scholar]
- 14.Lin YC, Lu CC, Su HJ, Shen CY, Lei HY, Guo YL. The association between tumor necrosis factor, HLA-DR alleles, and IgE- mediated asthma in Taiwanese adolescents. Allergy 2002;57:831–834. [DOI] [PubMed] [Google Scholar]
- 15.Louis R, Leyder E, Malaise M, Bartsch P, Louis E. Lack of association between adult asthma and the tumour necrosis factor α-308 polymorphism gene. Eur Respir J 2000;16:604–608. [DOI] [PubMed] [Google Scholar]
- 16.Moffatt MF, James A, Ryan G, Musk AW, Cookson WO. Extended tumour necrosis factor/HLA-DR haplotypes and asthma in an Australian population sample. Thorax 1999;54:757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trabetti E, Patuzzo C, Malerba G, Galavotti R, Martinati LC, Boner AL, Pignatti PF. Association of a lymphotoxin α gene polymorphism and atopy in Italian families. J Med Genet 1999;36:323–325. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, Chan-Yeung M, Becker AB, Dimich-Ward H, Ferguson AC, Manfreda J, Watson WT, Pare PD, Sandford AJ. Polymorphisms of the IL-4, TNF-α, and Fcε RIβ genes and the risk of allergic disorders in at-risk infants. Am J Respir Crit Care Med 2000;161:1655–1659. [DOI] [PubMed] [Google Scholar]
- 19.Albuquerque RV, Hayden CM, Palmer LJ, Laing IA, Rye PJ, Gibson NA, Burton PR, Goldblatt J, Lesouef PN. Association of polymorphisms within the tumour necrosis factor (TNF) genes and childhood asthma. Clin Exp Allergy 1998;28:578–584. [DOI] [PubMed] [Google Scholar]
- 20.Shin HD, Park BL, Kim LH, Jung JH, Wang HJ, Kim YJ, Park HS, Hong SJ, Choi BW, Kim DJ, et al. Association of tumor necrosis factor polymorphisms with asthma and serum total IgE. Hum Mol Genet 2004;13:397–403. [DOI] [PubMed] [Google Scholar]
- 21.Messer G, Spengler U, Jung MC, Honold G, Blomer K, Pape GR, Riethmuller G, Weiss EH. Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-β gene correlates with a variant amino acid in position 26 and a reduced level of TNF-β production. J Exp Med 1991;173:209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proc Natl Acad Sci USA 1997;94:3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardaba B, Moffatt MF, Fernandez E, Jurado A, Rojo M, Garcia M, Ansotegui IJ, Cortegano I, Arrieta I, Etxenagusia MA, et al. Allergy to Dermatophagoides in a group of Spanish gypsies: genetic restrictions. Int Arch Allergy Immunol 2001;125:297–306. [DOI] [PubMed] [Google Scholar]
- 24.Immervoll T, Loesgen S, Dutsch G, Gohlke H, Herbon N, Klugbauer S, et al. Fine mapping and single nucleotide polymorphism association results of candidate genes for asthma and related phenotypes. Hum Mutat 2001;18:327–336. [DOI] [PubMed] [Google Scholar]
- 25.Izakovicova HL, Vasku A, Izakovic V, Znojil V. The interaction of the polymorphisms in transporter of antigen peptides (TAP) and lymphotoxin α (LT-α) genes and atopic diseases in the Czech population. Clin Exp Allergy 2001;31:1418–1423. [DOI] [PubMed] [Google Scholar]
- 26.Noguchi E, Yokouchi Y, Shibasaki M, Inudou M, Nakahara S, Nogami T, Kamioka M, Yamakawa-Kobayashi K, Ichikawa K, Matsui A, et al. Association between TNFA polymorphism and the development of asthma in the Japanese population. Am J Respir Crit Care Med 2002;166:43–46. [DOI] [PubMed] [Google Scholar]
- 27.Randolph AG, Lange C, Silverman EK, Lazarus R, Weiss ST. Association of tumor necrosis factor gene polymorphisms with asthma and asthma related phenotypes [abstract]. Am J Respir Crit Care Med 2005;2:A790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 2000;343:1054–1063. [DOI] [PubMed] [Google Scholar]
- 29.Childhood Asthma Management Program Research Group. Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 30.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001;68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF. Minimal haplotype tagging. Proc Natl Acad Sci USA 2003;100:9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange C, Laird NM. On a general class of conditional tests for family-based association studies in genetics: the asymptotic distribution, the conditional power, and optimality considerations. Genet Epidemiol 2002;23:165–180. [DOI] [PubMed] [Google Scholar]
- 34.Lazarus R, Raby BA, Lange C, Silverman EK, Kwiatkowski DJ, Vercelli D, Klimecki WJ, Martinez FD, Weiss ST. TOLL-like receptor 10 genetic variation is associated with asthma in two independent samples. Am J Respir Crit Care Med 2004;170:594–600. [DOI] [PubMed] [Google Scholar]
- 35.Randolph AG, Lange C, Silverman EK, Lazarus R, Silverman ES, Raby B, Brown A, Ozonoff A, Richter B, Weiss ST. The IL12B gene is associated with asthma. Am J Hum Genet 2004;75:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lange C, Blacker D, Laird NM. Family-based association tests for survival and times-to-onset analysis. Stat Med 2004;23:179–189. [DOI] [PubMed] [Google Scholar]
- 38.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet 2003;33:469–475. [DOI] [PubMed] [Google Scholar]
- 39.McArthur JA, Zhang Q, Quasney MW. Association between the A/A genotype at the lymphotoxin-α+250 site and increased mortality in children with positive blood cultures. Pediatr Crit Care Med 2002;3:341–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.