Abstract
The pattern recognition receptors Toll-like receptor 2 (TLR2) and Dectin-1 play key roles in coordinating the responses of macrophages and dendritic cells (DC) to fungi. Induction of pro-inflammatory cytokines is instructed by signals from both TLR2 and Dectin-1. A recent report identified a role for CARD9 in innate anti-fungal responses, demonstrating CARD9-Bcl10-mediated activation of NF-κB and pro-inflammatory cytokine induction in murine bone marrow-derived DC (bmDC) stimulated via Dectin-1. We now report that Dectin-1-CARD9 signals fail to activate NF-κB and drive TNF-α induction in murine bone marrow-derived macrophages (bmM). However, priming of bmM with GM-CSF or IFN-γ permits Dectin-1-CARD9-mediated TNF-α induction. Analysis of other macrophage/DC populations revealed further variation in the ability of Dectin-1-CARD9 signaling to drive TNF-α production. Resident peritoneal cells and alveolar macrophages produce TNF-α upon Dectin-1 ligation, while thioglycollate-elicited peritoneal macrophages and Flt3L-derived DC do not. We present data demonstrating that CARD9 is recruited to phagosomes via its CARD domain where it enhances TLR-induced cytokine production even in cells in which Dectin-1 is insufficient to drive cytokine production. In such cells, Dectin-1, CARD9 and Bcl10 levels are not limiting, and data indicate that these cells express additional factors that restrict Dectin-1-CARD9 signaling for TNF-α induction.
Keywords: Dendritic cells, Monocytes/Macrophages, Fungal infection, Phagocytosis, Signal transduction
Introduction
The innate immune response is controlled by integrating signals from multiple receptors at the surface, in the phagosome, or in the cytosol of myeloid phagocytes, including Toll-like receptors (TLRs), C-type lectins, scavenger receptors, and nucleotide-binding oligomerization domain-containing (Nod) proteins(1, 2). Initiation of an immune response against pathogenic fungi by macrophages and dendritic cells (DC) involves collaboration between TLR2 and the C-type lectin Dectin-1, and has been extensively characterized by us and others using zymosan (Saccharomyces cerevisiae yeast cell wall preparation), as well as live pathogenic fungi such as Candida albicans, Aspergillus fumigatus and Pneumocystis carinii (reviewed in (3)). Two groups recently demonstrated the importance of Dectin-1 for in vivo anti-fungal immunity using Dectin-1 knockout mice(4, 5).
Dectin-1 recognizes β-glucans in zymosan and the cell walls of live pathogenic fungi, and is a key phagocytic receptor for fungal internalization (reviewed in (3)). It has an immunoreceptor tyrosine-based activation motif (ITAM)-like motif in its intracellular tail, through which it signals via Src and Syk kinases to induce ROS production(6). TLR2 signals via the adaptor MyD88 to trigger NF-κB activation and cytokine production. Dectin-1 signaling contributes to the transcriptional response by promoting TLR-mediated NF-κB activation, as well as itself triggering activation of nuclear factor of activated T cells (NFAT) transcription factors(7–9). Both receptors play key roles in the induction of a variety of inflammatory mediators (reviewed in (3)), thereby orchestrating the recruitment and activation of other immune cells and driving the development of adaptive immunity. For example, recent data indicated that DC matured with a Dectin-1 ligand instruct the differentiation of Th1 and Th17 responses(10), while another report demonstrated that co-ligation of Dectin-1 and TLR2 results in the induction of regulatory T cells and immunological tolerance(11).
In addition to its role in promoting TLR-induced NF-κB activation, a recent report indicated that Dectin-1 signals can directly activate NF-κB in DC via the caspase recruitment domain (CARD)-containing adaptor protein CARD9(12). Gross et al. reported that CARD9 knockout mice are significantly more susceptible to Candida albicans infection than their wild type littermates(12). Furthermore, CARD9-deficient DC displayed defective NF-κB activation and impaired cytokine responses to zymosan stimulation. In addition to CARD9, activation of NF-κB in zymosan-stimulated DC required Bcl10 and MALT1. Bcl10 and MALT1 also mediate NF-κB activation downstream of lymphocyte antigen receptors by association with CARMA1/CARD11, a CARD-containing adaptor structurally related to CARD9(13–17). Since TLR2 responses were unaffected by CARD9 deletion, the authors proposed that CARD9-Bcl10-MALT1 signaling mediates NF-κB activation downstream of Dectin-1.
In contrast, our previous studies in macrophages have indicated that while Dectin-1 signals collaborate with TLR2 signals to enhance TLR2-mediated NF-κB activation and pro-inflammatory cytokine production, Dectin-1 signals alone do not appear to be sufficient for NF-κB activation and cytokine induction(8). In this study we report that while Dectin-1 signals via CARD9 and Bcl10 to induce NF-κB activation and TNF-α production in bmDC, this pathway does not activate NF-κB or induce TNF-α in bmM. The data indicate that CARD9 in macrophages is recruited to phagosomes where it coordinates signaling to p38 MAP kinases, even in the absence of a connection to the NF-κB pathway. However, priming of bmM with GM-CSF or IFN-γ permits Dectin-1-CARD9-induced TNF-α production. Furthermore, Dectin-1 ligation directly triggers TNF-α production by resident peritoneal cells and alveolar macrophages, but not by thioglycollate-elicited peritoneal macrophages or Flt3L-derived DC. Thus the contribution of Dectin-1 signals to pro-inflammatory cytokine induction is variable in different macrophage/DC populations. Finally, we present evidence that an inhibitor restricts Dectin-1-CARD9 signaling in certain myeloid cells.
Materials and Methods
Reagents
Pam3CSK4 and Salmonella minnesota LPS were from InvivoGen (San Diego, CA) and zymosan was from Sigma-Aldrich (St. Louis, MO). Depleted zymosan was prepared by boiling 250 μg zymosan in 1 ml 10 M sodium hydroxide for 30 min and washing three times with sterile PBS(8). Recombinant growth factors and IFN-γ were from PeproTech (Rocky Hill, NJ).
Mice and cells
HEK293 cells were maintained in complete Dulbecco’s modified Eagle medium (DMEM, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine; Mediatech, Herndon, VA). RAW264.7 murine macrophage cell lines were cultured in complete RPMI. RAW264.7 cells stably expressing streptavidin binding peptide (SBPc)-tagged Dectin-1 were described previously(6).
Wild type C57BL/6 mice were maintained at Cedars-Sinai Medical Center, Los Angeles, CA. Wild type and CARD9 knockout mice(18) were maintained at University of Texas, MD Anderson Cancer Center, Houston, TX. Bone marrow-derived macrophages (bmM) and DC (bmDC) were prepared from mouse femurs by culture of bone marrow cells for 6 days in complete RPMI (RPMI 1640, 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine; Mediatech) supplemented with 50 ng/ml recombinant human M-CSF (bmM) or 10 ng/ml recombinant murine GM-CSF (bmDC). Flow cytometry of adherent cells at day 6 verified that M-CSF cultures yielded a single population of CD11b+ F4/80+ cells, and GM-CSF cultures yielded a single population of CD11c+ cells; neither preparation was contaminated with 7/4+ neutrophils (data not shown).
Peritoneal macrophages were elicited by intraperitoneal injection of thioglycollate for 3 days. Thioglycollate-elicited peritoneal macrophages (TEPM) and resident peritoneal cells (RPC) were harvested by peritoneal lavage with PBS, and adherent cells were enriched by plastic adherence for 2 h. Alveolar macrophages (AvM) were obtained by bronchoalveolar lavage using a 24G catheter to wash out the lungs 10 times with 1 ml PBS plus 2mM EDTA per wash. Flt3L-derived DC were obtained by 6 day culture of bone marrow progenitors with 10 ng/ml Flt3L.
Plasmids and cell line generation
The expression vector pEF6-CARD9-V5 used for HEK293 cell transfection was generated by cloning the coding region for murine CARD9, amplified from bmM cDNA, into the pEF6/V5-His expression vector (Invitrogen, Carlsbad, CA) such that it is tagged at its C-terminus with the V5-His epitope. The expression vector pEF6-SBPc-Bcl10 was generated by cloning the coding region for murine Bcl10, amplified from bmM cDNA, into pEF6 such that it is tagged at its N-terminus with an epitope (SBPc) combining sequences of protein C recognized by the HPC4 monoclonal antibody (Amersham, Piscataway, NJ) and the streptavidin binding peptide(6). The expression vector pEF6-Dectin-SBPc was generated by cloning the SBPc-tagged murine Dectin-1(6) into pEF6. For transient expression of Dectin-1 in bmM, murine Dectin-1-SBPc was cloned into the pMAX-GFP (Amaxa, Gaithersburg, MD) such that it is also tagged at its N-terminus with GFP.
For retroviral expression of CARD9, used to generate RAW264.7 macrophages stably expressing CARD9 for immunofluorescence assays, the coding region for murine CARD9 was cloned into the retroviral expression vector pMXs-IRES-puro (pMXsip(19)) such that it is tagged at its C-terminus with the SBPc tag. Shorter constructs consisting of the amino terminal CARD (aa 1–110) and the c-terminal coiled-coil domain (aa 100–536) similarly tagged were also generated. RAW264.7 macrophages stably expressing these constructs were generated by retroviral infection as previously described(6, 20).
HEK293 cell luciferase reporter assays
HEK 293 cell reporter assays were performed as described previously(8) using the indicated plasmids. Briefly, HEK293 cells (American Type Culture Collection no. CRL-1573) in 96-well plates were transfected as indicated using Lipofectamine 2000 (Invitrogen) with 17 ng each of ELAM-luciferase and CMV-βgal reporter plasmids, 17 ng pEF6-SBPc-Dectin-1, 3 ng pEF6-CARD9-V5, and 3 ng pEF6-SBPc-Bcl10 per well. The following day, the cells were stimulated with the indicated ligands for 4 h, and luciferase activity was measured (Luciferase Assay System, Promega, Madison, WI). Values were normalized for transfection efficiency according to β-galactosidase expression (β-Gal Assay Kit, Stratagene, Cedar Creek, TX). In some experiments an SRE-luciferase reporter plasmid (pSRE-luciferase(21), kindly provided by Dr. Walter Born, University of Zurich) was used in place of the NF-κB reporter plasmid.
Immunofluorescence microscopy
RAW264.7 macrophages stably expressing SBPc-tagged CARD9, CARD domain or coiled-coil domain were coated on to coverslips and treated with 100 μg/ml zymosan particles for 10 min prior to fixing with 10% formaldehyde. Following permeabilization with ice-cold acetone, coverslips were blocked with PBS containing 5% FBS and 0.1% ovalbumin, and stained with an antibody against the SBPc tag (HPC4, Amersham) and TRITC-phalloidin to detect actin. Following staining, coverslips were washed, mounted and viewed using a confocal microscope.
Phagocytosis assay
TRITC-labelled zymosan particles were fed to macrophages or DC for 15 min, with brief centrifugation upon zymosan addition to ensure particle contact with the cells. Cells were lifted in PBS containing 1 mM EDTA, 1 mM sodium azide and 2.4 U/ml proteinase K (to remove bound but uninternalized zymosan particles) prior to analysis by flow cytometry.
ELISA
TNF-α levels in culture supernatants were assayed using ELISA kits from BD Biosciences (San Jose, CA) according to the manufacturer’s instructions.
Measurement of reactive oxygen production
The production of reactive oxygen species was assayed by luminol-enhanced chemiluminescence as described previously(6).
Western Blotting
Whole cell lysates or phagosome preparations were probed by Western Blotting using the NuPAGE system from Invitrogen according to manufacturer’s instructions, and antibodies against I-κB, total Erk1/2, dual phosphorylated p38 and total p38 MAP kinases from Cell Signaling Technologies (Danvers, MA), CARD9 from eBioscience (San Diego, CA), and Bcl10, LAMP1, GAPDH and β-actin from Santa Cruz Biotechnology (Santa Cruz, CA).
EMSA
Nuclear extracts were prepared using a kit from Active Motif. NF-κB binding to a biotinylated probe comprising an NF-κB consensus promoter sequence (5′-ATCAGGGACTTTCCGCTGGGGACTTTCCG-3′) was assessed using a LightShift Chemiluminescent EMSA kit from Pierce (Rockford, IL) according to the manufacturer’s instructions.
Flow cytometry
Dectin-1 surface expression was assessed by flow cytometry using FITC- and PE-conjugated monoclonal anti-Dectin-1 antibodies (2A11) from AbD Serotec (Raleigh, NC). PE-conjugated antibodies against CD11c and TLR2, FITC-conjugated antibodies against CD11b and CD11c, and FITC- and PE-conjugated isotype controls were from BD Biosciences. For measurement of intracellular TNF-α, cells were stimulated in the presence of brefeldin A (BioLegend, San Diego, CA) for 4 h. Cells were lifted with PBS containing 1 mM EDTA and 1 mM sodium azide, and Fc receptors were blocked with 2.4G2 cell supernatant for 5 min prior to fixing for 1 h with 2.5% formalin and 0.05% saponin in PBS. Cells were then incubated for 30 min with PE-conjugated anti-TNF-α, and FITC-conjugated anti-CD11b in permeabilization buffer (0.1% saponin, 1% FBS, 1 mM sodium azide in PBS), washed and analyzed by flow cytometry, gating for CD11b+ RPC and TEPM, and CD11b− AvM. For some experiments, bmM were transfected with pMAX-GFP or pMAX-GFP-Dectin-1-SBPc using the Nucleofector II (Amaxa) as per the manufacturer’s instructions, and cells were plated into 24-well plates. The following day, the cells were given fresh media prior to assessment of Dectin-1 expression and particle-induced TNF-α production.
Statistical Analysis
Statistical analysis was performed using a student’s t-test. Triplicate experimental samples were compared to unstimulated or control samples.
Results
Dectin-1 signals can activate NF-κB directly in a CARD9/Bcl10-dependent manner
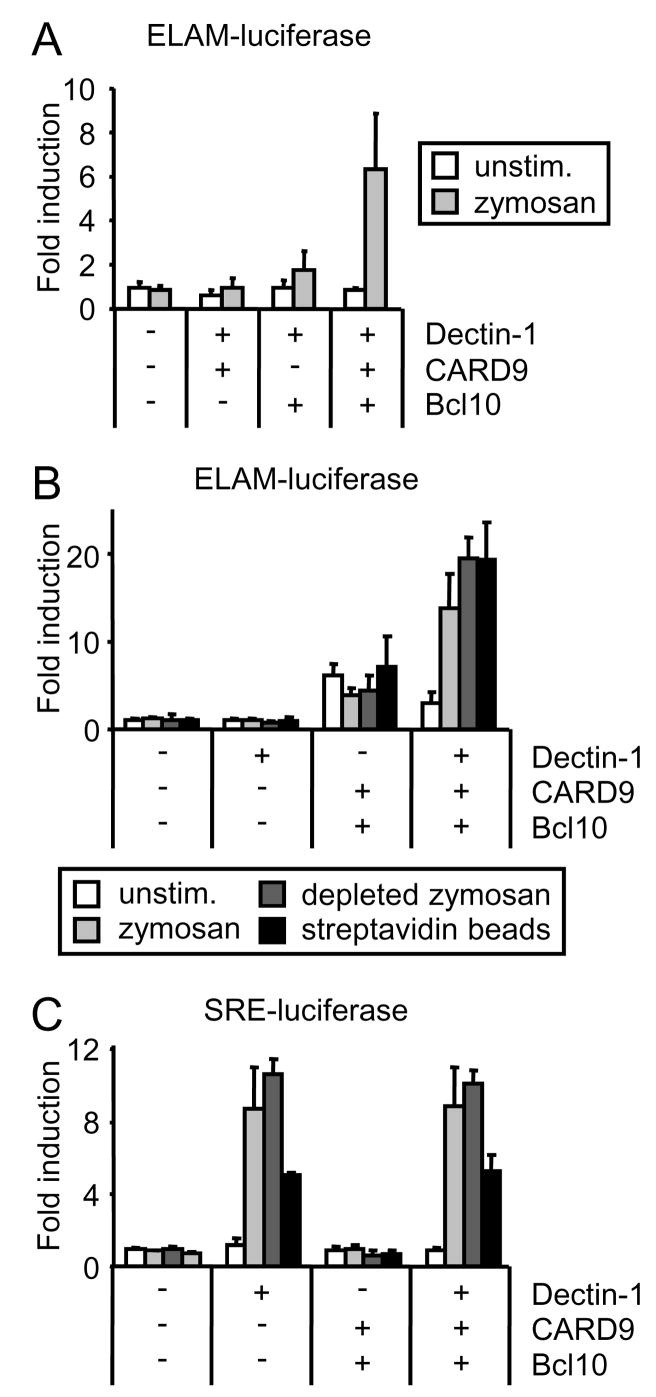
In their recent report, Gross et al. demonstrated that reconstitution of HEK293 cells with Dectin-1 in combination with CARD9 and Bcl10 is sufficient to permit NF-κB activation following exposure to zymosan(12). We further investigated Dectin-1-mediated NF-κB activation in this system to rule out the possible contribution of signals derived from other receptors involved in zymosan recognition. Consistent with the reported data from us and others(8, 12), transient transfection of HEK293 cells with Dectin-1 alone was not sufficient for NF-κB activation following zymosan stimulation. This is despite the fact that Dectin-1 clearly signals in these cells since Dectin-1 transfection of HEK293 cells is sufficient to trigger phagocytosis of zymosan particles(6). However, cotransfection of Dectin-1 with CARD9 and Bcl10 was sufficient to enable robust activation of an NF-κB-driven luciferase reporter (Figure 1A and B). Both CARD9 and Bcl10 were necessary to permit reporter activation (Figure 1A). As observed by Gross et al.(12), overexpression of CARD9 with Bcl10 was often sufficient to activate NF-κB in the absence of Dectin-1 (increased background, Figure 1B). Plasmid doses were therefore titrated to minimize the background.
Figure 1. Dectin-1 signals via CARD9 and Bcl10 to activate NF-κB.
HEK293 cells were transfected with ELAM- (A and B) or SRE- (C) firefly luciferase and CMV-β-galactosidase reporters (A–C), as well as plasmids encoding CARD9, Bcl10 and streptavidin binding peptide (SBPc)-tagged Dectin-1, as indicated. The following day cells were stimulated with 100 μg/ml zymosan or depleted zymosan, or 1:60 streptavidin beads for 4 h. ELAM/SRE-driven luciferase reporter activity was normalized to β-galactosidase activity and expressed as the mean plus standard deviation of triplicate culture.
We also observed CARD9-Bcl10-dependent NF-κB activation when Dectin-1-transfected cells were treated with β-glucan particles prepared by boiling zymosan particles in hot alkali (depleted zymosan; Figure 1B), which we have previously demonstrated destroys the TLR ligands without affecting their ability to signal via Dectin-1(8, 9). To confirm that Dectin-1 signals alone are sufficient to activate NF-κB via CARD9-Bcl10, we directly crosslinked streptavidin-binding peptide (SBPc)-tagged Dectin-1 with streptavidin-coated beads, which we have previously used extensively to study Dectin-1 signaling(6, 9), and observed NF-κB activation only in HEK293 cells transfected with SBPc-Dectin-1, CARD9 and Bcl10 (Figure 1B).
Dectin-1 activates additional transcriptional responses in addition to NF-κB. We have previously reported that Dectin-1 activates NFAT(9). However, Dectin-1 signaling to NFAT cannot be reconstituted in HEK293 cells with or without CARD9/Bcl10 (data not shown), suggesting that additional macrophage/DC factors are required to enable this signaling pathway. In contrast, zymosan, depleted zymosan and streptavidin-coated beads all stimulated the activity of a luciferase reporter driven by the binding of serum response factor (SRF) to the serum response element (SRE-luciferase) in SBPc-Dectin-1-transfected HEK293 cells, and this effect was neither dependent on nor influenced by co-transfection with CARD9 and Bcl10 (Figure 1C). Taken together, the above data demonstrate that Dectin-1 signals transduced by CARD9 and Bcl10 can trigger NF-κB activation, but that CARD9 is not required for all transcriptional responses triggered by Dectin-1 ligation.
CARD9 is recruited to Dectin-1 phagosomes, but is not required for Dectin-1-mediated phagocytosis
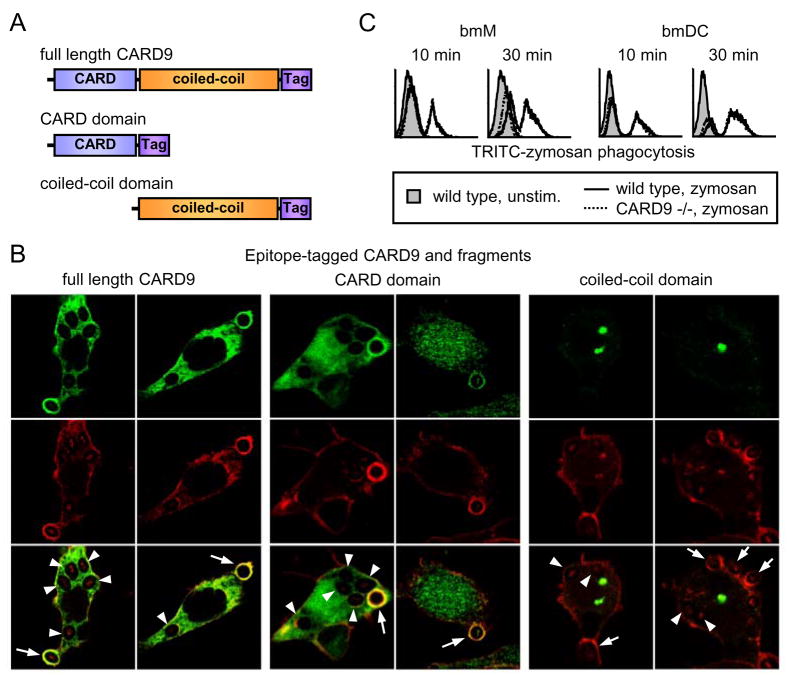
Since CARD9 can participate in signaling to NF-κB downstream of Dectin-1, we explored whether there is evidence that Dectin-1 signals to CARD9 in macrophages. Following binding of yeast particles by Dectin-1, actin polymerization coordinates yeast internalization by phagocytosis, a process which is dependent on Dectin-1 signaling(22). We therefore investigated whether CARD9 is recruited to Dectin-1 phagosomes. Using confocal immunofluorescence microscopy we observed that an SBPc-tagged form of CARD9 (Figure 2A) stably expressed in RAW264.7 macrophages was found diffusely in the cytosol (data not shown). Upon exposure to zymosan, CARD9 was strongly recruited to phagosomes (Figure 2B, arrows). Upon complete internalization of the particles, phagosomal staining was reduced or lost completely (Figure 2B, arrowheads), corresponding with the loss of actin from the maturing phagosome.
Figure 2. CARD9 is recruited to forming phagosomes during Dectin-1-mediated phagocytosis, but is not required for phagocytosis.
(A) Full length CARD9, the CARD domain of CARD9, and the CARD9 coiled-coil domain were tagged with streptavidin-binding peptide (SBPc). (B) RAW264.7 macrophages stably expressing SBPc-tagged full length CARD9, CARD domain, or coiled-coil domain were fed 100 μg/ml zymosan particles for 10 min, prior to fixing and staining with an antibody against the SBPc tag (CARD9 constructs) and TRITC-phalloidin (β-actin). Arrows identify actin-positive early phagosomes; arrowheads identify actin-negative later phagosomes. (C) Bone marrow-derived macrophages (bmM) and dendritic cells (bmDC) from wild type and CARD9-deficient mice were fed 100 μg/ml TRITC-labeled zymosan for the times indicated and phagocytosis was assessed by flow cytometry.
CARD9 comprises an N-terminal CARD domain and a C-terminal coiled-coil domain (Figure 2A), and we therefore examined which domain is responsible for phagosome recruitment. We found that a tagged version of the CARD domain alone was sufficient to reproduce the phagosome recruitment of the full-length protein (Figure 2B, arrows). In contrast, the coiled-coil domain formed aggregates in the cytosol and was not recruited to phagosomes (Figure 2B). Although we cannot exclude a role for the coiled-coil domain in promoting phagosomal localization, the data suggest that the CARD domain is sufficient to target the protein to Dectin-1 phagosomes.
Since CARD9 is found on Dectin-1 phagosomes, we investigated whether CARD9 recruitment and signaling are required for Dectin-1-mediated phagocytosis. Bone marrow-derived macrophages (bmM) and DC (bmDC) from CARD9-deficient mice(18) showed no defect in internalization of TRITC-labeled zymosan particles, indicating that CARD9 plays no role in phagocytosis in either cell type (Figure 2C). These data are consistent with HEK293 reconstitution experiments in which Dectin-1 transfection without addition of CARD9 is sufficient for phagocytosis(6).
β-glucan particles trigger TNF-α production by bmDC, but not bmM
Although we have previously demonstrated a role for Dectin-1 signaling in promoting TLR2-driven cytokine production in zymosan-stimulated macrophages by enhancing TLR2-mediated NF-κB activation, we have thus far failed to detect direct Dectin-1-mediated NF-κB activation in macrophages(8). In contrast, Gross et al. demonstrated that Dectin-1 can activate NF-κB in bmDC via CARD9, and that CARD9-deficient bmDC fail to produce TNF-α in response to zymosan stimulation(12). We therefore reassessed the ability of β-glucan particles to trigger TNF-α production by bmM and bmDC. We compared zymosan, which signals via both TLR2 and Dectin-1, and β-glucan particles (depleted zymosan), which do not activate TLR2.
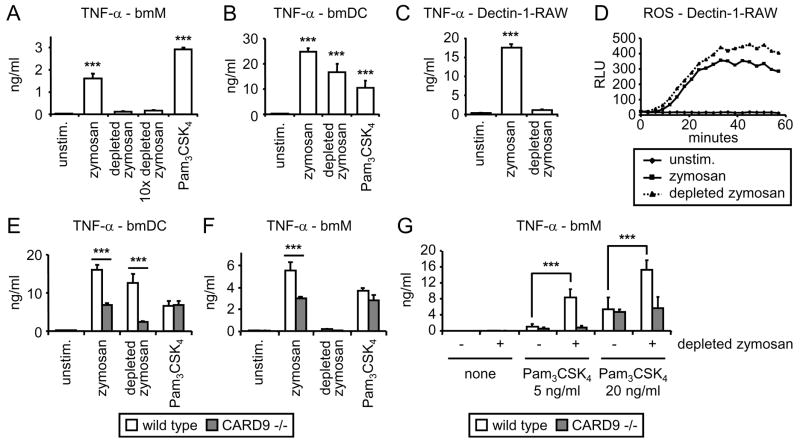
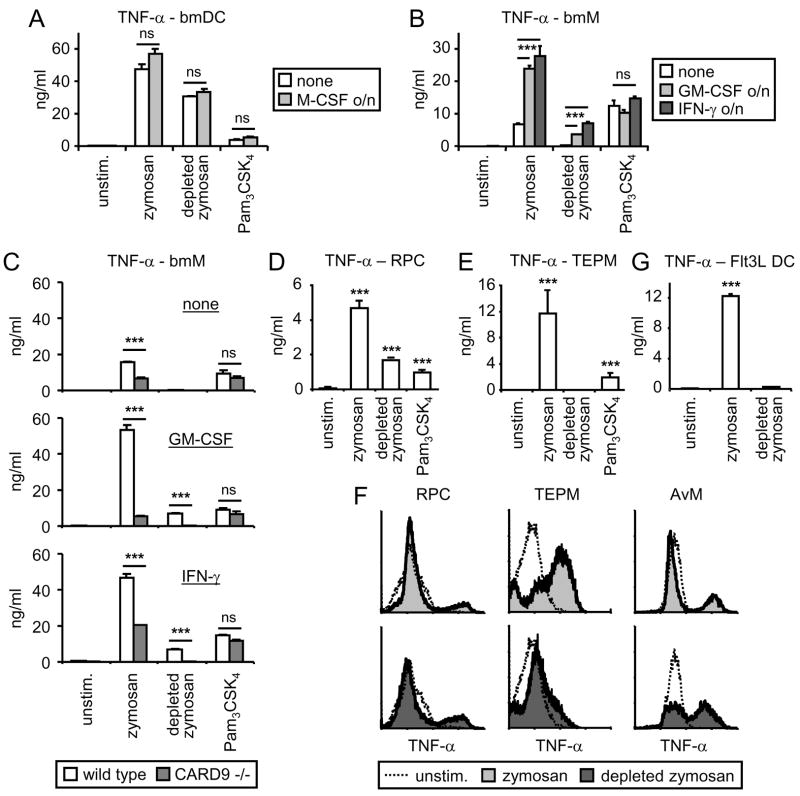
Zymosan triggered robust TNF-α production by both bmM and bmDC (Figure 3A and B), although TNF-α production by bmDC was on average approximately 5-fold greater (Figure 3A, B, E and F). Depleted zymosan triggered robust TNF-α production by bmDC but failed to elicit a response from bmM, even at a 10-fold higher dose. Similar results were obtained using RAW264.7 macrophages overexpressing SBPc-tagged Dectin-1; zymosan stimulated strong induction of TNF-α, while depleted zymosan had little effect (Figure 3C), despite the equivalent abilities of zymosan and depleted zymosan to induce reactive oxygen species (ROS) in these cells (Figure 3D). Specific ligation of SBPc-tagged Dectin-1 with streptavidin-coated beads also failed to induce TNF-α, despite triggering a robust oxidative burst (data not shown).
Figure 3. Dectin-1 signals directly trigger TNF-α production in bmDC, but not bmM.
(A – C) bmM (A), bmDC (B), and Dectin-1-expressing RAW264.7 cells (C) were stimulated with 100 μg/ml zymosan or depleted zymosan, 1 mg/ml depleted zymosan (10× depleted zymosan), or 100 ng/ml Pam3CSK4 for 24 h, and TNF-α levels in culture supernatants were assessed by ELISA. (D) Dectin-1-expressing RAW264.7 cells were stimulated with 100 μg/ml zymosan or depleted zymosan, and reactive oxygen production was measured by luminol-ECL. (E and F) bmDC (E) and bmM (F) from wild type and CARD9-deficient mice were stimulated with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for 24 h, and TNF-α levels in culture supernatants were assessed by ELISA. (G) bmM from wild type and CARD9-deficient mice were stimulated with Pam3CSK4 at the indicated concentrations in the presence or absence of 100 μg/ml depleted zymosan for 24 h, and TNF-α levels in culture supernatants were assessed by ELISA. All stimulations were performed in triplicate, and are presented as means (plus standard deviation for ELISA data). ***, p<0.001.
Having observed that Dectin-1 signaling is insufficient to trigger TNF-α production in macrophages, but that CARD9 is nevertheless recruited to Dectin-1 phagosomes in these cells, we looked specifically at whether CARD9 signaling influences zymosan-induced TNF-α production. We stimulated bmM and bmDC from wild type and CARD9-deficient mice(18) with either zymosan or depleted zymosan. In agreement with data published by Gross et al., TNF-α production triggered by zymosan was dramatically suppressed in CARD9 −/− bmDC, while induction by other TLR ligands (the pure TLR2 agonist Pam3CSK4 and the TLR4 agonist LPS) was not affected (Figure 3E and data not shown). TNF-α induction by depleted zymosan was also suppressed in bmDC lacking CARD9 (Figure 3E), supporting a role for CARD9 in the Dectin-1-mediated response. TNF-α induction by zymosan was also lower in CARD9 −/− bmM than wild type bmM (Figure 3F). Since Dectin-1 signals do not directly trigger TNF-α production in bmM, and TLR2-induced TNF-α induction is also CARD9-independent in these cells (Figure 3F), we hypothesized that Dectin-1 enhances the TLR2 signal via a CARD9-dependent mechanism, and that this collaboration effect is lost in the absence of CARD9. To verify this, we stimulated bmM with Pam3CSK4 in the presence of depleted zymosan, which we have previously shown enhances macrophage responses to the TLR2 ligand(8).
Pam3CSK4-induced TNF-α production by wild type bmM was enhanced in the presence of depleted zymosan, but this synergy was not seen in CARD9-deficient bmM (Figure 3G), demonstrating that despite the inability of Dectin-1 to activate NF-κB-driven transcription via CARD9 in bmM, these cells do use CARD9 signals to enhance TLR signaling.
Dectin-1 signals fail to activate NF-κB in bmM, but do trigger p38 MAP kinase activation
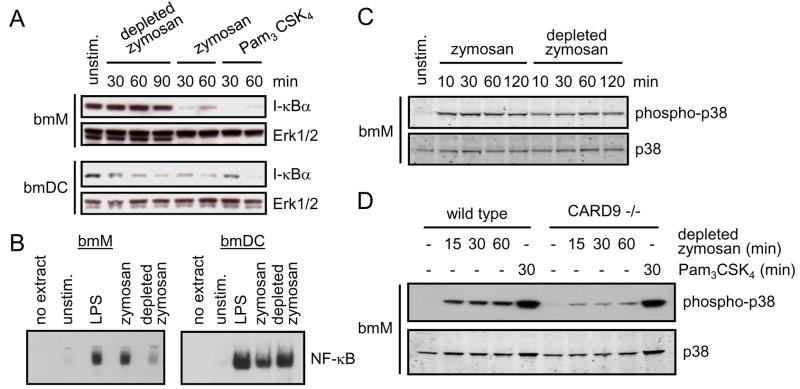
We next examined NF-κB activation in bmM and bmDC following Dectin-1 ligation. We first assessed I-κB degradation, which releases NF-κB to enter the nucleus and bind target promoters. Consistent with previous studies, stimulation of both bmM and bmDC with zymosan or the pure TLR2 agonist Pam3CSK4 triggered rapid degradation of I-κB (within 30 minutes of stimulation), followed by resynthesis at later timepoints (Figure 4A and data not shown). In contrast, depleted zymosan triggered rapid I-κB degradation in bmDC, but failed to alter I-κB levels in bmM (Figure 4A). Consistent with this, EMSA analysis of NF-κB binding to a consensus promoter binding site demonstrated strong NF-κB binding activity in nuclear extracts of zymosan-stimulated bmM, while depleted zymosan triggered little or no significant NF-κB binding activity (Figure 4B), as reflected in the failure of depleted zymosan to trigger TNF-α production by these cells (Figure 3A and F). In contrast, both zymosan and depleted zymosan induced strong NF-κB binding activity in bmDC nuclear extracts (Figure 4B).
Figure 4. Dectin-1-CARD9 signals trigger NF-κB activation in bmDC, but not bmM, but contribute to p38 MAP kinase activation in bmM.
(A) bmM and bmDC were stimulated with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for the times indicated, and I-κB levels were assessed by Western blotting, with Erk1/2 as a loading control. (B) bmM and bmDC were stimulated with 100 ng/ml LPS, or 100 μg/ml zymosan or depleted zymosan for 90 min, and nuclear translocation of NF-κB was assessed by EMSA. (C and D) bmM from wild type (C and D) and CARD9-deficient (D) mice were stimulated with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for the times indicated, and p38 phosphorylation was assessed by Western blotting, with total p38 as a loading control.
In contrast to the differential activation of NF-κB, both zymosan and depleted zymosan triggered p38 MAP kinase phosphorylation in bmM (Figure 4C). Specific ligation of SBPc-tagged Dectin-1 with streptavidin beads also induced p38 MAP kinase phosphorylation in RAW264.7 macrophages (data not shown). Depleted zymosan-induced p38 phosphorylation was reduced but not abolished in CARD9 −/− bmM (Figure 4D), indicating that Dectin-1-induced p38 activation is partially, but not entirely, dependent on CARD9. In contrast Pam3CSK4-induced p38 phosphorylation was CARD9-independent, consistent with the lack of an effect of CARD9 deletion on TLR-mediated pro-inflammatory cytokine production (Figure 3E and F and (12, 18)).
Collectively, our data indicate that while Dectin-1-CARD9-Bcl10 signals directly activate NF-κB and TNF-α production in bmDC, Dectin-1 signaling alone does not activate NF-κB in bmM. In contrast, CARD9 signals contribute to p38 MAP kinase activation in bmM and can enhance TLR2-activated TNF-α production.
GM-CSF and IFN-γ permit TNF-α induction by Dectin-1 in bmM
Considering the parallel differentiation of bmM and bmDC with M-CSF and GM-CSF respectively, we hypothesized that either 1) M-CSF signals restrict the ability of Dectin-1 to activate the CARD9-NF-κB pathway in bmM, or 2) GM-CSF signals permit the direct activation of NF-κB via CARD9 in bmDC. To address the first possibility we added M-CSF to GM-CSF-derived bmDC overnight prior to stimulation, and observed no effect of M-CSF on the ability of these cells to produce TNF-α upon stimulation with zymosan, depleted zymosan or the TLR2 ligand Pam3CSK4 (Figure 5A). In contrast, M-CSF-derived bmM exposed to GM-CSF overnight produced elevated levels of TNF-α in response to zymosan, but not upon stimulation with Pam3CSK4 (Figure 5B). Furthermore, these cells now produced robust TNF-α following exposure to depleted zymosan (Figure 5B). Overnight priming of bmM with IFN-γ had a similar effect (Figure 5B). Dectin-1-induced TNF-α production by GM-CSF/IFN-γ-primed bmM was completely CARD9-dependent (Figure 5C).
Figure 5. Analysis of Dectin-1-induced TNF-α production by GM –CSF- and IFN-γ-primed bmM, and other macrophage and DC populations.
(A and B) bmDC were pre-treated overnight with 50 ng/ml M-CSF (A), and bmM were pre-treated overnight with 10 ng/ml GM-CSF or 25 U/ml IFN-γ (B). bmDC and bmM were then stimulated with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for 24 h, and TNF-α in culture supernatants was assessed by ELISA. (C) bmM from wild type and CARD9-deficient bmM were pre-treated overnight with 10 ng/ml GM-CSF or 25 U/ml IFN-γ, prior to stimulation with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for 24 h, and TNF-α was assessed by ELISA. (D and E) Resident peritoneal cells (RPC; D) and thioglycollate-elicited peritoneal macrophages (TEPM; E) were stimulated with 100 μg/ml zymosan or depleted zymosan, or 100 ng/ml Pam3CSK4 for 24 h, and TNF-α was assessed by ELISA. (F) RPC, TEPM and alveolar macrophages (AvM) were stimulated with 100 μg/ml zymosan or depleted zymosan for 4 h in the presence of brefeldin A, and TNF-α production was assessed by intracellular flow cytometry, gating for CD11b+ RPC and TEPM, and CD11b− AvM. (G) DC derived from bone marrow with Flt3L (Flt3L DC) were stimulated with 100 μg/ml zymosan or depleted zymosan for 24 h and TNF-α in culture supernatants was assessed by ELISA. For ELISA measurements stimulations were performed in triplicate, and are presented as mean plus standard deviation. ***, p<0.001; ns, no significant difference.
The ability of Dectin-1 to directly drive TNF-α production in different macrophage and DC populations is variable
Our data have revealed an interesting difference in the ability of bmM and bmDC derived in vitro to respond to Dectin-1 ligation by β-glucan particles. To address whether in vivo populations of macrophages are similarly restricted in their ability to respond to β-glucan particles, we measured TNF-α production by resident peritoneal cells (RPC), thioglycollate-elicited peritoneal macrophages (TEPM), and alveolar macrophages (AvM). Like bmM, all of these macrophage subtypes produced robust TNF-α following zymosan stimulation (Figure 5D–F). However, we observed variable responsiveness of the different cell types to depleted zymosan (Figure 5D–F). Depleted zymosan stimulated TNF-α production by RPC, but failed to induce TEPM to produce TNF-α (Figure 5D–F). AvM also produced TNF-α in response to depleted zymosan exposure (Figure 5F). Thus it appears that there is considerable variation between in vivo-derived macrophage populations in the contribution of Dectin-1 signals to TNF-α induction.
We also measured TNF-α production by DC derived from bone marrow using Flt3L (Flt3L DC). These cells are thought to represent steady-state resident tissue DC(23). Zymosan stimulated robust TNF-α production, but in contrast to GM-CSF-derived bmDC, Flt3L DC failed to produce TNF-α upon exposure to depleted zymosan (Figure 5G).
Differential responsiveness to β-glucan particles is not due to variation in Dectin-1 and CARD9 expression
We hypothesized that the inability of β-glucan particles to directly trigger TNF-α production in certain macrophage and DC populations (unprimed bmM, TEPM and Flt3L-derived bmDC) indicates that these cells either lack a key component of the Dectin-1-CARD9-NF-κB signaling pathway, or that an inhibitor active in these cells blocks the signal.
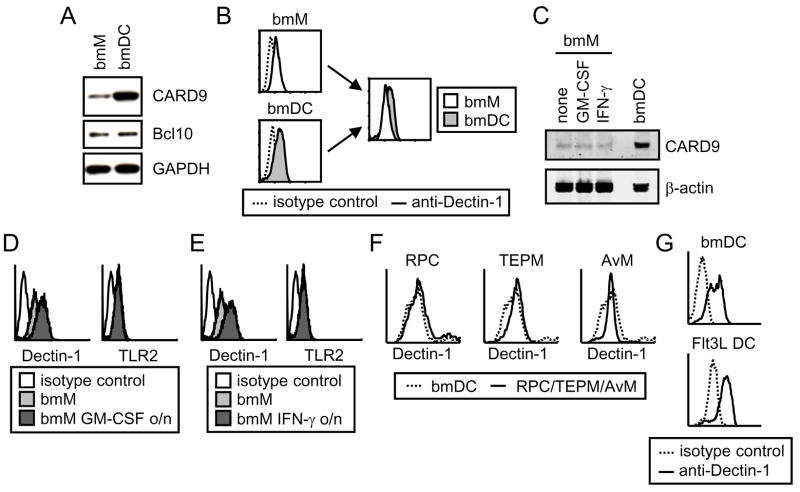
To address whether bmM are deficient in any of the key components of this pathway, we first compared the expression of CARD9 and Bcl10, as well as Dectin-1 itself, in bmM and bmDC. Consistent with our demonstration above that CARD9 is recruited to Dectin-1 phagosomes in macrophages (Figure 2B and C) and that CARD9 is responsible for Dectin-1 collaboration with TLR2 in bmM (Figure 3G), we observed CARD9 expression in bmM as well as GM-CSF-derived bmDC, although the DC had much higher levels of CARD9 than bmM (Figure 6A). Surface expression of Dectin-1 was also slightly higher on bmDC than bmM (Figure 6B). Bcl10 was detected in both cell types at similar levels and hence its expression level does not appear to correlate with the presence or absence of a response (Figure 6A).
Figure 6. There is no correlation between Dectin-1, CARD9 and Bcl10 expression levels and the ability of Dectin-1 to directly induce TNF-α.
(A) CARD9 and Bcl10 levels in bmM and bmDC were compared by Western blotting, with GAPDH as a loading control. (B) Surface expression of Dectin-1 on bmM and bmDC was assessed by flow cytometry. (C) CARD9 expression by bmM and bmM pre-treated overnight with 10 ng/ml GM-CSF or 25 U/ml IFN-γ was assessed by Western blotting, with β-actin as a loading control. (D and E) Surface expression of Dectin-1 and TLR2 on bmM and bmM pre-treated overnight with 10 ng/ml GM-CSF (D) or 25 U/ml IFN-γ (E) was assessed by flow cytometry. (F and G) Dectin-1 surface expression by RPC, TEPM, AvM, bmDC and Flt3L DC was assessed by flow cytometry.
We next examined whether GM-CSF and IFN-γ priming alter CARD9 levels or the surface expression of Dectin-1 on bmM. Neither GM-CSF nor IFN-γ had any effect on CARD9 expression (Figure 6C). Since these primed bmM respond robustly to depleted zymosan, we can conclude that CARD9 is not limiting in unprimed bmM. In contrast, both GM-CSF and IFN-γ enhanced Dectin-1 surface expression to levels comparable to those of bmDC, without affecting surface expression of TLR2 (Figure 6D and E). However, we also found that levels of Dectin-1 on RPC, TEPM, AvM and Flt3L DC were comparable to those on bmDC (Figure 6F and G), despite the varied responsiveness of these cells (Figure 5F and G). Hence there was no clear correlation between Dectin-1 surface expression levels and the ability of the cells to respond to depleted zymosan.
Macrophage unresponsiveness to β-glucan particles is likely due to the action of an inhibitor of the Dectin-1-CARD9 pathway
Although Dectin-1, CARD9 and Bcl10 levels do not appear to correlate with the differential responsiveness of the myeloid populations, it remains possible that these cells lack some other intrinsic component of the CARD9 pathway. Alternatively the signaling pathway may be fully intact, but expression of additional proteins may restrict its activation. To distinguish between these possibilities we overexpressed Dectin-1 in bmM. If these cells lack an intrinsic signaling component, no amount of enhanced Dectin-1 signaling would be sufficient to permit activation of the pathway. In contrast, if an inhibitor is blocking CARD9-mediated NF-κB activation, increased Dectin-1 signaling might be expected to overcome this blockade.
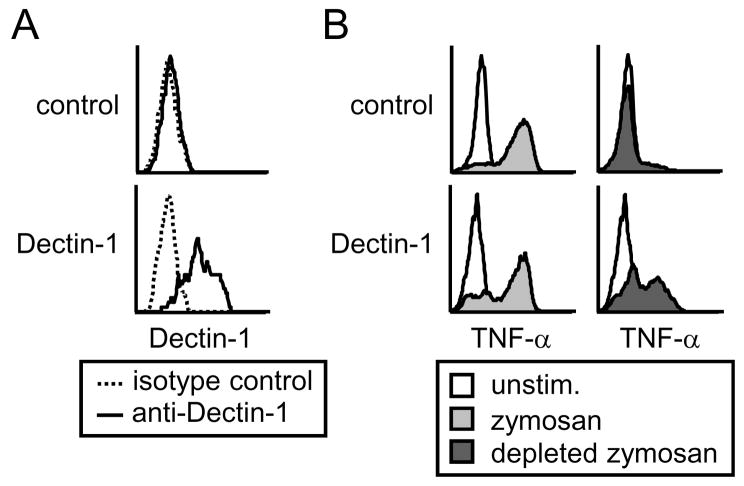
We therefore transfected bmM with a Dectin-1 expression vector, and verified Dectin-1 overexpression at the cell surface by flow cytometry (Figure 7A). We observed dramatically enhanced Dectin-1 surface expression following Dectin-1-GFP transfection compared to expression of endogenous Dectin-1 expression by control GFP transfected cells (which was poorly detected by the PE-conjugated anti-Dectin-1 antibody, in contrast to the FITC-conjugated antibody used in Figure 6). We then assessed the ability of zymosan and depleted zymosan to induce TNF-α production by control and Dectin-1-overexpressing bmM by intracellular flow cytometry. While bmM overexpressing either Dectin-1 or a control plasmid responded robustly to zymosan stimulation, only bmM overexpressing Dectin-1 produced TNF-α following exposure to depleted zymosan (Figure 7B). Thus it appears possible to activate CARD9 signaling directly downstream of Dectin-1 in these cells with a sufficiently large signal, supporting the notion that this pathway is intact but is held in check by expression of additional factors.
Figure 7. Overexpression of Dectin-1 in bmM overcomes blockade of direct Dectin-1-mediated TNF-α induction.
(A) bmM were transfected with either Dectin-1-GFP or a GFP control plasmid, and Dectin-1 surface expression was assessed by flow cytometry of unpermeabilized cells. (B) Control or Dectin-1-overexpressing bmM were stimulated for 4 h with 100 μg/ml zymosan or depleted zymosan in the presence of brefeldin A, and TNF-α production by GFP+ cells (control and Dectin-1 plasmids), was assessed by intracellular flow cytometry.
Discussion
Integration of signals derived from Dectin-1 and TLR2 directs the coordinated response of myeloid phagocytes to fungal particles. Previous studies indicated that the transcriptional induction of pro-inflammatory mediators is controlled by TLR2-MyD88-induced NF-κB activation, a signal that can be amplified by Dectin-1, and Dectin-1-mediated ITAM-like signaling which activates NFAT transcription factors(8, 9, 24, 25). A recent report also indicated that Dectin-1 signals can directly activate NF-κB in DC via the adaptor protein CARD9(12). In our current study we have revealed an additional layer of regulation of Dectin-1/TLR2-induced pro-inflammatory cytokine production.
Our data show that Dectin-1 signaling via CARD9 is differentially regulated in myeloid cells, resulting in the ability of only certain macrophage/DC populations to produce TNF-α upon Dectin-1 ligation alone. bmM, thioglycollate-elicited peritoneal macrophages (TEPM), and Flt3L-derived DC failed to produce TNF-α in response to Dectin-1 ligation, whereas bmDC, GM-CSF/IFN-γ-primed bmM, resident peritoneal cells and alveolar macrophages all displayed robust TNF-α production following Dectin-1 ligation (Figures 3 and 5). Expression of Dectin-1, CARD9 and its interacting partner Bcl10 did not correlate with the ability of the cells to produce TNF-α in response to Dectin-1 ligation (Figure 6). However, we demonstrated that overexpression of Dectin-1 in bmM is sufficient to permit CARD9-dependent TNF-α induction (Figure 7), indicating that these cells possess an inhibitor of Dectin-1-CARD9 signaling.
Meyer-Wentrup et al. recently demonstrated that the tetraspanin CD37 associates with Dectin-1 and limits pro-inflammatory cytokine induction(26). Induction of IL-6 by zymosan was 10-fold higher in TEPM from CD37 knockout mice compared to wild type TEPM. Furthermore, while wild type TEPM failed to produce IL-6 in response to the β-glucan curdlan, IL-6 was induced by curdlan treatment of CD37-deficient TEPM. Our data suggest that CD37 may block Dectin-1-CARD9-mediated cytokine induction in macrophage/DC populations unresponsive to depleted zymosan. Consistent with this, we observed higher CD37 expression by bmM than bmDC, and a reduction in CD37 expression following treatment of bmM with GM-CSF and IFN-γ (data not shown). Meyer-Wentrup et al. also observed a reduction in CD37 expression upon differentiation of DC from human monocytes(26).
Tetraspanins are small transmembrane scaffold proteins that anchor proteins in membrane microdomains(27). Hence CD37 may sequester Dectin-1 into a microdomain that selectively modulates the recruitment of key signaling molecules downstream of CARD9. Lower CD37 expression in bmDC/primed bmM, coupled with higher Dectin-1 expression in these cells, would be predicted to result in a larger pool of Dectin-1 that is not associated with and limited by CD37. Further studies are required to determine whether a CD37-based mechanism is responsible and sufficient for the differential CARD9 signaling we have observed in this study.
Dectin-1 has also been reported to associate with another tetraspanin, CD63, in human monocyte-derived DC, although the function of this interaction is unknown(28). Hence the relative levels of different tetraspanins may also determine the type of signal generated. Detailed investigation of the regulation of Dectin-1 signaling by CD37 and CD63 is required to further assess the ability of tetraspanins to differentially regulate Dectin-1 signals.
Gross et al. had demonstrated that Dectin-1-CARD9 signaling results in NF-κB activation in bmDC(12), which contrasted with our previous failure to observe NF-κB activation in RAW264.7 macrophages and bmM(8). We have now shown that Dectin-1 can indeed activate NF-κB directly in certain myeloid cells, but that this signaling connection is not always made. This is in contrast to other Dectin-1 signals, including Src-Syk activation(6), p38 phosphorylation (Figure 4C), and NFAT activation(9), which are triggered robustly in bmM (and RAW264.7 macrophages) as well as bmDC. Whether other signals downstream of CARD9 are also absent in the “unresponsive” macrophages, contributing to the inability of Dectin-1 signals to directly induce TNF-α, remains to be established.
The cell type-specific variability of CARD9-mediated NF-κB activation downstream of Dectin-1 ligation stands in sharp contrast to MyD88-mediated NF-κB activation downstream of TLRs. As far as we are aware TLR signaling via MyD88 always results in NF-κB activation, regardless of cell type. In contrast, we have shown that Dectin-1 signaling via CARD9 does not always result in NF-κB activation. Variability in Dectin-1 signaling in different cell types represents a novel mechanism by which myeloid cells can be fine-tuned to regulate anti-fungal immunity, with different myeloid populations having distinct inflammatory and anti-microbial properties. Whereas alveolar macrophages are inherently ready to release cytokines and initiate an inflammatory response to β-glucan particles, other macrophages may require information about an ongoing infection, such as IFN-γ production, to become primed for this response. Furthermore, as we have observed fundamentally different pro-inflammatory signaling in GM-CSF- and Flt3L-derived bmDC, there may be significant further heterogeneity in how different DC subsets prime adaptive immunity. Future studies will define the consequences of this variability on cell function and the contribution of differentially responsive cells to anti-fungal immunity.
Despite the apparent restricted ability of CARD9 to activate NF-κB in bmM, CARD9 is not completely silent in these cells, as indicated by the defect in Dectin-1-induced p38 phosphorylation in CARD9-deficient bmM (Figure 4D). Furthermore, CARD9 appears to be responsible for the collaboration of Dectin-1 signals with TLR2 signals, as evidenced by the defect in TNF-α production by zymosan-stimulated CARD9-deficient bmM (Figure 3F and G). These data are consistent with our observations of CARD9 recruitment to Dectin-1 phagosomes even in macrophages where Dectin-1 signaling is insufficient to drive TNF-α production (Figure 2).
Hara et al. recently showed that CARD9 mediates signaling downstream of the FcγR, which, like Dectin-1, is an ITAM-containing phagocytic receptor(29). Hence we predict that CARD9 is similarly recruited to FcγR phagosomes. That the CARD domain of CARD9 is enough to drive the protein to the Dectin-1 phagosome suggests that phagosomes might be an important scaffold for coordinating CARD-CARD signaling interactions. Future studies will have to define how the presence or absence of additional signaling molecules on the phagosomes of different cell types regulates inflammation and immunity.
Footnotes
This work was supported by grants to DMU from the National Institutes of Health (AI071116, GM085796) and the American Heart Association (0640100N). HSG holds a Research Fellowship Award from the Crohn’s and Colitis Foundation of America.
References
- 1.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 3.Goodridge HS, Underhill DM. Fungal Recognition by TLR2 and Dectin-1. Handb Exp Pharmacol. 2008:87–109. doi: 10.1007/978-3-540-72167-3_5. [DOI] [PubMed] [Google Scholar]
- 4.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, Akira S, Adachi Y, Ohno N, Kinjo T, Nakamura K, Kawakami K, Iwakura Y. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8:39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 6.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106:2543–2550. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennehy KM, Ferwerda G, Faro-Trindade I, Pyz E, Willment JA, Taylor PR, Kerrigan A, Tsoni SV, Gordon S, Meyer-Wentrup F, Adema GJ, Kullberg BJ, Schweighoffer E, Tybulewicz V, Mora-Montes HM, Gow NA, Williams DL, Netea MG, Brown GD. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur J Immunol. 2008;38:500–506. doi: 10.1002/eji.200737741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gantner BN, Simmons RM, Canavera SJ, Akira S, Underhill DM. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J Exp Med. 2003;197:1107–1117. doi: 10.1084/jem.20021787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 10.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 11.Dillon S, Agrawal S, Banerjee K, Letterio J, Denning TL, Oswald-Richter K, Kasprowicz DJ, Kellar K, Pare J, van Dyke T, Ziegler S, Unutmaz D, Pulendran B. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J Clin Invest. 2006;116:916–928. doi: 10.1172/JCI27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 13.Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, Domashenz H, Hong NA, Glynne RJ, Nelms KA, Goodnow CC. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18:751–762. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 14.Pomerantz JL, Denny EM, Baltimore D. CARD11 mediates factor-specific activation of NF-kappaB by the T cell receptor complex. Embo J. 2002;21:5184–5194. doi: 10.1093/emboj/cdf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaide O, Favier B, Legler DF, Bonnet D, Brissoni B, Valitutti S, Bron C, Tschopp J, Thome M. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:836–843. doi: 10.1038/ni830. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nat Immunol. 2002;3:830–835. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 17.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 18.Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, Qin XF, Dong C, Lin X. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura T, Koshino Y, Shibata F, Oki T, Nakajima H, Nosaka T, Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 20.Nakayama M, Underhill DM, Petersen TW, Li B, Kitamura T, Takai T, Aderem A. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178:4250–4259. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- 21.Fluhmann B, Zimmermann U, Muff R, Bilbe G, Fischer JA, Born W. Parathyroid hormone responses of cyclic AMP-, serum- and phorbol ester-responsive reporter genes in osteoblast-like UMR-106 cells. Mol Cell Endocrinol. 1998;139:89–98. doi: 10.1016/s0303-7207(98)00070-7. [DOI] [PubMed] [Google Scholar]
- 22.Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, Wong SY, Gordon S. Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179:7577–7584. doi: 10.4049/jimmunol.179.11.7577. [DOI] [PubMed] [Google Scholar]
- 24.Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. Dectin-1 mediates the biological effects of beta-glucans. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Schweighoffer E, Williams DL, Gordon S, Tybulewicz VL, Brown GD, Reis e Sousa C. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Meyer-Wentrup F, Figdor CG, Ansems M, Brossart P, Wright MD, Adema GJ, van Spriel AB. Dectin-1 interaction with tetraspanin CD37 inhibits IL-6 production. J Immunol. 2007;178:154–162. doi: 10.4049/jimmunol.178.1.154. [DOI] [PubMed] [Google Scholar]
- 27.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 28.Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, Teillaud JL, Mordoh J. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood. 2004;104:1183–1190. doi: 10.1182/blood-2004-01-0104. [DOI] [PubMed] [Google Scholar]
- 29.Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, Inui M, Takai T, Shibuya A, Saijo S, Iwakura Y, Ohno N, Koseki H, Yoshida H, Penninger JM, Saito T. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]