Abstract
Background
Artemether-lumefantrine (AL) is a major and highly effective artemisinin-based combination therapy that is becoming increasingly important as a new first-line therapy against Plasmodium falciparum malaria. However, recrudescences occurring after AL treatment have been reported. Identification of drug-specific parasite determinants that contribute to treatment failures will provide important tools for the detection and surveillance of AL resistance.
Method
The findings from a 42-day follow-up efficacy trial in Tanzania that compared AL with sulfadoxine-pyrimethamine (SP) were analyzed to identify candidate markers for lumefantrine tolerance/resistance in the chloroquine resistance transporter gene (pfcrt) and multidrug resistance gene 1 (pfmdr1). The findings were corroborated in vitro with genetically modified isogenic P. falciparum parasite lines.
Results
Treatment with AL selected for the chloroquine-susceptible pfcrt K76 allele (P < .0001) and, to a lesser extent, the pfmdr1 N86 (P = .048) allele among recurrent infections. These genotypes were not selected during SP treatment. No pfmdr1 gene amplifications were observed. Isogenic pfcrt-modified parasite lines demonstrated a 2-fold increase in susceptibility to lumefantrine, which was directly attributable to the K76T mutation.
Conclusions
Our findings suggest that the pfcrt K76T mutation is a drug-specific contributor to enhanced P. falciparum susceptibility to lumefantrine in vivo and in vitro, and they highlight the benefit of using AL in areas affected by chloroquine-resistant P. falciparum malaria.
The development and spread of Plasmodium falciparum antimalarial drug resistance has spurred a global change in policy from the use of the former first-line antimalarials chloroquine (CQ) and sulfadoxine-pyrimethamine (SP) to the use of artemisinin-based combination therapies (ACTs). In 2006, Tanzania changed its drug policy to the use of artemether-lumefantrine (AL; Coartem [Novartis]), which is currently the most favored ACT used in Africa.
AL is highly effective on the African continent [1–3]. Nevertheless, AL has been associated with a relatively high frequency of reinfection events during the post-treatment period, when lumefantrine, the longer-lasting partner drug (with a plasma half-life of 4–5 days vs. 3–11 h for artemether), remains in the blood in subtherapeutic concentrations. During this period, selection of less-susceptible parasites (which have the ability to cause infection in the presence of plasma drug concentrations higher than those associated with susceptible parasites causing infection) may occur.
One gene that has been found to influence susceptibility to lumefantrine is the P. falciparum multidrug resistance gene 1 (pfmdr1), which encodes a transporter located on the digestive vacuole (DV) membrane of the erythrocytic stages. Allelic exchange experiments with culture-adapted lines, as well as field studies of clinical isolates, have shown that pfmdr1 single-nucleotide polymorphisms (SNPs) influence the in vitro response to various antimalarial drugs, such as the artemisinin derivatives and the arylaminoalcohols lumefantrine, mefloquine (MQ), and halofantrine (HF) [4–7]. An increased pfmdr1 copy number has also been associated with treatment failure after short 4-dose AL therapy [8], as well as after MQ or artesunate-MQ therapy [9]. So far, these associations have been observed only in Southeast Asia. However, an increased number of pfmdr1 copies has been found in a few patient samples in Gabon, subsequent to a trial with low-dose MQ treatment, thereby suggesting the potential of selection of multicopy pfmdr1 in this region [10]. Hence, the pfmdr1 copy number may also become important on the African continent after wide-scale deployment of AL.
Our previous studies in Zanzibar showed that treatment with AL selected for reinfecting parasites harboring pfmdr1 alleles encoding the N86, 184F, and D1246 residues [11, 12]. These findings are consistent with reports from other East African field sites [13, 14] and suggest the selection of lumefantrine-tolerant parasites, which may constitute a first step toward full resistance and associated clinical failure [15]. Nonetheless, it remains unclear whether this selection is the result of specific influences on the drug target or is caused by nonspecific selection of parasites that are generally more fit and able to thrive in the presence of environmental stressors. This is an important distinction, considering that mutations at codons 1034, 1042, and 1246 in pfmdr1 have been reported to incur fitness costs in vitro [16].
Another question arising from our previous studies is the role of the chloroquine resistance transporter gene (pfcrt), which encodes a transmembrane protein that localizes to the membrane of the DV and is a key determinant of the chloroquine response of P. falciparum [17]. The pfcrt K76T mutation has been consistently associated with CQ resistance in vitro and in vivo [18 –20]. This putative transporter also appears to influence the parasite response to arylaminoalcohols. Accordingly, mutations at the pfcrt amino acid 76 position have been suggested to play a role in MQ susceptibility [19 –21]. In addition, in vitro selection of HF resistance has been associated with a novel pfcrt S163R mutation, which also has been shown to affect the in vitro response to MQ [22]. In light of these data, we hypothesized that pfcrt SNPs might also influence susceptibility to lumefantrine.
To evaluate the influence of pfmdr1 and pfcrt mutations on the mechanism of resistance to lumefantrine, we searched for selection of these mutations in samples collected during a clinical trial in Fukayosi, Tanzania, in which children with uncomplicated P. falciparum malaria were treated with AL or SP [2]. SP inhibits the synthesis of folate by targeting the dihydropteroate synthetase and dihydrofolate reductase enzymes, respectively. This treatment arm therefore enabled us to test whether any evidence of selection for pfcrt or pfmdr1 mutations in the AL arm was drug dependent and was not a result of general fitness issues associated with the parasite response to any antimalarial drug treatment. Furthermore, the availability of isogenic parasites resulting from allelic exchange experiments at pfcrt amino acid position 76 [19, 20] allowed us to evaluate the specific influence of mutant pfcrt alleles (in particular, the K76T mutation) on the response of P. falciparum to lumefantrine.
SUBJECTS, MATERIALS, AND METHODS
Subjects
The study was conducted in Fukayosi, Bagamoyo District, Tanzania, during April–July 2004, as reported elsewhere [2]. In brief, 106 children with uncomplicated P. falciparum malaria were enrolled and randomly allocated to receive either SP (Fansidar; Roche) or AL (Coartem). Both drugs were administered according to body weight. Children who were allocated to receive SP and who had body weights categorized as 5–10 kg, 11–20 kg, and 21–30 kg were given a single dose of sulfadoxine/pyrimethamine of 250 mg/12.5 mg, 500 mg/25 mg, and 750 mg/37.5 mg, respectively, whereas children who were allocated to receive AL and who had body weights categorized as 5–14 kg, 15–24 kg, and 25–34 kg were given a twice-daily dose of artemether/lumefantrine of 20 mg/120 mg, 40 mg/240 mg, and 60 mg/360 mg, respectively, for 3 days. After enrollment in the study and initiation of treatment, the children were checked routinely on days 1, 2, 3, 7, 14, 21, 28, 35, and 42 or on any day of recurrent illness occurring during the 42-day follow-up. Blood samples were collected on filter paper (3MM; Whatman) for molecular analysis. Written informed consent was obtained from the parents or guardians of the children. The study was approved by the ethics committees of the Muhimbili University College of Health and Allied Sciences (Dar Es Salaam, Tanzania) and Karolinska Institutet (Stockholm, Sweden).
Molecular analysis
DNA was extracted from blood samples on filter paper by use of the ABI Prism 6100 Nucleic Acid PrepStation (Applied Biosystems). The status of recurrent infections (recrudescence or reinfection) was available from a previous study [2] that used stepwise genotyping of the polymorphic genetic markers P. falciparum merozoite surface protein (pfmsp)–2 and –1 to classify treatment outcomes. In the present study, we analyzed the first baseline day (D0) and the initial day of recurrent infection (R0).
Mutations in pfmdr1 and pfcrt were investigated to explore their association with AL tolerance. All infections observed at baseline were examined, as were all infections that recurred after AL treatment. The pfmdr1 N86Y and D1246Y SNPs, as well as the pfcrt K76T and S163R SNPs, were analyzed by polymerase chain reaction (PCR) restriction fragment–length polymorphism (RFLP) analysis, as described elsewhere [12, 23]. The pfmdr1 Y184F SNP was assayed using pyrosequencing (Pyro-Gold Reagents PSQ 96MA and PyroMark MD System; Biotage AB) with the primer 5'-CCAGTTCCTTTTTAGGTT-3'. The pfmdr1 S1034C and N1042D SNPs were examined through direct sequencing of PCR products (Macrogen). For infections recurring after SP treatment, only the pfmdr1 N86Y and pfcrtK76T mutations were assessed. The pfmdr1 gene copy number was assessed through the use of TaqMan probe quantitative PCR–based protocols [12]. All primers were synthesized by Thermo Electron. RFLP analysis products were separated by electrophoresis in 2% agarose gels with ethidium bromide and were visualized under UV transillumination (GelDoc System; Bio-Rad).
Statistical analysis
Data from patient samples were analyzed using GraphPad QuickCalcs software (GraphPad Software). Proportions were compared using Fisher’s exact test for the association of treatment type with SNPs. The complete set of baseline data (D0 for the AL arm + D0 for the SP arm) was used as a common comparator group for both arms of the drug trial. Statistical significance was denoted by P ≤ .05. When mixed infections were evaluated, all calculations were performed by comparing carriers of the selected allele (including pure allele carriers and mixed infections) with parasites carrying the alternative allele in pure form.
P. falciparum propagation in vitro
All parasite lines were grown at 37°C in human red blood cells at a hematocrit of 3%–4%, by use of RPMI 1640 medium supplemented with 0.5% Albumax, 0.25% sodium bicarbonate, and 0.01 mg/mL gentamicin. Cultures were maintained in an atmosphere of 90% N2, 5% O2, and 5% CO2.
In vitro drug assays
Parasite susceptibilities to lumefantrine were measured in vitro using 72-h 3H-hypoxanthine assays with serial drug dilutions, as described elsewhere [24]. IC50 and IC90 values were determined by linear extrapolation. For statistical analyses, 2-tailed unpaired Student’s t tests, as well as nonparametric Mann-Whitney U tests, were performed.
RESULTS
In total, 38 of the 50 children who were treated with AL experienced recurrent infections during the 42-day follow-up. Of these 38 children, 37 had sufficient parasite material to be included in the analysis. Of the 56 children included in the SP treatment arm, 39 had recurrent infections [2].
In the AL arm, only 2 recrudescences were recorded, which precluded a meaningful analysis of recrudescence versus reinfection. Hence, all breakthrough infections in the AL arm were simply considered to be recurrent and were treated equally. As for the SP arm, 16 recrudescences were identified. No selection for either pfcrt or pfmdr1 variants was observed when SP recrudescences were compared with reinfections. These infections were also grouped together as recurrent infections.
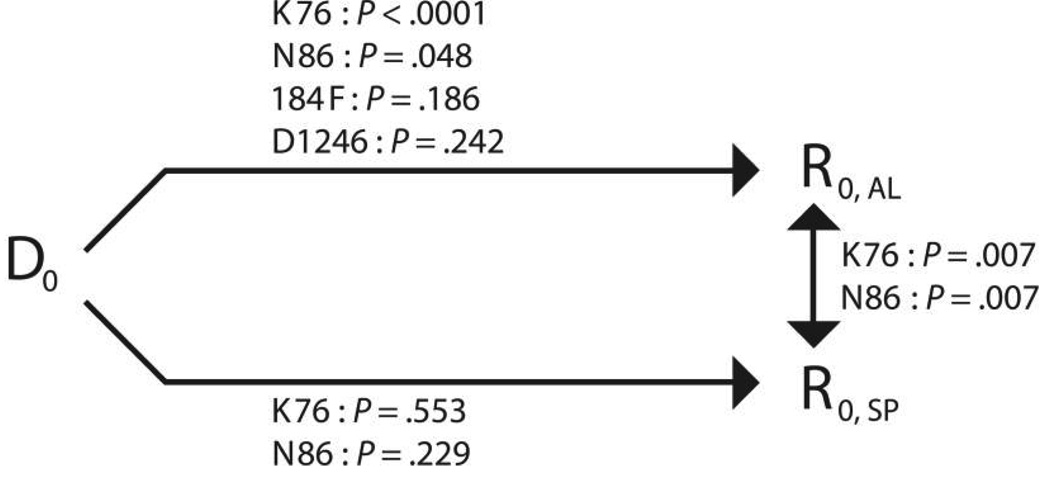
The frequency of parasites carrying the pfcrt K76 allele (in pure form or mixed with 76T) increased from a baseline prevalence of 48.0% to 86.5% (P < .0001) among all recurrent infections in persons in the AL arm (table 1 and figure 1) and to 85.7% (P = .0001) among only reinfections occurring in persons in the AL arm. A statistically significant selection was also observed for pfmdr1 N86, which displayed an increase in frequency from 55.9% at baseline to 75.7% among all recurrent infections (P = .048) (table 1 and figure 1), whereas the frequency increased to 74.3% among only reinfections occurring in the AL arm (P = .071). The frequencies of the pfmdr1 184F and D1246 alleles remained unaltered (table 1 and figure 1). The SP arm revealed no changes in the frequency of pfcrt K76T or pfmdr1 N86Y in recurrent infections (table 1). The pfmdr1 1034 and 1042 SNPs, as well as the pfcrt 163 SNPs, showed no variation in either treatment.
Table 1.
Frequencies of the analyzed single-nucleotide polymorphisms (SNPs), both before the administration of the drug (D0) and in recurrent infections (R0).
| Frequency, (pure + mix)/all [95% CI]a | |||
|---|---|---|---|
| R0 | |||
| SNP | D0 | AL | SP |
| pfcrt K76 | (26 + 23)/102 = 0.480 [0.386–0.576] | (22 + 10)/37 = 0.865 [0.716–0.946] | (12 + 7)/34 = 0.559 [0.394–0.711] |
| pfmdr1 N86 | (27 + 30)/102 = 0.559 [0.462–0.651] | (21 + 7)/37 = 0.757 [0.597–0.868] | (8 + 6)/33 = 0.424 [0.272–0.592] |
| pfmdr1 184F | (0 + 25)/100 = 0.250 [0.175–0.344] | (3 + 10)/34 = 0.382 [0.239–0.550] | NA |
| pfmdr1 D1246 | (50 + 24)/98 = 0.755 [0.661–0.830] | (21 + 9)/35 = 0.857 [0.702–0.942] | NA |
NOTE. The D0 data for both treatment arms were pooled at baseline, because no significant differences were found between the 2 groups for any of the analyzed SNPs. AL, artemether-lumefantrine; CI, confidence interval; NA, not analyzed; SP, sulfadoxine-pyrimethamine.
“All” denotes the total no. of successful analyses. The 95% CI was determined using the modified Wald method (GraphPad Software; GraphPad).
Figure 1.
Set of comparisons (conducted using Fisher’s exact test [see Subjects, Materials, and Methods]) between infections at baseline (D0) and all recurrent infections (R0) concerning the chloroquine resistance transporter gene (pfcrt) K76 allele and the multidrug resistance gene 1 (pfmdr1) N86, 184F, and D1246 alleles. The pfcrt S163 residue, as well as the pfmdr1 S1034 and N1042 residues, had a prevalence of 100% in this setting, as did single-copy pfmdr1. AL, artemether-lumefantrine; SP, sulfadoxine-pyrimethamine.
Our sequencing procedures also revealed a novel nonsynonymous pfmdr1 W1031R SNP (TGG→CGG) located in a predicted transmembrane domain [25]. The new SNP, which was observed in 2 separate PCR amplification and sequencing studies, was present in 1 infection at baseline but was not detected among the recurrent infections in the group given AL. Finally, an increased number of pfmdr1 gene copies was not detected in any of the 89 infections noted at baseline or in the 29 recurrent infections noted in the AL treatment arm that were successfully analyzed in this Tanzanian cohort.
To directly test the contribution of mutant pfcrt to susceptibility to lumefantrine under controlled in vitro conditions, we assayed P. falciparum lines that had been genetically engineered to express distinct pfcrt alleles, rather than the endogenous pfcrt allele in isogenic background. One set was produced in the CQ-susceptible GC03 line—a progeny of the HB3 × Dd2 genetic cross [26]. This set enabled us to compare clones expressing the wild-type CQ-susceptible allele (C2GC03), the CQ-resistant Dd2 allele typical of Asian and African CQ-resistant strains (C4Dd2), and the CQ-resistant 7G8 allele observed in South America, the Oceanic region, and India (C67G8) [19]. The Dd2 and 7G8 alleles contain 8- and 5-point mutations (M74I/N75E/K76T/A220S/Q271E/N326S/I356T/R371I and C72S/K76T/A220S/N326D/I356L), respectively, compared with the wild-type allele. To test the observed clinical association between the K76T mutation and susceptibility to lumefantrine, we also assayed clones generated in the Dd2 and 7G8 lines, which expressed either the parental allele (C-1Dd2 and C-17G8) or an allele in which the mutant 76T codon had been replaced with the wild-type K76 codon (known as the T76K-1Dd2 and T76K-17G8 “back mutant” clones, respectively). Recombinant clones were assayed in vitro on 5–8 separate occasions, and parental controls (GC03, Dd2, and 7G8) were assayed on 7–12 separate occasions.
These in vitro assays revealed that CQ-resistant lines with mutant pfcrt alleles carrying the K76T mutation were uniformly more susceptible to lumefantrine (table 2). C4Dd2 and C67G8, expressing the 2 most prevalent alleles of mutant pfcrt worldwide, both showed a ~35% decrease in lumefantrine IC50 values, compared with C2GC03 (P < .05 and P < .01, respectively, by the Mann-Whitney U test). The results achieved using the back-mutant clones demonstrated that the K76T mutation itself is a large mediator of this increased susceptibility. Both the T76K-1Dd2 and T76K-17G8 clones were 2-fold less susceptible to lumefantrine than were their recombinant counterparts (for both, P < .01, by the Mann-Whitney U test). In the parental lines, we also observed substantially lower lumefantrine IC50 values in Dd2 and 7G8, compared with GC03.
Table 2.
Lumefantrine susceptibility levels in chloroquine resistance transporter gene (pfcrt)–modified and control lines.
| Line | LMF IC50, nmol/La |
Recombinant | Parental line |
PfCRT haplotype | CQb | ARTM IC50,b nmol/L |
MQ IC50,b nmol/L |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 72 | 74 | 75 | 76 | 220 | 271 | 326 | 356 | 371 | IC50, nmol/L | Phenotype | ||||||
| GC03 | 85.5 ± 7.2 | No | … | C | M | N | K | A | Q | N | I | R | 29.2 ± 2.0 | Susc. | 39.4 ± 6.9 | 52.6 ± 5.4 |
| Dd2 | 55.2 ± 9.6 | No | … | C | I | E | T | S | E | S | T | I | 195.1 ± 25.3 | Res. | 46.3 ± 7.6 | 46.8 ± 2.0 |
| 7G8 | 27.0 ± 4.1 | No | … | S | M | N | T | S | Q | D | L | R | 140.4 ± 15.0 | Res. | 21.6 ± 6.3 | 9.6 ± 1.3 |
| C2GC03 | 74.0 ± 7.7 | Yes | GC03 | C | M | N | K | A | Q | N | I | R | 22.9 ± 2.8 | Susc. | 39.1 ± 7.0 | 54.7 ± 1.5 |
| C4Dd2 | 48.7 ± 4.3c | Yes | GC03 | C | I | E | T | S | E | S | T | I | 147.4 ± 13.3d | Res. | 12.6 ± 1.8d | 33.1 ± 3.1d |
| C67G8 | 46.3 ± 4.6d | Yes | GC03 | S | M | N | T | S | Q | D | L | R | 126.9 ± 17.2d | Res. | 17.2 ± 3.0d | 27.3 ± 3.4d |
| T76K-1Dd2 | 73.2 ± 10.5d | Yes | Dd2 | C | I | E | K | S | E | S | T | I | 22.1 ± 2.1d | Susc. | 40.4 ± 11.7 | 27.2 ± 8.0c |
| C-1Dd2 | 33.7 ± 5.0 | Yes | Dd2 | C | I | E | T | S | E | S | T | I | 160.7 ± 28.9 | Res. | 32.9 ± 6.6 | 36.9 ± 4.2 |
| T76K-17G8 | 68.3 ± 11.7d | Yes | 7G8 | C | I | E | K | S | Q | D | L | R | 23.9 ± 4.6e | Susc. | 13.8 ± 3.2 | 3.6 ± 0.7 |
| C-17G8 | 33.6 ± 5.5 | Yes | 7G8 | S | M | N | T | S | Q | D | L | R | 108.8 ± 15.9 | Res. | 7.4 ± 2.1 | 3.1 ± 0.9 |
NOTE. ARTM, artemisinin; CQ, chloroquine; LMF, lumefantrine; MQ, mefloquine; Res., resistant; Susc., susceptible.
LMF IC50 values are expressed as the mean value (±SE) for pfcrt–modified and control lines. Data were derived from 5–12 independent assays performed in duplicate. (Results from Mann-Whitney U tests compared mutants and recombinant controls).
CQ, ARTM, and MQ IC50 values (from at least 3 independent assays performed in duplicate) are expressed as the mean value (±SE) and are reproduced from [19] for the top 6 lines and from [20] for the bottom 4 lines.
P < .05.
P < .01.
P < .001.
DISCUSSION
Coartem, a fixed formulation of artemether and lumefantrine, is currently the most successful and widely implemented ACT and recently has been adopted as first- or second-line chemotherapy for the treatment of uncomplicated malaria by a large number of national malaria control programs in sub-Saharan Africa [27]. This combination, referred to as “AL” in the present study, has consistently demonstrated high efficacy. Nevertheless, the high levels of parasite transmission in most parts of sub-Saharan Africa dictate that reinfections will often occur after cessation of AL treatment [1–3]. This creates a risk for selection of reinfecting lumefantrine-tolerant/-resistant parasites after treatment, when the slowly eliminated lumefantrine remains at subtherapeutic concentrations. Previous data from some of the investigators involved in the present study [11]—data subsequently confirmed by others [13, 14]—have shown a consistent pattern of detectable increases in the frequency of drug resistance–associated pfmdr1 haplotypes among recurrent infections after AL treatment. Because these recurrent parasites represent reinfections in all but a few cases, the data have been interpreted as a lumefantrine-driven selection for variant pfmdr1 alleles.
In the present study, we report significant selection of the pfcrt K76 allele after AL treatment, which, to our knowledge, represents the first report of a significant in vivo association of this gene with an ACT containing an arylaminoalcohol (a group of drugs that includes lumefantrine, MQ, and HF). This selection was not observed in our preceding studies, probably because of the rarity of the K76 allele in Zanzibar [1, 11, 12], as opposed to the present study site on mainland Tanzania, where ~50% of the analyzed infections harbored K76 allele carriers. As discussed elsewhere [12], the high efficacy of AL precludes the collection of statistically meaningful numbers of recrudescent parasites for analysis. Hence, the conclusions of the present study are mainly considered to be applicable to parasites causing reinfection (i.e., formally “lumefantrine-tolerant” parasites, as opposed to “lumefantrine-resistant” parasites [15]).
The selection for pfcrt K76 in the recurrent infections observed in the present study is consistent with earlier in vitro data supporting a role for this gene in P. falciparum susceptibility to arylaminoalcohols. After allele exchange at the pfcrt amino acid 76 position, isogenic clones have been shown to acquire different susceptibilities to MQ and HF. In the first of these experiments, exchanging the K76 for 76T (in the CQ-susceptible GC03 background) increased parasite in vitro susceptibility to MQ by 2- to 3-fold [19]. In a set of reverse experiments, the replacement of 76T with K76 decreased the susceptibility of the parasites to MQ, albeit to a lesser extent (1.5- to 2-fold) [20] (table 2). In another study, P. falciparum lines selected in vitro for HF resistance were shown to have acquired pfcrt mutations that included S163R, which was also associated with MQ resistance [22]. Finally, in vitro development of resistance to CQ in the Sudanese 106/1 clone (which already harbored 7 of the 8 Dd2 pfcrt mutations, lacking only K76T) resulted in novel pfcrt SNPs at position 76 (K76I/N) [17, 21]. These novel K76I/N SNPs were shown to significantly increase susceptibility to MQ and HF [21].
Our present research extends these observations by documenting a 2-fold increase in susceptibility to lumefantrine in parasites expressing the K76T mutation (compare C-1Dd2 with T76K-1Dd2 and compare C-17G8 with T76K-17G8). Interestingly, 7G8 was 2-fold more susceptible than Dd2, a finding that, on the basis of our data, cannot be attributed to differences between these pfcrt alleles. These lines also differ in their pfmdr1 point mutations (with Dd2 having the 86Y/Y184/S1034/N1042/D1246 haplotype and with 7G8 having the N86/184F/1034C/1042D/1246Y haplotype) as well as their copy number (3–4 copies in our Dd2 line, compared with 1 copy in 7G8). Both types of genetic changes in pfmdr1 are known to affect susceptibility to lumefantrine. Other, as yet undefined mediators may also differ between 7G8 and Dd2. Overall, our data clearly define the pfcrt K76T mutation as a major determinant that contributes to decreased susceptibility to lumefantrine on distinct genetic backgrounds.
Our results also show that the 2 globally most prevalent mutant pfcrt alleles, Dd2 and 7G8, both increase susceptibility to lumefantrine (compare C4Dd2 and C67G8 with the isogenic CQ-susceptible C2GC03 line). In this regard, lumefantrine is an ideal ACT partner drug in areas of high resistance to CQ. Interestingly, mutant pfcrt alleles have also previously been found to significantly increase susceptibility to artemisinin [19]. This finding indicates an important dual benefit of using AL in areas with a high prevalence of mutant pfcrt conferring resistance to CQ.
Mutant PfCRT is thought to efflux compounds out of the DV into the cytoplasm. For CQ, which interferes with the detoxification of heme moieties inside the DV, resistance is thought to be achieved by mutant PfCRT-mediated export of drug away from its hematin target [25]. CQ resistance has been shown to critically depend on the pfcrt K76T mutation [20]. The observation that parasites carrying the pfcrt K76T mutation are more susceptible to lumefantrine and MQ suggests that those compounds might also be transported by mutant PfCRT out of the DV and, consequently, might exert their action in the parasite cytoplasm.
For pfmdr1 and its encoded ABC-transporter Pgh-1, transport studies have shown that the pfmdr1 copy number is associated with Fluo-4 accumulation inside the DV [28]. Moreover, it has been shown that the pfmdr1 gene expressed in Saccharomyces cerevisiae functionally complements an exporter in the plasma membrane [29], with the nucleotide-binding domains on the cytosolic side [30]. In the DV membrane, Pgh-1 is situated with the nucleotide-binding domains in the cytoplasm [31, 32], indicating a transport direction from the cytoplasm into the DV. Thus, Pgh-1 may function as a drug importer. For drugs such as CQ that have their target inside the DV, an increased number of pfmdr1 copies might therefore be expected to result in increased drug susceptibility. In line with this hypothesis, selection of high-level CQ resistance has been observed to result in pfmdr1 deamplification [33]. For drugs that have their target outside the DV, increased numbers of pfmdr1 copies might therefore result in decreased susceptibility. The observed association between an increased pfmdr1 copy number and treatment failures after AL [8], MQ, or artesunate-MQ therapy [9] is therefore another indication that the target of lumefantrine and MQ might lie outside the DV.
The CQ susceptibility markers pfcrt K76 and pfmdr1 N86 [18] and increases in pfmdr1 copy number have all been associated with a decreased susceptibility to both lumefantrine and MQ [7–9, 11, 19–21, 34, 35]. Thus, substantial evidence points to cross-resistance between the 2 arylaminoalcohols. In Southeast Asia, decreased efficacy of artesunate-MQ treatment has been reported [36], presumably because of MQ resistance caused by an increased pfmdr1 copy number [37]. Although an increased pfmdr1 copy number seems to be rare in Africa, it has previously been detected in Gabon in a subset of patients treated with a low dose of MQ. This suggests a risk for selection and spread of multicopy pfmdr1 in Africa after the ongoing wide-scale deployment of AL [10]. This finding, combined with our observation of selection of pfcrt K76 and pfmdr1 N86 alleles, suggests that AL tolerance or resistance could emerge in Africa. This scenario would have major public health implications for AL as a malaria treatment, in terms of its ability to effectively and sustainably reduce disease-associated morbidity and mortality.
In the present study, we compared selection of pfmdr1 and pfcrt mutations that occurred in association with AL treatment with selection that occurred in association with treatment with SP, a drug for which resistance is established to be unrelated to genetic alterations in these genes. Because no selection was observed for the analyzed SNPs in the SP arm, the present study supports the argument that pfmdr1 N86 and, now, pfcrt K76 are specifically associated with a decreased susceptibility to lumefantrine, allowing a more successful reinfection of the human host in the period corresponding to slowly decreasing lumefantrine concentrations [11, 15].
The lack of pfcrt K76 selection in the SP treatment arm is in apparent contrast to the documented replacement of the 76T allele by K76 in regions where SP has been introduced to replace CQ, as has been illustrated by reports from Malawi [38, 39]. In this regard, we speculate that harboring pfcrt K76 presents a relatively small fitness advantage, compared with harboring pfcrt 76T. In the absence of CQ pressure over several years, the progressive replacement becomes detectable in the overall population, ultimately overtaking the parasite population harboring the K76T mutation [38–41].
In conclusion, we identified the pfcrt K76 allele as a further potential marker of decreased susceptibility to lumefantrine in vivo, in addition to the established pfmdr1 SNPs [10–12]. Importantly, pfcrt encoding the K76T mutation, which confers CQ resistance in vitro and is a highly sensitive marker of CQ treatment failure in vivo, also increases in vitro susceptibility to artemisinin [19]. These findings highlight a substantial benefit of using AL in areas with a high prevalence of CQ-resistant P. falciparum malaria. Defining the interrelationships between drug-determinant polymorphisms and changes in susceptibility to the various antimalarials will be important in ongoing efforts to best manage the implementation of different ACTs and reduce the dissemination and influence of drug-resistant malaria.
Acknowledgments
Financial support: Swedish Development Cooperation Agency–Department for Research Cooperation (grants SWE 2005–017 and SWE 2005–004596); Research and Development Unit, Sörmland Count Council, Sweden (project grant 82090); Goljes Foundation (project grants 248/04 and 317/05); Fundacão para a Ciência e Tecnologia, Portugal (PhD grant SFRH/BD/28393/2006 to M.I.V.); US National Institutes of Health (grant R01 AI50234 to D.A.F.).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: Molecular Approaches to Malaria–2008, Lorne, Victoria, Australia, 3–7 February 2008 (abstract 132).
References
- 1.Martensson A, Strömberg J, Sisowath C, et al. Efficacy of artesunate plus amodiaquine versus that of artemether-lumefantrine for the treatment of uncomplicated childhood Plasmodium falciparum malaria in Zanzibar, Tanzania. Clin Infect Dis. 2005;41:1079–1086. doi: 10.1086/444460. [DOI] [PubMed] [Google Scholar]
- 2.Martensson A, Ngasala B, Ursing J, et al. Influence of consecutive-day blood sampling on polymerase chain reaction–adjusted parasitological cure rates in an antimalarial-drug trial conducted in Tanzania. J Infect Dis. 2007;195:597–601. doi: 10.1086/510910. [DOI] [PubMed] [Google Scholar]
- 3.Zongo I, Dorsey G, Rouamba N, et al. Randomized comparison of amodiaquine plus sulfadoxine-pyrimethamine, artemether-lumefantrine, and dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in Burkina Faso. Clin Infect Dis. 2007;45:1453–1461. doi: 10.1086/522985. [DOI] [PubMed] [Google Scholar]
- 4.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 5.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 6.Anderson TJ, Nair S, Qin H, et al. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 8.Price RN, Uhlemann AC, van Vugt M, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlemann AC, Ramharter M, Lell B, Kremsner PG, Krishna S. Amplification of Plasmodium falciparum multidrug resistance gene 1 in isolates from Gabon. J Infect Dis. 2005;192:1830–1835. doi: 10.1086/497337. [DOI] [PubMed] [Google Scholar]
- 11.Sisowath C, Strömberg J, Mårtensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 12.Sisowath C, Ferreira PE, Bustamante LY, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 13.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphreys GS, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastings IM, Ward SA. Coartem (artemether-lumefantrine) in Africa: the beginning of the end? J Infect Dis. 2005;192:1303–1304. doi: 10.1086/432554. author reply, 1304–5. [DOI] [PubMed] [Google Scholar]
- 16.Hayward R, Saliba KJ, Kirk K. pfmdr1 mutations associated with chloroquine resistance incur a fitness cost in Plasmodium falciparum. Mol Microbiol. 2005;55:1285–1295. doi: 10.1111/j.1365-2958.2004.04470.x. [DOI] [PubMed] [Google Scholar]
- 17.Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djimdé A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 19.Sidhu AB, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lakshmanan V, Bray PG, Verdier-Pinard D, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper RA, Ferdig MT, Su XZ, et al. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 22.Johnson DJ, Fidock DA, Mungthin M, et al. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veiga MI, Ferreira PE, Björkman A, Gil JP. Multiplex PCR-RFLP methods for pfcrt, pfmdr1 and pfdhfr mutations in Plasmodium falciparum. Mol Cell Probes. 2006;20:100–104. doi: 10.1016/j.mcp.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- 25.Valderramos SG, Fidock DA. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellems TE, Panton LJ, Gluzman IY, et al. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 27.World Health Organization. [Accessed 20 May 2008];Treatment policies. Available at http://www.who.int/malaria/treatmentpolicies.html.
- 28.Rohrbach P, Sanchez CP, Hayton K, et al. Genetic linkage of pfmdr1 with food vacuolar solute import in Plasmodium falciparum. EMBO J. 2006;25:3000–3011. doi: 10.1038/sj.emboj.7601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkman SK, Cowman AF, Wirth DF. Functional complementation of the ste6 gene of Saccharomyces cerevisiae with the pfmdr1 gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1995;92:8921–8925. doi: 10.1073/pnas.92.19.8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaelis S. STE6, the yeast α-factor transporter. Semin Cell Biol. 1993;4:17–27. doi: 10.1006/scel.1993.1003. [DOI] [PubMed] [Google Scholar]
- 31.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to anti-malarial drug-resistance. Acta Trop. 2005;94:181–190. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Karcz SR, Galatis D, Cowman AF. Nucleotide binding properties of a P-glycoprotein homologue from Plasmodium falciparum. Mol Biochem Parasitol. 1993;58:269–276. doi: 10.1016/0166-6851(93)90048-3. [DOI] [PubMed] [Google Scholar]
- 33.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 1992;11:3067–3075. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price RN, Cassar C, Brockman A, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White NJ. Qinghaosu (artemisinin): the price of success. Science. 2008;320:330–334. doi: 10.1126/science.1155165. [DOI] [PubMed] [Google Scholar]
- 37.Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14:716–769. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kublin JG, Cortese JF, Njunju EM, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 39.Mita T, Kaneko A, Lum JK, et al. Recovery of chloroquine sensitivity and low prevalence of the Plasmodium falciparum chloroquine resistance transporter gene mutation K76T following the discontinuance of chloroquine use in Malawi. Am J Trop Med Hyg. 2003;68:413–415. [PubMed] [Google Scholar]
- 40.Mita T, Kaneko A, Lum JK, et al. Expansion of wild type allele rather than back mutation in pfcrt explains the recent recovery of chloroquine sensitivity of Plasmodium falciparum in Malawi. Mol Biochem Parasitol. 2004;135:159–163. doi: 10.1016/j.molbiopara.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Laufer MK, Thesing PC, Eddington ND, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]