Abstract
Osteopetrosis is a disease characterised by a generalized skeletal sclerosis resulting from a reduced osteoclast-mediated bone resorption. Several spontaneous mutations lead to osteopetrotic phenotypes in animals. Moutier et al. (1974) discovered the osteopetrosis (op) rat as a spontaneous, lethal, autosomal recessive mutant. op rats have large non-functioning osteoclasts and severe osteopetrosis [1]. Dobbins et al. (2002) localised the disease causing gene to a 1.5 cM genetic interval on rat chromosome 10, which we confirm in the present report [2]. We also refined the genomic localisation of the disease gene and provide statistical evidence for a disease causing gene in a small region of rat chromosome 10. Three strong functional candidate genes are within the delineated region. Clcn7 was previously shown to underlie different forms of osteopetrosis, both in human and mice. ATP6v0c encodes a subunit of the vacuolar H(+)-ATPase or proton-pump. Mutations in TCIRG1, another subunit of the proton-pump, are known to cause a severe form of osteopetrosis. Given the critical role of proton pumping in bone resorption, the Slc9a3r2 gene, a sodium/hydrogen exchanger, was also considered as a candidate for the op mutation. RT-PCR showed that all 3 genes are expressed in osteoclasts, but sequencing found no mutations in either the coding regions nor in intron splice junctions. Our ongoing mutation analysis of other genes in the candidate region will lead to the discovery of a novel osteopetrosis gene and further insights into osteoclast functioning.
1. Introduction
Osteopetroses are a heterogeneous group of bone disorders characterised by a generalized skeletal sclerosis resulting from a reduced osteoclast-mediated bone resorption. The impaired bone resorption results usually in retarded bone growth and abnormally shaped bones. Despite their increased mass, osteopetrotic bones tend to be more brittle than normal [3]. The bones are shorter and broader and bone marrow cavities fail to develop in the long bones, which often results in immunological malfunctioning. In humans, the osteopetroses are clinically and radiologically heterogeneous. Based on age of onset, inheritance, severity and secondary clinical features, different subforms are defined: autosomal recessive infantile malignant osteopetrosis (ARO), autosomal recessive intermediate mild osteopetrosis (IARO) and autosomal dominant adult onset benign osteopetrosis (ADO). Seven genes are known to cause osteopetrosis in humans. Five of them are involved in the effector function of the osteoclast (TCIRG1, CLCN7, PLEKHM1, OSTM1, CA2) and two in the osteoclast differentiation and formation (RANK, RANKL) [4–11]. In most cases, the disease can (partly) be cured with hematopoietic stem cell transplantation (HSCT)[12]. However, when the cellular defect is not intrinsic to the osteoclast (i.e. RANKL) HSCT will not cure the osteopetrosis because the osteoclast precursors lack the right microenvironment necessary for osteoclastogenesis [10].
A large part of our understanding of osteoclast differentiation and functioning emerged from the investigation of osteopetrotic mutations in animals. These can either be induced experimentally or occur spontaneously. In the rat, 4 naturally occurring mutants with osteopetrosis have been described. The toothless (tl) rat was described in 1974 by Cotton and Gaines as a rat model suffering from severe osteopetrosis with absence of tooth eruption due to a reduction in osteoclast number [13]. Much later, we and others were able to show that a mutation in the Csf1 gene is causative for the disease[14, 15]. The second rat model, the incisors absent (ia) rat, suffers from a mild osteopetrosis that resolves with age [16]. We recently reported the involvement of mutations in Plekhm1 in osteopetrosis in the ia rat and two patients with intermediate autosomal recessive osteopetrosis [9]. Plekhm1 encodes a protein with a currently unknown function in the osteoclast. However, based on the presence of certain functional domains, the subcellular localisation of the protein, and the osteoclast phenotypes of ia rats and the human patients, it likely plays a role in osteoclastic vesicular transport and ruffled border formation. A third osteopetrotic rat model is microphthalmia blanc (mib) [17], which has a mild and transient form of osteopetrosis due to a large deletion that truncates the encoded microphthalmia transcription factor [18]. The fourth and only remaining unknown, osteopetrotic mutation in the rat is osteopetrosis (op), first discovered by Moutier et al. (1974) as a spontaneous, lethal mutation [1]. The affected mutants exhibit a persistent, generalized sclerosis and show no signs of remission with age. The animals fail to develop marrow cavities and the impaired resorption gives rise to bones with a typical club-shaped appearance (Figure 1). In op/op rats, osteoclasts are significantly reduced in number but are larger and more vacuolated than in normal littermates. Despite their foamy appearance, mutant osteoclasts can form ruffled borders and clear zones, but their ability to resorb bone is greatly impaired [19]. The ability to cure these rats by bone-marrow transplantation from normal littermates indicates that the primary problem lies within the osteoclast itself [20, 21]. Remmers et al. localized the gene responsible for the op defect on chromosome 10, while Dobbins et al. (2002) localised the disease causing gene to a 1.5 cM genetic interval on rat chromosome 10 [2, 22].
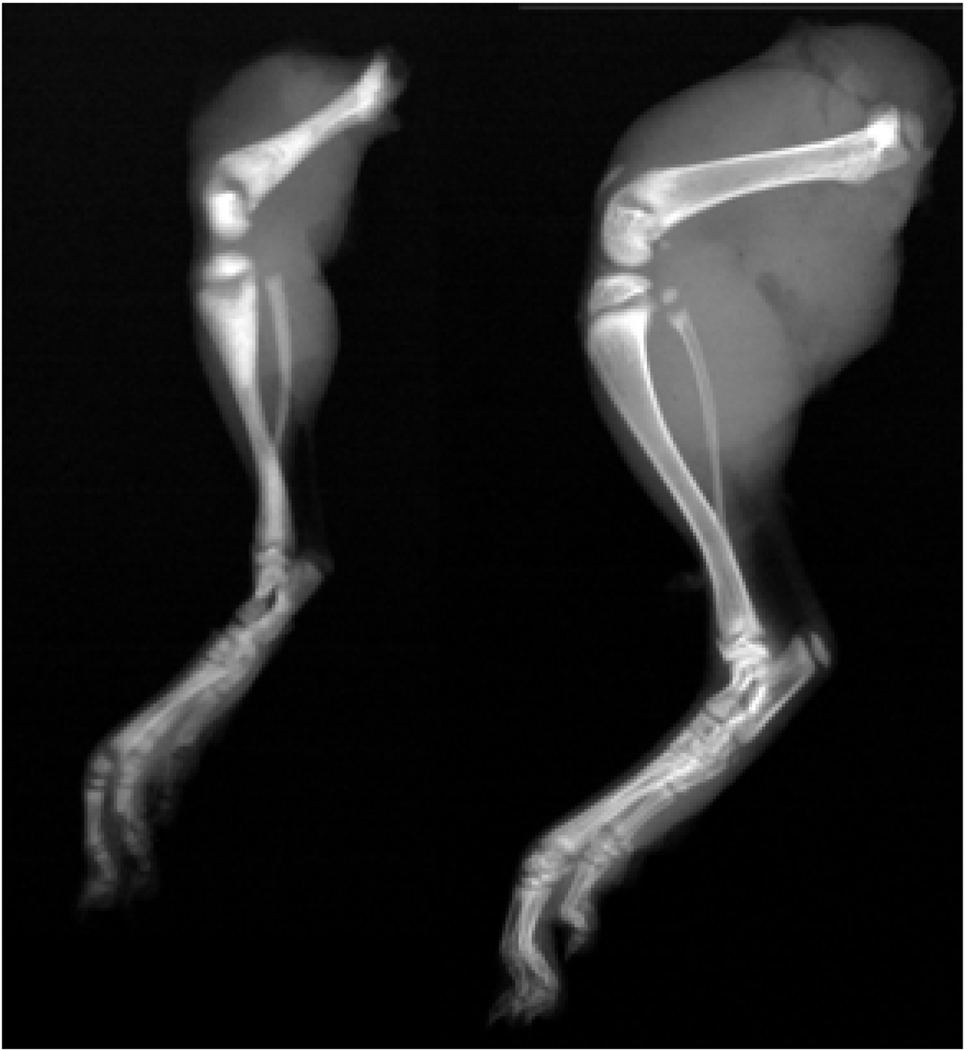
Figure 1.
Hind limbs of a three week-old op/op (left) rat and normal littermate (right). Note the typical osteosclerotic features in the affected rat. The mutants exhibit a persistent, generalized sclerosis. The club-shaped bones lack bone marrow cavities
This report describes our efforts towards the identification of the gene responsible for the osteopetrotic phenotype of the op rat, the last osteopetrotic ratmodel with an unknown genetic cause. We confirmed the chromosomal localisation of Dobbins et al. and were able to significantly reduce the candidate region. Furthermore, we present evidence that excludes three functional and positional candidate genes from this region (Atp6v0c, Clcn7 and Slc9a3r2) as the disease causing genes.
2. Materials and Methods
Breeding Strategy
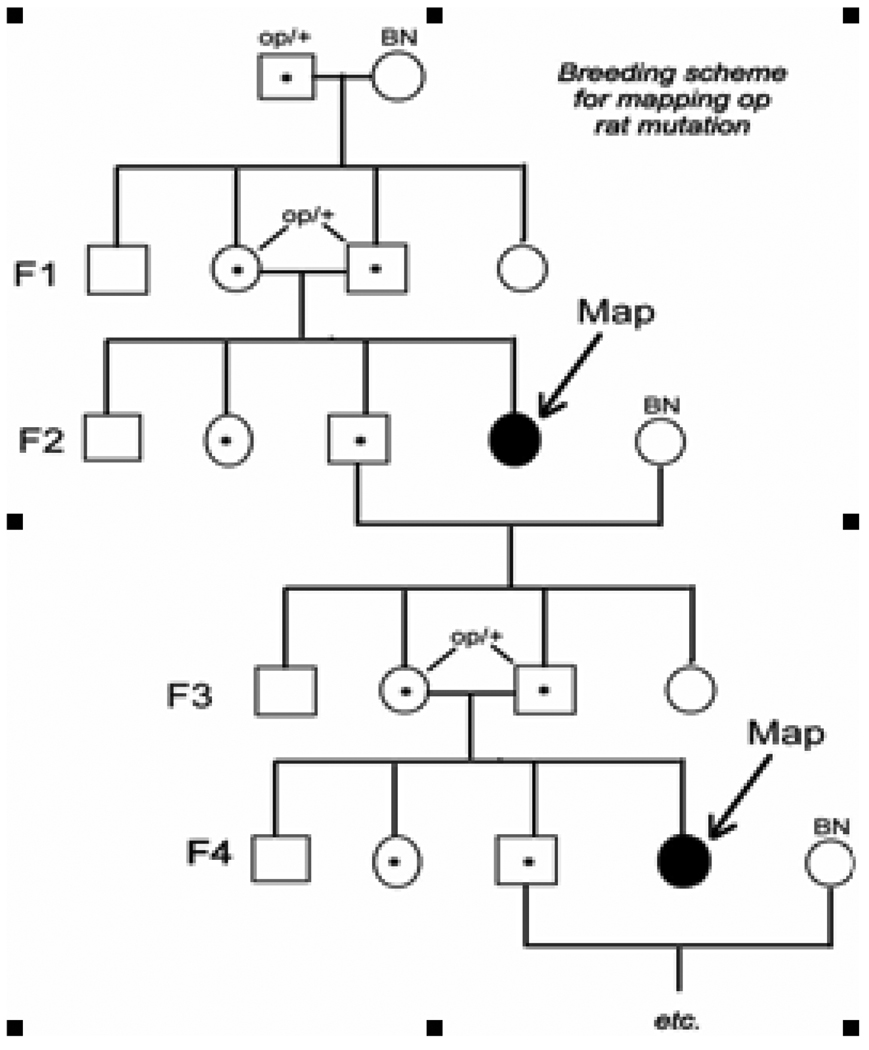
All animal procedures were in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health and were approved by the Institutional Animal Care and Committee of the University of Massachusetts Medical School. Mutant op rats, which are on a LEW/Ssn background, were obtained from inbred colonies maintained at the University of Massachusetts Medical School. Male LEW/SsN. +/op were crossed with normal BN/SsN rats (Figure 2), because these strains differ for a high percentage of polymorphisms (60 %). The progeny from this mating are all phenotypically normal, since it is inherited as an autosomal recessive trait. We intercrossed the F1 generation and selected the affected F2 pups. Radiography within 3 days of birth was used to identify mutant animals by the failure to develop bone marrow cavities. Genomic DNA from the tails of the affected pups was purified by standard methods. This strategy was repeated with subsequent even and odd generations to continue and refine the search for informative recombinations.
Figure 2.
Breeding strategy. Male LEW/SsN. +/op were crossed with normal BN/SsN rats. The progeny from this mating are all phenotypically normal, since it is inherited as an autosomal recessive trait. We intercrossed the F1 generation and selected the affected F2 pups. Radiography within 3 days of birth was used to identify mutant animals by the failure to develop bone marrow cavities. The breeding strategy was continued in successive odd and even generations.
Mapping Strategy
Variation in simple sequence repeat length was used to identify the LEW and BN alleles of each SSLP marker in 43 affected rats. Markers for loci near the candidate region proposed by Remmers et al. were selected (D10mit6, D10Rat198, D10Rat258, D10Got28, D10Rat51), analysed by PCR using fluorescent labelled primers and fragments were separated on an ABI3100 Analyzer (Applied Biosystems).
Expression Analysis
RNA was isolated from human osteoclasts generated from peripheral blood of healthy volunteers by culturing with CSF-1 and RANKL as previously desribed (Taylor et al. 2007). SuperScript ™ III First-Strand Synthesis System for RT-PCR was used to synthesize first strand cDNA (Invitrogen). One set of primers, spanning at least one intron, was constructed for each gene: clcn7, Atp6v0c and Slc9a3r2. The predicted product lengths at the cDNA level are 150 (Clcn7), 277 (Atp6v0c) and 417 base pairs (Slc9a3r2).
Mutation analysis
To detect the disease causing mutation, sequence analysis on three different DNA samples was performed: DNA extracted from the op/op rat, DNA from a BN/BN normal litter mate (nlm), and DNA from a healthy rat from the Lewis strain. PCR fragments spanning exons and exon/intron boundaries were analysed with the Big Dye terminator v3.1 Cycle sequencing kit (Applied Biosystems). The fragments were analysed on an ABI3100 analyzer (Applied Biosystems).
3. Results
Fine mapping
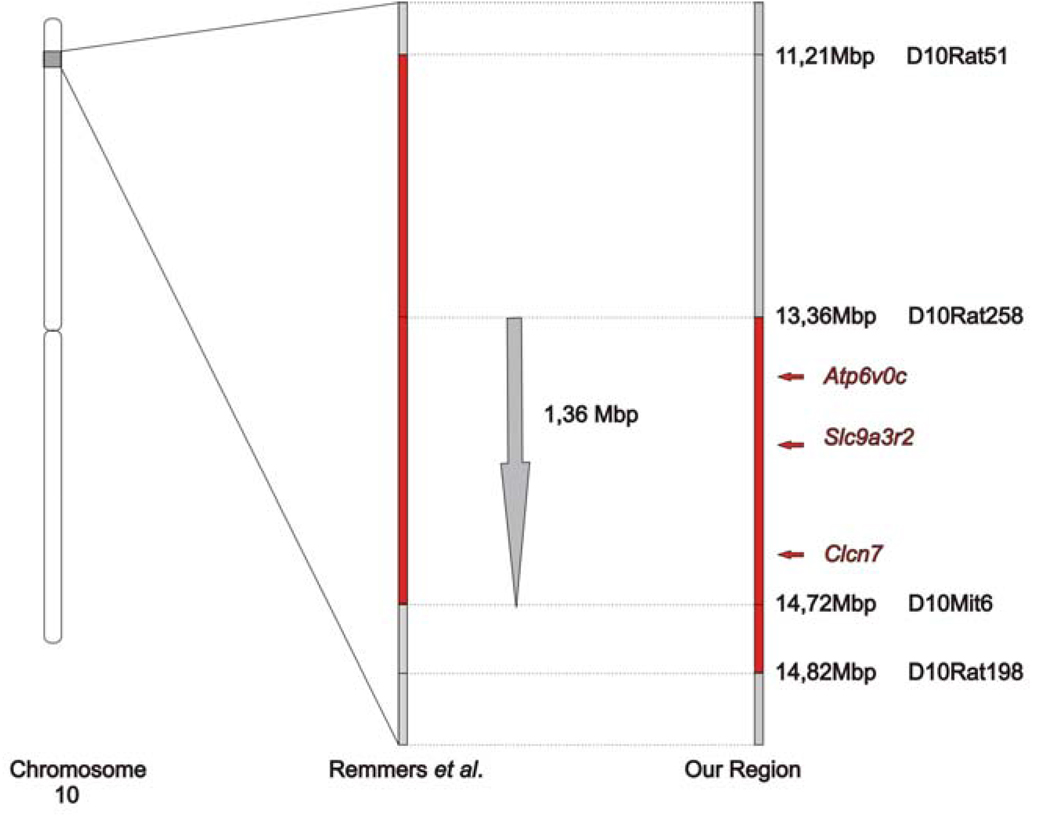
Dobbins et al. identified a candidate region of 1.5 cM on rat chromosome 10 [2]. We obtained DNA from 43 affected animals generated in our breeding to confirm this localisation and to perform fine mapping analysis. The results are shown in Figure 3. All affected (op/op) animals were homozygous for the LEW/SsN allele of one of their flanking markers, D10Mit6. This indicates a clear co-segregation between this chromosomal region and the disease, confirming the localisation of the disease-causing gene to this chromosomal region. Next, we tried to narrow the candidate region. D10Rat51, the flanking marker of Remmers et al. on the distal side was homozygous for the lew/ssn allele in all affected animals with the exception of two that were heterozygous lew/ssn- bn/ssn. These two animals were subjected to further fine mapping analysis in an effort to reduce the proposed candidate region. As one of them also recombined with the more proximal marker, D10Rat258, the candidate region at the proximal side was reduced by 2.15 Mbp. The marker proximal to D10Rat258, D10Got28, was homozygous for all our rats.
Figure 3.
By segregation analysis we narrowed the candidate region that carries the op mutation. In combination with the region defined by Remmers et al. we delineated a candidate region between the markers D10rat258 and D10Mit6 [22].
At the proximal end, no reduction of the candidate region could be obtained as there was no recombination with the flanking marker D10Mit6. The first recombination event in our animals was detected for marker D10Rat198. Therefore, our data delineate a region between D10Rat258 and D10Rat198. In combination with the data from Remmers et al., we provide evidence for a disease causing gene in a region of 1.36 Mbp on rat chromosome 10 (Figure 3).
Expression analysis
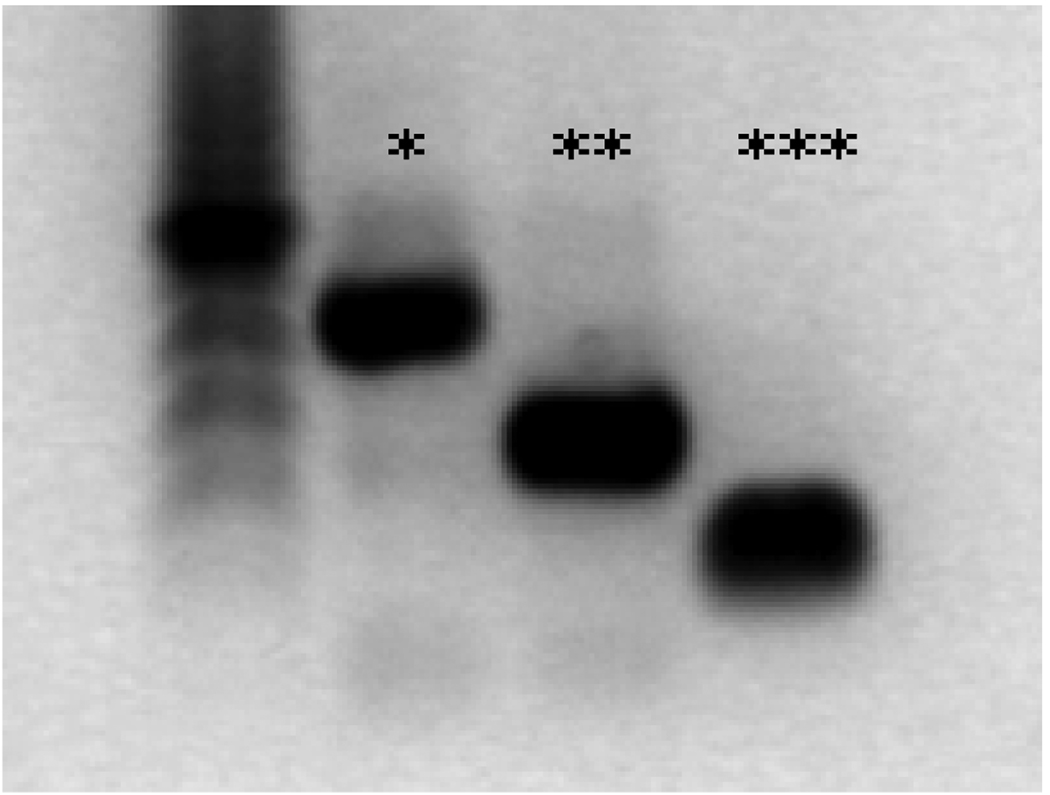
70 genes are located within the delineated candidate region among which three strong functional candidate genes: Atp6v0c, Clcn7 and Slc9a3r2. Expression analysis of these three genes on osteoclast cDNA showed that these genes are expressed in the osteoclast (Figure 4), making them of interest for further analysis.
Figure 4.
expression analysis of the three candidate genes by RT-PCR on osteoclast cDNA. The product lengths are 417* (Slc9a3r2), 277** (Atp6v0c) and 150** (Clcn7) basepairs.
Mutation analysis
Sequencing of the 25 exons of clcn7, the 3 exons of atp6v0c, the 7 exons of Slc9a3r2 and all of the exon/intron boundaries of these genes, showed no sequence difference between the op/op rat and the normal rats (nlm and Lewis).
4. Discussion
The results of this study suggest that identification of the mutation in the op rat will reveal a gene with a previously unknown role in osteoclast biology. The fact that no abnormalities were found in any of the three strongest candidate genes in the region supports this notion. The Clcn7 gene is known to cause severe osteopetrosis in humans and in knockout mice, and mutations in a subunit of the vacuolar ATPase proton pump are similarly involved in severe osteopetrosis. Our hypothesis that the sodium/proton exchanger may carry the op mutation was based on the well-known role of proton transport and pH regulation in bone resorption. Previous identification of osteopetrosis genes by us and by others has been a rich source of new information about osteoclast regulation and activity. For example, the op mouse was shown to carry a mutation in the Csf1 gene [23] which demonstrated the critical requirement of that growth factor in osteoclast ontogeny. Later, we and others showed that the tl rat also carried an inactivating mutation in Csf1 [24, 25]. The gl mouse, a naturally-occurring osteopetrotic mutant, was found to carry a mutation in a previously unknown gene [6] now called Ostm1. Mutational screening of this gene in patients with ARO revealed this gene as an osteopetrotic gene in humans [6, 26, 27]. Investigation of the mutation underlying the ia rat phenotype identified the previously unknown gene, Plekhm1, as an osteopetrotic gene in rat and human [9]. The precise function of this gene is currently not well understood, but phenotypic and microscopic data suggest an important role in vesicular transport and secretion in osteoclasts.
With our current mapping efforts in the op rat, significant reduction (roughly by half) of the candidate region and the number of remaining potential disease causing genes was obtained.
This region contained what we considered to be 3 strong candidate genes: Atp6v0c, is a subunit of the H+-transporting V-ATPase, as is the osteopetrosis gene Tcirg1; This proton pump is responsible for the acidification of the resorption lacuna that is required for demineralization of the bone matrix. Mutations in TCIRG1 are responsible for about half of the human cases of ARO [28]. The Tcirg1−/− mouse and the spontaneous osteosclerotic (oc) mouse are two animal models in which the Tcirg1 gene is disrupted [29, 30]. Both show typical, severe osteopetrotic features in the presence of abundant osteoclasts. The activity of the proton pump in acidification of resorption lacuna during bone resorption requires the chloride channel CLC7 to maintain electroneutrality. Loss-of-function mutations in the CLCN7 gene are known to cause autosomal recessive osteopetrosis (ARO (MIM 259700)), which correlates with the severe form of ostepetrosis of the Clcn7−/−. Dominant negative mutations in the CLCN7 gene are known to cause autosomal dominant osteopetrosis (ADO (MIM 166600)), which is a more benign form of osteopetrosis.
Finally, given the critical role of proton pumping in bone resorption, the Slc9a3r2 gene, a sodium/hydrogen exchanger, was considered by us to be an important candidate for the op mutation.
Although clcn7 and Atp6v0c were both strong candidate genes, the exclusion of them was not entirely surprising. The number and the characteristics of osteoclasts in the op rat differ from those of the animal models with mutations in these genes. The Clcn7−/− and Tcirg1−/− mice don’t have reduced numbers of osteoclasts in bone tissues, and the typical foamy appearance of op osteoclasts is not observed in the osteoclasts from animals with mutations in Clcn7 or Tcirg1. As was the case for these two genes, analysis of the Slc9a3r2 gene revealed no mutations in the coding region or in splice junctions of introns. There is currently no knockout model available for this gene that would allow us to compare the phenotype with the op rat.
Although no mutations were found in the coding regions of these genes or in intron splice junctions, theoretically they can’t be fully excluded as disease causing genes. There remains the formal possibility that deep-intronic mutations or mutations in regulatory regions of one of these 3 genes might be the underlying genetic lesion causing the osteopetrotic phenotype in the op rat.
With the most probable exclusion of the obvious candidate genes, the elucidation of the causative mutation of the op rat is likely to deliver a novel osteopetrotic gene with a currently unknown function in osteoclasts. Therefore, further molecular genetic mapping and sequencing experiments are currently underway. Further breeding and positional cloning will enlarge the population of affected rats and may reduce the candidate region even further. After identification of the osteopetrotic causing gene, we will screen the human gene in a set of osteopetrotic patients with no known mutation in any of the currently known osteopetrosis genes. This approach has proven to be a fruitful way to identify osteopetrotic genes in humans, as proven by Ostm1 and plekhm1, two genes first identified in osteopetrotic animals and now known to cause osteopetrosis in human [6, 9, 26, 27].
In conclusion, these data indicate that the osteopetrosis seen in the op rat is not the result of a mutation within the coding region of the Slc9a3r2, Atp6v0c and Clcn7 genes. Analysis of additional functional and positional candidate genes in the 1.36 Mbp candidate region will be required to identify the causative mutation in this animal model of disturbed bone homeostasis.
Acknowledgment
Supported by grant DE07444 to PRO. BP holds a PhD studentship from the European Calcified Tissue Society. This work was supported by the F.W.O. Vlaanderen (grant G.0117.06) and by a grant from the Special Research Fund (B.O.F.) of the University of Antwerp both to WVH
References
- 1.Moutier R, Toyama K, Charrier MF. Genetic study of osteopetrosis in the Norway rat. J Hered. 1974;65:373–375. doi: 10.1093/oxfordjournals.jhered.a108554. [DOI] [PubMed] [Google Scholar]
- 2.Dobbins DE, Joe B, Hashiramoto A, Salstrom JL, Dracheva S, Ge L, Wilder RL, Remmers EF. Localization of the mutation responsible for osteopetrosis in the op rat to a 1.5-cM genetic interval on rat chromosome 10: identification of positional candidate genes by radiation hybrid mapping. J Bone Miner Res. 2002;17:1761–1767. doi: 10.1359/jbmr.2002.17.10.1761. [DOI] [PubMed] [Google Scholar]
- 3.de Vernejoul MC. Sclerosing bone disorders. Best Pract Res Clin Rheumatol. 2008;22:71–83. doi: 10.1016/j.berh.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Kornak U, Kasper D, Bosl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–215. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 5.Sly WS, Hewett-Emmett D, Whyte MP, Yu YS, Tashian RE. Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc Natl Acad Sci U S A. 1983;80:2752–2756. doi: 10.1073/pnas.80.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalhoub N, Benachenhou N, Rajapurohitam V, Pata M, Ferron M, Frattini A, Villa A, Vacher J. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat Med. 2003;9:399–406. doi: 10.1038/nm842. [DOI] [PubMed] [Google Scholar]
- 7.Kornak U, Schulz A, Friedrich W, Uhlhaas S, Kremens B, Voit T, Hasan C, Bode U, Jentsch TJ, Kubisch C. Mutations in the a3 subunit of the vacuolar H(+)-ATPase cause infantile malignant osteopetrosis. Hum Mol Genet. 2000;9:2059–2063. doi: 10.1093/hmg/9.13.2059. [DOI] [PubMed] [Google Scholar]
- 8.Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, Notarangelo LD, Vezzoni P, Villa A. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 9.Van Wesenbeeck L, Odgren PR, Coxon FP, Frattini A, Moens P, Perdu B, Mackay CA, Van Hul E, Timmermans JP, Vanhoenacker F, Jacobs R, Peruzzi B, Teti A, Helfrich MH, Rogers MJ, Villa A, Van Hul W. Involvement of PLEKHM1 in osteoclastic vesicular transport and osteopetrosis in incisors absent rats and humans. J Clin Invest. 2007;117:919–930. doi: 10.1172/JCI30328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobacchi C, Frattini A, Guerrini MM, Abinun M, Pangrazio A, Susani L, Bredius R, Mancini G, Cant A, Bishop N, Grabowski P, Del Fattore A, Messina C, Errigo G, Coxon FP, Scott DI, Teti A, Rogers MJ, Vezzoni P, Villa A, Helfrich MH. Osteoclast-poor human osteopetrosis due to mutations in the gene encoding RANKL. Nat Genet. 2007;39:960–962. doi: 10.1038/ng2076. [DOI] [PubMed] [Google Scholar]
- 11.Guerrini MM, Sobacchi C, Cassani B, Abinun M, Kilic SS, Pangrazio A, Moratto D, Mazzolari E, Clayton-Smith J, Orchard P, Coxon FP, Helfrich MH, Crockett JC, Mellis D, Vellodi A, Tezcan I, Notarangelo LD, Rogers MJ, Vezzoni P, Villa A, Frattini A. Human osteoclast-poor osteopetrosis with hypogammaglobulinemia due to TNFRSF11A (RANK) mutations. Am J Hum Genet. 2008;83:64–76. doi: 10.1016/j.ajhg.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Driessen GJ, Gerritsen EJ, Fischer A, Fasth A, Hop WC, Veys P, Porta F, Cant A, Steward CG, Vossen JM, Uckan D, Friedrich W. Long-term outcome of haematopoietic stem cell transplantation in autosomal recessive osteopetrosis: an EBMT report. Bone Marrow Transplant. 2003;32:657–663. doi: 10.1038/sj.bmt.1704194. [DOI] [PubMed] [Google Scholar]
- 13.Cotton WR, Gaines JF. Unerupted dentition secondary to congenital osteopetrosis in the Osborne-Mendel rat. Proc Soc Exp Biol Med. 1974;146:554–561. doi: 10.3181/00379727-146-38146. [DOI] [PubMed] [Google Scholar]
- 14.Van Wesenbeeck L, Odgren PR, MacKay CA, D'Angelo M, Safadi FF, Popoff SN, Van Hul W, Marks SC., Jr The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: Evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification. Proc Natl Acad Sci U S A. 2002;99:14303–14308. doi: 10.1073/pnas.202332999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobbins DE, Sood R, Hashiramoto A, Hansen CT, Wilder RL, Remmers EF. Mutation of macrophage colony stimulating factor (Csf1) causes osteopetrosis in the tl rat. Biochem Biophys Res Commun. 2002;294:1114–1120. doi: 10.1016/S0006-291X(02)00598-3. [DOI] [PubMed] [Google Scholar]
- 16.Marks SC., Jr Pathogenesis of osteopetrosis in the ia rat: reduced bone resorption due to reduced osteoclast function. Am J Anat. 1973;138:165–189. doi: 10.1002/aja.1001380204. [DOI] [PubMed] [Google Scholar]
- 17.Moutier R, Ostrowski K, Lamendin H. Microphthalmia: a new recessive mutation in the Norway rat. J Hered. 1989;80:76–78. doi: 10.1093/oxfordjournals.jhered.a110798. [DOI] [PubMed] [Google Scholar]
- 18.Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J Exp Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marks SC, Jr, Popoff SN. Osteoclast biology in the osteopetrotic (op) rat. Am J Anat. 1989;186:325–334. doi: 10.1002/aja.1001860402. [DOI] [PubMed] [Google Scholar]
- 20.Milhaud G, Labat ML, Graf B, Juster M, Balmain N, Moutier R, Toyama K. [Kinetic, radiographic, and histologic demonstration of the curing of congenital osteopetrosis in rats] C R Acad Sci Hebd Seances Acad Sci D. 1975;280:2485–2488. [PubMed] [Google Scholar]
- 21.Nisbet NW, Waldron SF, Marshall MJ. Failure of thymic grafts to stimulate resorption of bone in the Fatty/Orl-op rat. Calcif Tissue Int. 1983;35:122–125. doi: 10.1007/BF02405017. [DOI] [PubMed] [Google Scholar]
- 22.Remmers EF, Du Y, Ding YP, Kotake S, Ge L, Zha H, Goldmuntz EA, Hansen C, Wilder RL. Localization of the gene responsible for the op (osteopetrotic) defect in rats on chromosome 10. J Bone Miner Res. 1996;11:1856–1861. doi: 10.1002/jbmr.5650111205. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 24.Dobbins DE, Sood R, Hashiramoto A, Hansen CT, Wilder RL, Remmers EF. Mutation of macrophage colony stimulating factor (Csf1) causes osteopetrosis in the tl rat. Biochem Biophys Res Commun. 2002;294:1114–1120. doi: 10.1016/S0006-291X(02)00598-3. [DOI] [PubMed] [Google Scholar]
- 25.Van Wesenbeeck L, Odgren PR, MacKay CA, D'Angelo M, Safadi FF, Popoff SN, Van Hul W, Marks SC., Jr The osteopetrotic mutation toothless (tl) is a loss-of-function frameshift mutation in the rat Csf1 gene: Evidence of a crucial role for CSF-1 in osteoclastogenesis and endochondral ossification. Proc Natl Acad Sci U S A. 2002;99:14303–14308. doi: 10.1073/pnas.202332999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pangrazio A, Poliani PL, Megarbane A, Lefranc G, Lanino E, Di Rocco M, Rucci F, Lucchini F, Ravanini M, Facchetti F, Abinun M, Vezzoni P, Villa A, Frattini A. Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. J Bone Miner Res. 2006;21:1098–1105. doi: 10.1359/jbmr.060403. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez A, Faupel J, Goebel I, Stiller A, Beyer S, Stockle C, Hasan C, Bode U, Kornak U, Kubisch C. Identification of a novel mutation in the coding region of the grey-lethal gene OSTM1 in human malignant infantile osteopetrosis. Hum Mutat. 2004;23:471–476. doi: 10.1002/humu.20028. [DOI] [PubMed] [Google Scholar]
- 28.Del Fattore A, Peruzzi B, Rucci N, Recchia I, Cappariello A, Longo M, Fortunati D, Ballanti P, Iacobini M, Luciani M, Devito R, Pinto R, Caniglia M, Lanino E, Messina C, Cesaro S, Letizia C, Bianchini G, Fryssira H, Grabowski P, Shaw N, Bishop N, Hughes D, Kapur R, Datta H, Taranta A, Fornari R, Migliaccio S, Teti A. Clinical, genetic and cellular analysis of forty-nine osteopetrotic patients: implications for diagnosis and treatment. J Med Genet. 2005 doi: 10.1136/jmg.2005.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seifert MF, Marks SC., Jr Morphological evidence of reduced bone resorption in the osteosclerotic (oc) mouse. Am J Anat. 1985;172:141–153. doi: 10.1002/aja.1001720204. [DOI] [PubMed] [Google Scholar]
- 30.Li YP, Chen W, Liang Y, Li E, Stashenko P. Atp6i-deficient mice exhibit severe osteopetrosis due to loss of osteoclast-mediated extracellular acidification. Nat Genet. 1999;23:447–451. doi: 10.1038/70563. [DOI] [PubMed] [Google Scholar]