Fig. 2.
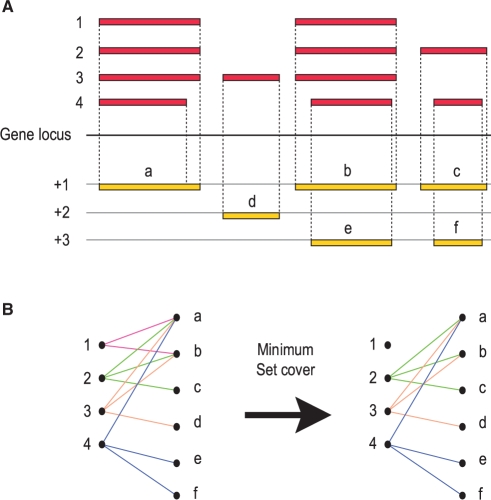
Subsequence delineation and merging procedure. In order to demonstrate how subsequences are generated, shown is a hypothetical query protein sequence with regions of local similarity to a set of target proteins (or match partners) labelled 1–6. On this basis, the query protein sequence is divided into non-overlapping subsequences such that each individual subsequence matches a set of partners (indicated at the top of each subsequence) that is different from the set matched by its adjacent subsequences. Some of these adjacent subsequences are merged (indicated by arrows) if they do not match mutually exclusive partners. Merging occurs iteratively until no further adjacent subsequences can be merged. At the end of this process, the query protein is divided into regions which show different signals of homology that are indicative of a modular composition.